Abstract
Host range factor 1 (HRF-1) of Lymantria dispar multinucleocapsid nucleopolyhedrovirus promotes Autographa californica MNPV replication in nonpermissive Ld652Y cells derived from L. dispar. Here we demonstrate that restricted Hyphantria cunea NPV replication in Ld652Y cells was not due to apoptosis but was likely due to global protein synthesis arrest that could be restored by HRF-1. Our data also showed that HRF-1 promoted the production of progeny virions for two other baculoviruses, Bombyx mori NPV and Spodoptera exigua MNPV, whose replication in Ld652Y cells is limited to replication of viral DNA without successful production of infectious progeny virions. Thus, HRF-1 is an essential viral factor required for productive infection of NPVs in Ld652Y cells.
Nucleopolyhedroviruses (NPVs) belong to the family Baculoviridae and possess a double-stranded, circular DNA genome of 80 to 180 kbp within a large rod-shaped capsid (4). These viruses are insect-specific viruses reported from more than 520 insect species (1). NPVs generally exhibit a narrow host range property (1). Lethal infection is commonly restricted to insect species from which the NPVs are originally isolated and to closely related insect species. Analyses with cell cultures have revealed that while budded virions (BV) of NPVs are capable of entering a phylogenetically broad range of insect cells, NPV replication is often restricted at a step after viral entry that differs depending on the specific combinations of NPVs and insect cell lines (20, 27, 29, 33, 34).
The molecular mechanisms underlying the host specificity of NPVs are not clear. Recent studies have identified several viral genes that are involved in host range determination of Autographa californica multinucleocapsid NPV (AcMNPV) in insect cell systems (6, 7, 18, 21, 22, 24, 31). One of these genes, host range factor 1 (hrf-1) from the gypsy moth NPV, Lymantria dispar MNPV (LdMNPV), was identified as a factor that promoted AcMNPV replication in nonpermissive cell line Ld652Y (12), derived from L. dispar. AcMNPV infection of Ld652Y cells results in global protein synthesis shutoff and yields no progeny virions (8, 13, 14, 23, 32). The recombinant AcMNPV that possesses hrf-1 restores viral protein synthesis and replicates successfully in Ld652Y cells and L. dispar larvae (5, 7, 31). Thus, HRF-1 protein precludes global protein synthesis shutoff and promotes production of progeny virions in AcMNPV-infected Ld652Y cells. Analyses of whole-genome sequences from several NPVs (2, 3, 11, 15, 16, 19, 28) revealed that hrf-1 was specifically located on the genome of LdMNPV and Orgyia pseudotsugata MNPV that could replicate in Ld652Y cells. In this study, we demonstrate that HRF-1 is an essential factor required for NPVs to replicate successfully in Ld652Y cells.
HycuNPV replication is restricted in Ld652Y cells by a mechanism other than apoptosis.
It was previously shown that infection of Ld652Y cells with Hyphantria cunea NPV (HycuNPV) resulted in induction of severe cellular apoptosis in which no progeny virions were produced (17). To determine if the defects in HycuNPV replication in Ld652Y cells were due to virus-induced apoptosis, Ld652Y cells were infected with HycuNPV at a multiplicity of infection (MOI) of 5 PFU/cell. Infected cells were cultured in TC100 medium (Invitrogen) only or in medium containing a pancaspase inhibitor Z-VAD-FMK (Sigma) at a concentration of 20 μM. Microscopic examination showed that Z-VAD-FMK had no adverse effect on uninfected Ld652Y cells and effectively blocked apoptosis of Ld652Y cells induced by HycuNPV infection (Fig. 1A). However, the Z-VAD-FMK-treated HycuNPV-infected Ld652Y cells produced no polyhedra, even at 96 h postinfection (pi) (Fig. 1A).
FIG. 1.
