Abstract
The L2 minor capsid proteins enter the nucleus twice during viral infection: in the initial phase after virion disassembly and in the productive phase when, together with the L1 major capsid proteins, they assemble the replicated viral DNA into virions. In this study we investigated the interactions between the L2 protein of high-risk human papillomavirus type 16 (HPV16) and nuclear import receptors. We discovered that HPV16 L2 interacts directly with both Kapβ2 and Kapβ3. Moreover, binding of Ran-GTP to either Kapβ2 or Kapβ3 inhibits its interaction with L2, suggesting that the Kapβ/L2 complex is import competent. In addition, we found that L2 forms a complex with the Kapα2β1 heterodimer via interaction with the Kapα2 adapter. In agreement with the binding data, nuclear import of L2 in digitonin-permeabilized cells could be mediated by either Kapα2β1 heterodimers, Kapβ2, or Kapβ3. Mapping studies revealed that HPV16 L2 contains two nuclear localization signals (NLSs), in the N terminus (nNLS) and C terminus (cNLS), that could mediate its nuclear import. Together the data suggest that HPV16 L2 interacts via its NLSs with a network of karyopherins and can enter the nucleus via several import pathways mediated by Kapα2β1 heterodimers, Kapβ2, and Kapβ3.
In the United States, infections caused by human papillomaviruses (HPVs) are more prevalent than all other sexually transmitted diseases combined. High-risk HPV infections are associated with more than 95% of cervical cancer, which is the second leading cause of cancer death among women. In addition, a high percentage of anal, perianal, vulvar, and penile cancers, as well as some non-melanoma skin cancers, are linked to oncogenic HPV infections. The main high-risk HPV types are HPV type 16 (HPV16), HPV18, HPV31, and HPV45, with HPV16 the most prevalent type in cervical cancers (29).
HPVs are small, nonenveloped, icosahedral DNA viruses that specifically infect the basal squamous epithelial cells. The virion particles (55 to 60 nm in diameter) consist of a single molecule of 8-kb double-stranded circular DNA contained within an icosahedral capsid comprising the L1 major and L2 minor capsid proteins (7). The L1 major capsid protein forms pentamers (capsomeres), and 72 capsomeres assemble into T-7d icosahedral lattice (11). The L2 minor structural protein is at about 1/30 the abundance of L1 (21), and it interacts with the L1 pentamers (2). Expression of L1 with a vaccinia virus or baculovirus system results in the formation of virus-like particles similar to viral capsids (12, 13). During the late phase of infection, the L1 and L2 proteins are synthesized in the cytoplasm of the differentiated cells of the infected epithelium and then transported to the nucleus for assembly of the replicated viral DNA into virions. Although L1 expressed alone in mammalian cells harboring episomal DNA forms virions (26), the L2 minor capsid protein dramatically increases the efficiency of DNA encapsidation (21, 24). Recent studies show that, at least for HPV33, expression and nuclear import of L2 precede those of L1 during the productive phase of viral infection in natural lesions (9a). Interestingly, bovine papillomavirus type 1 (BPV1) pseudovirions containing wild-type L1 without L2 protein or with mutant L2 protein lacking either an N-terminal or a C-terminal positively charged domain were noninfectious, despite efficient binding to the cell surface (20). This suggests that L2 also plays an essential role(s) in the infectious process via its N and C termini at a step after the binding of virions to the cells.
We have previously established that the L1 major capsid proteins of both high-risk HPV16 and -45 and low-risk HPV11 form complexes with Kapα2β1 heterodimers via interaction with the Kapα2 adapter and enter the nucleus via a classical Kapα2β1-mediated import pathway (14, 16, 17). Virtually nothing is known about the nuclear import pathways for the L2 minor capsid proteins of different papillomaviruses.
Nuclear import of NLS-containing proteins is mediated by soluble import receptors belonging to the karyopherin β/importin β (Kapβ/Impβ) superfamily that interact with nucleoporins at the nuclear pore complex to transport the proteins into the nucleus (10, 15). Binding of Ran-GTP to the Kapβs (importins) causes dissociation of the import complexes, causing or leading to release of the proteins inside the nucleus. It is thought that there are more than 20 members of the karyopherin β superfamily in higher eukaryotes, with some of them being better characterized than others. Kapβ1/Impβ can function together with a Kapα/Impα adapter in the nuclear import of proteins that contain classic monopartite or bipartite NLSs or without adapters in the import of proteins containing basic stretches such as ribosomal proteins, core histones, and several viral proteins. Mammalian Kapβ2/transportin mediates nuclear import of hnRNPs A1 and A2 via interaction with their Gly/Asn-rich NLS called M9 and of ribosomal proteins and core histones via interaction with their NLSs rich in basic amino acids. Interestingly, both the ribosomal proteins and core histones can interact with several import receptors and enter the nucleus via multiple redundant pathways (10, 15).