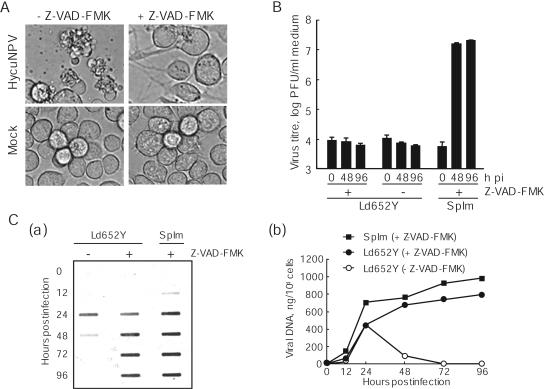
Cytopathology, BV yields, and viral DNA accumulation in HycuNPV-infected and Z-VAD-FMK-treated Ld652Y cells. Monolayer cultures of Ld652Y cells were infected with HycuNPV at an input MOI of 5 PFU/cell and were cultured in medium only or in medium containing 20 μM Z-VAD-FMK. (A) Cytopathology of HycuNPV-infected and Z-VAD-FMK-treated Ld652Y cells at 96 hpi. Mock-infected and Z-VAD-FMK-treated Ld652Y cells were incorporated as controls. (B) BV yields from HycuNPV-infected and Z-VAD-FMK-treated Ld652Y cells were determined by plaque assay on SpIm cells. Vertical bars indicate standard deviations of averages from three determinations. (C) Slot blot hybridization analysis of viral DNA in HycuNPV-infected and Z-VAD-FMK-treated Ld652Y cells. The viral DNAs were blotted onto a Hybond-N+ membrane and were hybridized with fluorescein-labeled hycu-iap3 gene probe. The probe was visualized by gene images (a) and was quantified with a Lumi Imager by comparing the signal intensities in infected cells with those of serially diluted HycuNPV DNAs of known amount (b). (B and C) HycuNPV-infected and Z-VAD-FMK-treated SpIm cells (conventional host cells) are also shown for comparison.
To examine yields of BVs, culture media were harvested from HycuNPV-infected Ld652Y cells at 0, 48, and 96 hpi and were subjected to plaque assay on SpIm cells (25) from the mulberry tiger moth, Spilosoma imparilis, cultured in MM medium (26). The results showed no significant BV yield, even in HycuNPV-infected Ld652Y cells whose apoptosis was blocked by Z-VAD-FMK treatment (Fig. 1B). In contrast, HycuNPV-infected SpIm cells cultured in the medium containing Z-VAD-FMK produced high titers of BV (Fig. 1B), indicating that Z-VAD-FMK did not interfere with HycuNPV replication.
Slot blot hybridization analysis showed that the absence of BV production in HycuNPV-infected Ld652Y cells was not due to the absence of viral DNA production (Fig. 1C). Our results showed that a considerable amount of viral DNA was produced in HycuNPV-infected Ld652Y cells cultured in the medium containing Z-VAD-FMK, with a changing pattern similar to that in HycuNPV-infected SpIm cells (Fig. 1C). Slot blot analysis also showed that viral DNA in HycuNPV-infected Ld652Y cells without Z-VAD-FMK treatment decreased sharply from 24 hpi, suggesting that the decrease of viral DNA was likely due to apoptosis in HycuNPV-infected Ld652Y cells. Consistent with previous results (17) for HycuNPV-infected Ld652Y cells without Z-VAD-FMK treatment, immunoblot analysis with antisera against polyhedrin and viral structural proteins showed that there was no detectable production of virus-encoded proteins in HycuNPV-infected Ld652Y cells, irrespective of Z-VAD-FMK treatment (data not shown). These results suggested that HycuNPV replication was restricted in Ld652Y cells at a step after viral DNA replication by a mechanism other than apoptosis.
HRF-1 promotes HycuNPV replication in Ld652Y cells.
It has been shown that HRF-1 of LdMNPV promotes AcMNPV replication in nonpermissive Ld652Y cells (7, 31). To examine whether HRF-1 also promotes HycuNPV replication in Ld652Y cells, a mutant HycuNPV, vHycuΔhr6/HA-HRF1 (Fig. 2A), which possessed a hemagglutinin (HA) tag-fused hrf-1 gene under the control of the hycu-ie1 promoter, was generated by homologous recombination between vHycuΔhr6/lacZ (Fig. 2A) genome DNA and a transfer vector, pHycuΔhr6/HA-HRF1, in SpIm cells. vHycuΔhr6/lacZ was generated by homologous recombination between wild-type (wt) HycuNPV (Fig. 2A) genome DNA and pHycuΔhr6/lacZ in SpIm cells. To construct pHycuΔhr6/HA-HRF1 and pHycuΔhr6/lacZ, p74-76.7 (Fig. 2A), which included a segment ranging from 74.0 to 76.7 map units (mu) of the wt HycuNPV genome, was constructed by inserting the SacII-O fragment (74.5 to 76.7 mu) of wt HycuNPV genome into SacII-treated p2.2-BstXI (9, 10) that contained a 2.2-kbp BstXI fragment (74.0 to 74.5 mu) of wt HycuNPV genome cloned into the SmaI site of pBluescript (Stratagene). p74-76.7 was then digested with BstXI and was blunt-ended by T4 DNA polymerase (New England Biolabs), into which the blunt-ended fragments containing the hycu-ie1 promoter-driven HA-fused hrf-1 gene isolated from EcoRI- and XbaI-digested pHyHr6IE1/HA-HRF1 (see Fig. 4A) and the SeMNPV polyhedrin promoter-driven lacZ gene isolated from SalI- and XbaI-digested pBKblue (Nihon Nosan Kogyo) were subcloned, generating pHycuΔhr6/HA-HRF1 and pHycuΔhr6/lacZ, respectively. Insertion of hrf-1 and lacZ genes into the genomes of vHycuΔhr6/HA-HRF1 and vHycuΔhr6/lacZ, respectively, was confirmed by PCR with primers specific to each gene (data not shown).
FIG. 2.
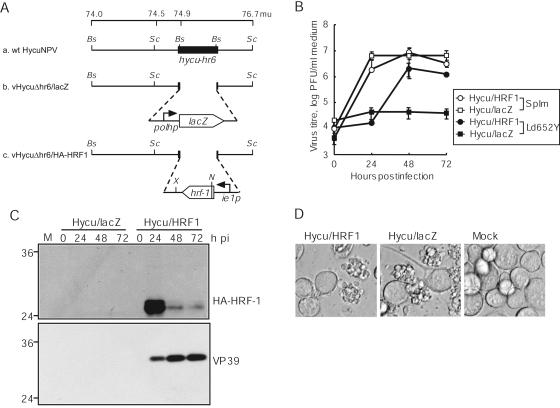
Viral replication in vHycuΔhr6/HA-HRF1- and vHycuΔhr6/lacZ-infected Ld652Y cells. (A) Schematic representation of wt HycuNPV, vHycuΔhr6/lacZ, and vHycuΔhr6/HA-HRF1. The schematics represent the 74.0- to 76.7-mu region of wt HycuNPV (a) as well as the genetic modification of vHycuΔhr6/lacZ (b) and vHycuΔhr6/HA-HRF1 (c), in which hycu-hr6 was replaced by the lacZ gene under the control of the SeMNPV polyhedrin promoter (polhp) and the HA tag-fused hrf-1 gene under the control of the hycu-ie1 promoter (ie1p), respectively. Bs, BstXI; N, NcoI; Sc, SacII; X, XbaI. (B) BV yields in culture medium determined by plaque assay on SpIm cells. (C) Accumulation of HA-HRF-1 and VP39 in infected cells monitored by immunoblotting using antisera against HA and VP39, respectively. (D) Photographs of infected cells at 72 hpi. (B, C, and D) Monolayer cultures of Ld652Y cells were infected with vHycuΔhr6/HA-HRF1 (Hycu/HRF1) and vHycuΔhr6/lacZ (Hycu/lacZ) at an input MOI of 5 PFU/cell. Vertical bars in panel B represent standard deviations of averages from three determinations. The numbers on the left of panel C indicate molecular masses (kilodaltons) of marker proteins (BenchMark Prestained Protein Ladder; Invitrogen). M, mock-infected cells.
FIG. 4.