Here we investigated the interactions of the HPV16 L2 minor capsid protein with nuclear import receptors and dissected the pathways used by L2 to enter the nucleus. We found that L2 forms a complex with Kapα2β1 heterodimers via interaction with the Kapα2 adapter. In addition, we discovered that L2 interacts with both Kapβ2 and Kapβ3 nuclear import receptors. Significantly, Ran-GTP inhibits the interactions between L2 and either Kapβ2 or Kapβ3, suggesting that the Kapβ/L2 complex is import competent. In agreement with the binding data, nuclear import of L2 in digitonin-permeabilized cells could be mediated by either Kapα2β1, Kapβ2, or Kapβ3 pathways. Together the data suggest that HPV16 L2 may enter the nuclei of host cells via several pathways mediated by either Kapα2β1 heterodimers, Kapβ2, or Kapβ3.
MATERIALS AND METHODS
Preparation of recombinant human nuclear import factors.
His-tagged Kapα2 (27) and His-tagged Kapβ1 (3) were expressed in Escherichia coli BL21 Star and E. coli BL21(DE3), respectively (3 h of induction with 2 mM isopropyl-β-d-thiogalactopyranoside [IPTG] at 30°C), and the soluble His-tagged proteins were purified in their native state on Talon beads by a standard procedure. Glutathione S-transferase (GST)-Kapβ1 (3) and GST-Kapβ2 (4) were expressed in E. coli BL21(DE3), and GST-Kapβ3 (28) was expressed in E. coli BL21-CodonPlus (3 h of induction with 1 mM IPTG at 30°C), and the soluble GST-Kapβ fusion proteins were purified in their native state on glutathione-Sepharose beads by a standard procedure. Human Ran (5) was prepared as previously described (8). Ran was loaded with GTP by incubation with 100 mM GTP-0.5 M EDTA-1 M dithiothreitol (DTT)-1 M HEPES (pH 7.4) for 30 min at room temperature, and the reaction was stopped with 1.5 M magnesium chloride (pH 7.4). All proteins were checked for purity and lack of proteolytic degradation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. The purified proteins were dialyzed in transport buffer (20 mM HEPES-KOH [pH 7.3], 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM DTT, protease inhibitors) and stored in aliquots at −80°C until use. HeLa cytosol from National Cell Culture Center (Minneapolis, Minn.) was centrifuged and stored in small aliquots at −80°C.
Preparation of HPV16 L2 protein and mutant L2 proteins.
The pProEX HTb plasmid vector containing wild-type HPV16 L2 (22) was a kind gift from Richard Roden. To prepare the L2 deletion mutant proteins (L2ΔN lacking amino acids [aa] 1 to 9, L2ΔM lacking aa 297 to 313, and L2ΔC lacking aa 456 to 461), the L2ΔNΔC double mutant protein, and the L2ΔNΔMΔC triple mutant protein, the QuikChange site-directed mutagenesis kit from Stratagene was used in accordance with the instructions in the manual. All of the constructs were transformed into XL1 blue bacteria and confirmed by automated sequence analysis (Massachusetts General Hospital DNA Sequencing Department). His-tagged wild-type HPV16 L2 (22) and the mutant L2 proteins were expressed in E. coli BL21-CodonPlus (2 h of induction with 0.125 mM IPTG at 30°C) and purified from inclusion bodies with the Novagen Protein Refolding kit. The bacteria were lysed by sonication in 20 mM Tris-HCl (pH 7.5) buffer containing 10 mM EDTA, 1% Triton X-100, and 100 μg of lysozyme per ml and then subjected to centrifugation. The pellets were solubilized in 50 mM 3-cyclohexylamino-propanesulfonic acid (CAPS) (pH 11.0) containing 0.3% N-lauroylsarcosine. Refolding of the protein was achieved by extensive dialysis against 20 mM Tris-HCl (pH 8.5) containing 0.1 mM DTT. Final dialysis was performed against transport buffer (pH 7.3). His-tagged L2 was specifically bound to Talon beads via its His tag and subsequently eluted to be used in overlay blot assays.
For nuclear import assays, the wild-type and mutant L2 proteins were purified in urea on Talon beads as previously described (QIAGEN Inc.), followed by extensive dialysis in transport buffer to allow refolding of the proteins.
Preparation of GST-NLS fusion proteins.
The N-terminal sequence (1MRHKRSAKRTKRA13), the middle sequence (296SRRTGIRYSRIGNKQTLRTRSGK318), and the C-terminal sequence (455LRKRRKRL462) of HPV16 L2 were separately fused to a GST reporter protein to obtain the GST-nNLS, GST-mNLS, and GST-cNLS constructs. To make the GST-NLS constructs, the corresponding forward and reverse oligonucleotides containing EcoRI and XhoI restriction enzyme cutting sites were annealed and inserted directly into the pGEX4T-1 vector, which had been double cut with the same enzymes. The GST-NLS constructs were transformed into XL1 blue bacteria and confirmed by automated sequence analysis (Massachusetts General Hospital DNA Sequencing Department). For protein expression, the GST-nNLS, GST-mNLS, and GST-cNLS constructs were used to transform E. coli BL21-CodonPlus. After induction of the bacteria with 1 mM IPTG for 2 h at 30°C, the GST-NLS fusion proteins were purified in their native state on glutathione-Sepharose by a standard procedure. GST-NLSHPV16L1 containing the monopartite NLS (AKRKKRKL) of the HPV16 L1 major capsid protein was prepared as previously described (17).
Antibodies.
Rabbit polyclonal antiserum was raised against HPV16 L2 as previously described (23). A mouse antibody to Kapα2/Rch1 was from Transduction Laboratories; a mouse antibody to the six-His tag and a goat anti-GST antibody were from Amersham Biosciences. Horseradish peroxidase -conjugated secondary antibodies (anti-rabbit, anti-mouse, and anti-goat) were from Santa Cruz Biotechnology.
Overlay blot assays.
Wild-type and mutant HPV16 L2 proteins, different GST-NLS fusion proteins, and GST (all of the proteins were used at 2 μg/lane) were subjected to SDS-PAGE and transferred to nitrocellulose membrane. The blots were stained with Ponceau, and the individual lanes were cut and then blocked overnight at 4°C with 5% nonfat milk in phosphate-buffered saline. The individual blots were incubated with different Kaps as indicated in the figure legends. Bound Kapα2 was detected with an anti-Kapα2 antibody (1:500 dilution), and bound GST-Kap (Kapβ1, Kapβ2, or Kapβ3) or GST was detected with an anti-GST antibody (1:1,000 dilution), followed by the corresponding horseradish peroxidase-conjugated secondary antibodies (1:1,000 dilution). The signal was detected with an ECL Detection Kit (Amersham Biosciences).
In-solution binding assays.
Binding assays were performed as previously described (17). Briefly, the different GST-NLSs, or GST itself, immobilized on glutathione-Sepharose beads (2 μg/10 μl of beads) were incubated under rotation for 1 h at room temperature with the purified Kaps in binding buffer (transport buffer containing 0.01% Tween 20) as indicated in the figure legends. The bound proteins were eluted with SDS-PAGE sample buffer and analyzed by SDS-PAGE and Coomassie blue staining.
In vitro nuclear import assays.
The nuclear import assays were carried out as previously described (17). Briefly, subconfluent HeLa cells, grown on poly-l-lysine-coated glass coverslips for 24 h, were permeabilized with 70 μg of digitonin per ml for 5 min on ice and washed with transport buffer. All import reaction mixtures contained an energy-regenerating system (1 mM GTP, 1 mM ATP, 5 mM phosphocreatine, 0.4 U creatine phosphokinase) plus either HeLa cytosol or various transport factors (1 μM Kapα2, 0.5 μM Kapβ1, and 3 μM Ran-GDP) plus the different GST-NLS fusion proteins (0.25 μM) or L2 proteins. The final import reaction mixture volume was adjusted to 20 μl with transport buffer. For visualization of nuclear import, the GST-NLS fusion proteins were detected by immunofluorescence assay with an anti-GST antibody as previously described (17). Nuclear import of wild-type and mutant L2 proteins was detected with a polyclonal anti-L2 antibody. The nuclei were identified by 4′,6′-diamidino-2-phenylindole (DAPI) staining. Nuclear import was analyzed with a Nikon Eclipse TE 300 microscope that has a fluorescence attachment and a Sony DKC-5000 charge-coupled device camera.
RESULTS
The HPV16 L2 minor capsid protein forms a complex with Kapα2β1 heterodimers via interaction with the Kapα2 adapter.