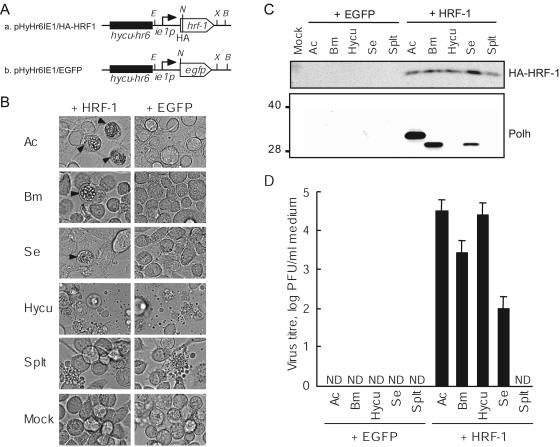
Replication of five different NPVs in HRF-1-expressing Ld652Y cells. (A) Schematic representation of the pGL3-Basic vector (Promega)-based construct used for transfection. pHyHr6IE1/HA-HRF1 (a) and pHyHr6Ie1/EGFP (b) contained the HA-fused hrf-1 gene and the egfp gene, respectively, both regulated by the hycu-ie1 promoter enhanced by hycu-hr6. B, BamHI; E, EcoRI; N, NcoI; X, XbaI. (B) Monolayer cultures of Ld652Y cells (106 cells) were transfected with 1 μg of pHyHr6IE1/HA-HRF1 (HRF-1) or pHyHr6IE1/EGFP (EGFP). At 24 h posttransfection, transfected cells were infected with AcMNPV (Ac), BmNPV (Bm), SeMNPV (Se), HycuNPV (Hycu), and SpltMNPV (Splt) at an MOI of 1 PFU/cell and were examined for cytopathology at 96 hpi. Arrowheads indicate the cells containing polyhedra. (C) Accumulation of HA-fused HRF-1 and polyhedrin in transfected and infected cells were monitored by immunoblotting with antisera against HA (HA-HRF-1; upper panel) and BmNPV polyhedrin (Polh; lower panel). The numbers on the left indicate the molecular masses (kilodaltons) of marker proteins. (D) Ld652Y cells were transfected with genomes of five different NPVs together with either pHyHr6IE1/HA-HRF1 (HRF-1) or pHyHr6IE1/EGFP (EGFP). BV yields in culture medium from transfected cells at 96 h posttransfection were determined by plaque assay using cell lines permissive for each NPV: Sf9, BmN4, Se301, SpIm, and SpLi cells for AcMNPV, BmNPV, SeMNPV, HycuNPV, and SpltMNPV, respectively. Vertical bars indicate the standard deviations from three determinations. ND, less than 1 PFU/ml.
Ld652Y and SpIm cells were infected with vHycuΔhr6/HA-HRF1 and vHycuΔhr6/lacZ at an MOI of 10 PFU/cell, and at various times pi, BV yields were examined by plaque assay on SpIm cells. The results showed that the BV titer increased to 1.4 × 106 PFU/ml at 72 hpi in vHycuΔhr6/HA-HRF1-infected Ld652Y cells, while there was no detectable BV production in vHycuΔhr6/lacZ-infected Ld652Y cells (Fig. 2B). In SpIm cells, no significant differences in levels of BV production were detected between vHycuΔhr6/HA-HRF1 and vHycuΔhr6/lacZ (Fig. 2B). Immunoblot analysis using anti-HA antibody (BABCO) showed that the HA-fused HRF-1 protein with an approximate molecular mass of 27 kDa was clearly detected at 24 hpi and decreased at 48 and 72 hpi in Ld652Y cells infected with vHycuΔhr6/HA-HRF1 (Fig. 2C, upper panel). Consistent with the results of BV yields, immunoblot analysis showed substantial accumulation of a viral major capsid protein, VP39, only in vHycuΔhr6/HA-HRF1-infected Ld652Y cells (Fig. 2C, lower panel). In vHycuΔhr6/HA-HRF1-infected Ld652Y cells, however, polyhedra were not observed under a microscope (Fig. 2D), and accumulation of polyhedrin was not detected by immunoblot analysis in either vHycuΔhr6/HA-HRF1- or vHycuΔhr6/lacZ-infected Ld652Y cells (data not shown). Addition of Z-VAD-FMK to inhibit apoptosis did not rescue polyhedrin synthesis in vHycuΔhr6/HA-HRF1-infected Ld652Y cells (data not shown). These results indicated that HRF-1 was an essential factor for productive HycuNPV infection of Ld652Y cells. However, HRF-1 did not promote polyhedrin production.
HRF-1 precludes global protein synthesis shutoff in HycuNPV-infected Ld652Y cells.
It has been shown that HRF-1 precludes global protein synthesis shutoff in AcMNPV-infected Ld652Y cells and leads to the productive infection of AcMNPV in Ld652Y cells (8, 31, 32). By analogy, it is possible that HRF-1 precludes virus-induced global protein synthesis shutoff in HycuNPV-infected Ld652Y cells as well. To explore this possibility, protein synthesis was comparatively analyzed in Ld652Y cells infected with vHycuΔhr6/HA-HRF1 and vHycuΔhr6/lacZ. Ld652Y cells (106) in 35-mm-diameter culture dishes (Falcon 3001) were infected with these recombinant viruses at an MOI of 10 PFU/cell and were cultured in medium containing or lacking Z-VAD-FMK. At 0, 12, 24, 36, and 48 hpi, infected cells were labeled with 1.8 MBq of [35S]methionine (specific activity, 43.5 TBq/mmol) (American Radiolabeled Chemicals) for 60 min, and labeled proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by fluorography, as described previously (30). The results showed that vHycuΔhr6/lacZ infection of Ld652Y cells resulted in global protein synthesis shutoff by 24 hpi, while the bands of presumable virus-specific proteins were detected by 48 hpi in vHycuΔhr6/HA-HRF1-infected Ld652Y cells (Fig. 3A). Z-VAD-FMK did not affect the pattern of protein synthesis in infected cells (Fig. 3B), indicating that global protein synthesis shutoff in vHycuΔhr6/lacZ-infected Ld652Y cells was not due to apoptosis. These results indicated that HRF-1 precluded global protein synthesis shutoff in Ld652Y cells infected with HycuNPV and promoted synthesis of HycuNPV-encoded proteins, suggesting that HRF-1 rescued HycuNPV replication by precluding global protein synthesis shutoff.
FIG. 3.
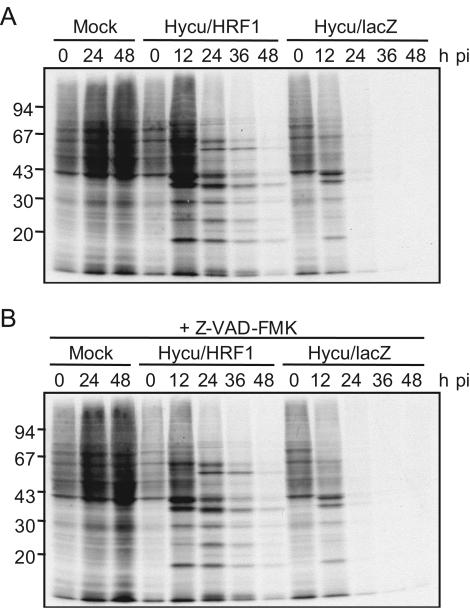
Profiles of protein synthesis in vHycuΔhr6/HA-HRF1- and vHycuΔhr6/lacZ-infected Ld652Y cells. Monolayer cultures of Ld652Y cells (106) were infected with vHycuΔhr6/HA-HRF1 (Hycu/HRF1) and vHycuΔhr6/lacZ (Hycu/lacZ) at an MOI of 10 PFU/cell and were cultured in medium only (A) or in medium containing 20 μM Z-VAD-FMK (B). At indicated times pi, infected cells were pulse-labeled with 1.8 MBq of [35S]methionine for 1 h, harvested, and lysed. Labeled polypeptides were resolved by SDS-PAGE and visualized by fluorography of the dried gels. Mock-infected cells were processed in the same manner. The numbers on the left indicate the molecular masses (kilodaltons) of marker proteins.
HRF-1 is an essential viral factor required for NPV productive infection of Ld652Y cells.
Previous studies have shown that infection of Ld652Y cells with Bombyx mori NPV (BmNPV), Spodoptera exigua MNPV (SeMNPV), and Spodoptera litula MNPV (SpltMNPV) result in nonproductive infections (20, 33). To determine if HRF-1 rescues replication of BmNPV, SeMNPV, and SpltMNPV in Ld652Y cells, BV yields and production of virus-encoded proteins were examined in Ld652Y cells transiently expressing HRF-1. To construct an hrf-1 expression vector, an HA tag-fused hrf-1 open reading frame amplified by PCR was subcloned into NcoI- and XbaI-digested pHyHr6IE1/Luc (9), generating pHyHr6IE1/HA-HRF1 (Fig. 4A), which contained an HA-fused hrf-1 gene regulated by the HycuNPV ie1 promoter enhanced by hycu-hr6. Ld652Y cells (106) were transfected with 1 μg of pHyHr6IE1/HA-HRF1 or pHyHr6IE1/EGFP (Fig. 4A) containing the egfp gene instead of HA-fused hrf-1, and at 24 h posttransfection transfected Ld652Y cells were infected with BmNPV, SeMNPV, SpltMNPV, AcMNPV, or HycuNPV, each at an MOI of 1 PFU/cell. Microscopic observation showed that Ld652Y cells transfected with pHyHr6IE1/HA-HRF1 prior to infection with AcMNPV, BmNPV, or SeMNPV produced polyhedra (Fig. 4B); in contrast, there was no detectable polyhedron formation in Ld652Y cells transfected with pHyHr6IE1/EGFP prior to infection with those NPVs (Fig. 4B). Immunoblot analysis with anti-HA antibody showed that Ld652Y cells transfected with pHyHr6IE1/HA-HRF1 prior to infection with NPVs accumulated substantial amounts of HA-fused HRF-1 protein (Fig. 4C, upper panel). Accumulation of polyhedrin was also observed in Ld652Y cells transfected with pHyHr6IE1/HA-HRF1 and then subsequently infected with AcMNPV, BmNPV, or SeMNPV (Fig. 4C, lower panel). Production of polyhedrin was not rescued by HRF-1 in Ld652Y cells infected with HycuNPV and SpltMNPV (Fig. 4C, lower panel).
Ld652Y cells were cotransfected with pHyHr6IE1/HA-HRF1 or pHyHr6IE1/EGFP and genomic DNA from AcMNPV, BmNPV, HycuNPV, SeMNPV, or SpltMNPV. Plaque assay on appropriate cell lines permissive for the respective NPVs showed that BV yields at 72 hpi in culture medium from pHyHr6IE1/HA-HRF1-transfected Ld652Y cells were 3.1 × 104, 2.7 × 103, 2.5 × 104, and 8.6 × 101 PFU/ml upon transfection with genomic DNAs from AcMNPV, BmNPV, HycuNPV, and SeMNPV, respectively, whereas there were no detectable BV yields in Ld652Y cells cotransfected with pHyHr6IE1/EGFP and NPV genomes (Fig. 4D). SpltMNPV BV production in Ld652Y cells was not rescued by HRF-1 (Fig. 4D). These results indicated that HRF-1 promoted the replication of BmNPV, HycuNPV, SeMNPV, and AcMNPV in Ld652Y cells.
Conclusions.
Our results demonstrated that BmNPV, HycuNPV, SeMNPV, and AcMNPV, which produced viral DNA in nonpermissive Ld652Y cells (14, 17, 33), were able to replicate with the aid of HRF-1, while replication of SpltMNPV, which produced no detectable viral DNA in Ld652Y cells (20), was not rescued by HRF-1. These results suggest that HRF-1 is required for an event that occurs after viral DNA replication to yield progeny virions in NPV-infected Ld652Y cells. Consistent with the results of AcMNPV-infected Ld652Y cells (8, 31, 32), our results also demonstrated that HRF-1 precluded global protein synthesis shutoff in HycuNPV-infected Ld652Y cells. It is thus concluded from the results presented in this study that global protein synthesis shutoff is the major factor that restricts NPV replication in Ld652Y cells and that HRF-1 is a crucial viral factor that copes with this antiviral mechanism operating in NPV-infected Ld652Y cells.
Acknowledgments
We thank T. Yaginuma and T. Niimi of the Laboratory of Sericulture and Entomoresources, Nagoya University, for their helpful discussions during this study.
This work was supported in part by grants in aid (13660059 and 14206007) from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Adams, J. R., and J. T. McClintock. 1991. Baculoviridae. Nuclear polyhedrosis viruses. Part 1. Nuclear polyhedrosis viruses of insects, p. 89-204. In J. R. Adams and J. R. Bonami (ed.), Atlas of invertebrate viruses. CRC Press, Boca Raton, Fla.
- 2.Ahrens, C. H., R. L. Q. Russell, C. J. Funk, J. T. Evans, S. H. Harwood, and G. F. Rohrmann. 1997. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology 229:381-399. [DOI] [PubMed] [Google Scholar]
- 3.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 4.Blissard, G., B. Black, N. Crook, B. A. Keddie, R. Possee, G. Rohrmann, D. Theilmann, and L. E. Volkman. 2000. Family Baculoviridae, p. 195-202. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the international committee on taxonomy of viruses. Academic Press, San Diego, Calif.
- 5.Chen, C.-J., M. E. Quentin, L. A. Brennan, C. Kukel, and S. M. Thiem. 1998. Lymantria dispar nucleopolyhedrovirus hrf-1 expands the larval host range of Autographa californica nucleopolyhedrovirus. J. Virol. 72:2526-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croizier, G., L. Croizier, O. Argaud, and D. Poudevigne. 1994. Extension of Autographa californica nuclear polyhedrosis virus host range by interspecific replacement of a short DNA sequence in the p143 helicase gene. Proc. Natl. Acad. Sci. USA 91:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du, X., and S. M. Thiem. 1997. Characterization of host range factor 1 (hrf-1) expression in Lymantria dispar M nucleopolyhedrovirus- and recombinant Autographa californica M nucleopolyhedrovirus-infected IPLB-Ld652Y cells. Virology 227:420-430. [DOI] [PubMed] [Google Scholar]
- 8.Du, X., and S. M. Thiem. 1997. Responses of insect cells to baculovirus infection: protein synthesis shutdown and apoptosis. J. Virol. 71:7866-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felipe Alves, C. A., M. Ikeda, and M. Kobayashi. 2002. Characterization of Hyphantria cunea nucleopolyhedrovirus gp64 gene and analysis of elements regulating its early promoter activity. J. Insect Biotechnol. Sericol. 71:141-150. [Google Scholar]
- 10.Felipe Alves, C. A., M. Ikeda, and M. Kobayashi. 2002. Identification and characterization of Hyphantria cunea nucleopolyhedrovirus homologous repeated regions. Virus Genes 25:281-290. [DOI] [PubMed] [Google Scholar]
- 11.Gomi, S., K. Majima, and S. Maeda. 1999. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 80:1323-1337. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin, R. H., G. J. Tompkins, and P. McCawley. 1978. Gypsy moth cell lines divergent in viral susceptibility. I. Culture and identification. In Vitro 14:485-494. [DOI] [PubMed] [Google Scholar]
- 13.Guzo, D., E. M. Dougherty, D. E. Lynn, S. K. Braun, and R. M. Weiner. 1991. Changes in macromolecular synthesis of gypsy moth cell line IPLB-Ld652Y induced by Autographa californica nuclear polyhedrosis virus infection. J. Gen. Virol. 72:1021-1029. [DOI] [PubMed] [Google Scholar]
- 14.Guzo, D., H. Rathburn, K. Guthrie, and E. Dougherty. 1992. Viral and host cellular transcription in Autographa californica nuclear polyhedrosis virus-infected gypsy moth cell lines. J. Virol. 66:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reily. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ijkel, W. F. J., E. A. van Strien, J. G. M. Heldens, R. Broer, D. Zuidema, R. W. Goldbach, and J. M. Vlak. 1999. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J. Gen. Virol. 80:3289-3304. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa, H., M. Ikeda, K. Yanagimoto, C. A. Felipe Alves, Y. Katou, B. A. Laviña-Caoili, and M. Kobayashi. 2003. Induction of apoptosis in an insect cell line, IPLB-Ld652Y, infected with nucleopolyhedroviruses. J. Gen. Virol. 84:705-714. [DOI] [PubMed] [Google Scholar]
- 18.Kamita, S. G., and S. Maeda. 1997. Sequencing of the putative DNA helicase-encoding gene of the Bombyx mori nuclear polyhedrosis virus and fine-mapping of a region involved in host range expansion. Gene 190:173-179. [DOI] [PubMed] [Google Scholar]
- 19.Kuzio, J., M. N. Pearson, S. H. Harwood, C. J. Funk, J. T. Evans, J. M. Slavicek, and G. F. Rohrmann. 1999. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology 253:17-34. [DOI] [PubMed] [Google Scholar]
- 20.Laviña, B. A., L. E. Padua, F. Q. Wu, N. Shirata, M. Ikeda, and M. Kobayashi. 2001. Biological characterization of a nucleopolyhedrovirus of Spodoptera litura (Lepidoptera: Noctuidae) isolated from the Philippines. Biol. Control 20:39-47. [Google Scholar]
- 21.Lu, A., and L. K. Miller. 1996. Species-specific effects of the hcf-1 gene on baculovirus virulence. J. Virol. 70:5123-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, S., S. G. Kamita, and A. Kondo. 1993. Host range expansion of Autographa californica nuclear polyhedrosis virus (NPV) following recombination of a 0.6-kilobase-pair DNA fragment originating from Bombyx mori NPV. J. Virol. 67:6234-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzacano, C. A., X. Du, and S. M. Thiem. 1999. Global protein synthesis shutdown in Autographa californica nucleopolyhedrovirus-infected Ld652Y cells is rescued by tRNA from uninfected cells. Virology 260:222-231. [DOI] [PubMed] [Google Scholar]
- 24.Miller, L. K., and A. Lu. 1997. The molecular basis of baculovirus host range, p. 217-235. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 25.Mitsuhashi, J., and H. Inoue. 1988. Obtainment of a continuous cell line from the larval fat bodies of the mulberry tiger moth, Spilosoma imparilis (Lepidoptera: Arctiidae). Appl. Entomol. Zool. 23:488-490. [Google Scholar]
- 26.Mitsuhashi, J., and K. Maramorosch. 1964. Leafhopper tissue culture: embryonic, nymphal, and imaginal tissues from aseptic insects. Contrib. Boyce Thompson Inst. 22:435-460. [Google Scholar]
- 27.Morris, T. D., and L. K. Miller. 1993. Characterization of productive and non-productive AcMNPV infection in selected insect cell lines. Virology 197:339-348. [DOI] [PubMed] [Google Scholar]
- 28.Pang, Y., J. Yu, L. Wang, X. Hu, W. Bao, G. Li, C. Chen, H. Han, S. Hu, and H. Yang. 2001. Sequence analysis of the Spodoptera litura multicapsid nucleopolyhedrovirus genome. Virology 287:391-404. [DOI] [PubMed] [Google Scholar]
- 29.Shirata, N., M. Ikeda, K. Kamiya, S. Kawamura, Y. Kunimi, and M. Kobayashi. 1999. Replication of nucleopolyhedroviruses of Autographa californica (Lepidoptera: Noctuidae), Bombyx mori (Lepidoptera: Bombycidae), Hyphantria cunea (Lepidoptera: Arctiidae), and Spodoptera exigua (Lepidoptera: Noctuidae) in four lepidopteran cell lines. Appl. Entomol. Zool. 34:507-516. [Google Scholar]
- 30.Sugimori, H., T. Nagamine, and M. Kobayashi. 1991. Protein synthesis in Bm-N cells infected with Bombyx mori nuclear polyhedrosis virus. J. Invertebr. Pathol. 58:257-268. [DOI] [PubMed] [Google Scholar]
- 31.Thiem, S. M., X. Du, M. E. Quentin, and M. M. Berner. 1996. Identification of a baculovirus gene that promotes Autographa californica nuclear polyhedrosis virus replication in a nonpermissive insect cell line. J. Virol. 70:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiem, S. M., and N. Chejanovsky. 2004. The role of baculovirus apoptotic suppressors in AcMNPV-mediated translation arrest in Ld652Y cells. Virology 319:292-305. [DOI] [PubMed] [Google Scholar]
- 33.Wu, F. Q., B. A. Laviña, M. Ikeda, N. Shirata, Y. X. Cai, S. X. Pan, and M. Kobayashi. 2000. Cloning and biological characterization of Spodoptera exigua nucleopolyhedroviruses isolated in China. J. Sericult. Sci. Jpn. 69:177-189. [Google Scholar]
- 34.Yanase, T., C. Yasunaga, and T. Kawarabata. 1998. Replication of Spodoptera exigua nucleopolyhedrovirus in permissive and non-permissive lepidopteran cell lines. Acta Virol. 42:293-298. [PubMed] [Google Scholar]