To analyze the interactions between HPV16 L2 and the karyopherins, we used overlay blot assays. Purified, His-tagged L2 was subjected to SDS-PAGE and transferred to nitrocellulose membranes, and the L2 blots were either detected with anti-HPV16 L2 and anti-His antibodies (lanes 1 and 2 in Fig. 1, 2A, and 3A) or incubated with increasing concentrations of different karyopherins. As HPV16 L2 contains potential classical monopartite NLSs, we first investigated the interaction of L2 with the Kapα2 adapter. The L2-containing blots were incubated with Kapα2 in the absence (Fig. 1, lanes 3 to 5) or presence (Fig. 1, lanes 6 to 11) of GST-Kapβ1, and bound Kapα2 and GST-Kapβ1 were detected with an anti-Kapα2 antibody and an anti-GST antibody, respectively. We found that Kapα2 bound to L2 at concentrations of 2.5 and 5 μg of Kapα2 per ml either in the absence (Fig. 1, lanes 4 and 5) or in the presence (Fig. 1, lanes 7 and 8) of the Kapβ1 nuclear import receptor. At a lower concentration, 1 μg/ml, Kapα2 did not interact with L2 (Fig. 1, lanes 3 and 6). GST-Kapβ1 bound to L2 only in the presence of the Kapα2 adapter (Fig. 1, lanes 10 and 11) and not in its absence (Fig. 1, lane 12). GST, the negative control, did not bind L2 (Fig. 1, lane 13). The data suggest that HPV16 L2 forms a complex with Kapα2β1 heterodimers via interaction with the Kapα2 adapter.
FIG. 1.
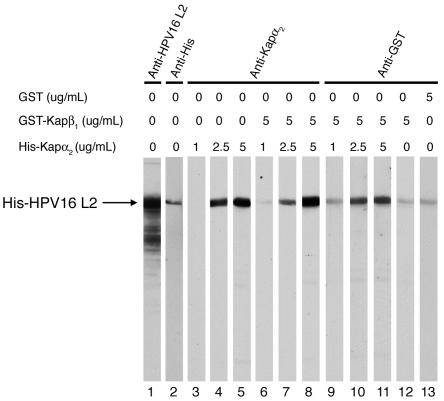
HPV16 L2 forms a complex with the Kapα2β1 heterodimer via interaction with the Kapα2 adapter. Purified L2 was subjected to SDS-PAGE and transferred onto a nitrocellulose membrane. Two L2 blots were detected with anti-HPV16 L2 (lane 1) and anti-His (lane 2) antibodies. Other L2 blots were incubated with increasing concentrations of Kapα2 in the absence (lanes 3 to 5) or the presence (lanes 6 to 11) of 5 μg of GST-Kapβ1 per ml. Separate L2 blots were incubated with GST-Kapβ1 alone (lane 12) or with GST as a negative control (lane 13). Bound Kapα2 was detected with an anti-Kapα2 antibody (lanes 3 to 8), and bound GST-Kapβ1 and GST were detected with an anti-GST antibody (lanes 9 to 13).
FIG. 2.
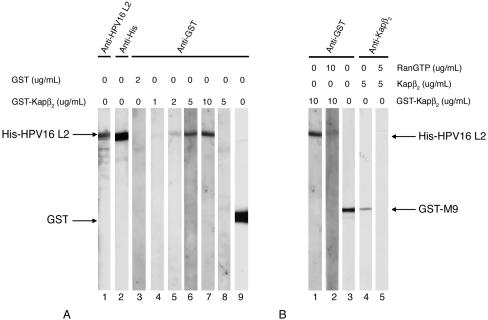
HPV16 L2 interacts specifically with Kapβ2, and Ran-GTP inhibits the interaction. (A) L2 (lanes 1 to 7), BSA (lane 8), and GST (lane 9) were subjected to SDS-PAGE and transferred onto nitrocellulose membrane. Two L2 blots were detected with anti-HPV16 L2 (lane 1) and anti-His (lane 2) antibodies. Other L2 blots were incubated with increasing concentrations of GST-Kapβ2 (lanes 4 to 7) or with 2 μg of GST per ml (lane 3). The BSA blot was incubated with 5 μg of GST-Kapβ2 per ml (lane 8). Bound GST-Kapβ2 and GST were detected with an anti-GST antibody. (B) L2 blots were incubated with GST-Kapβ2 in the absence (lane 1) or presence of Ran-GTP (lane 2), and bound GST-Kapβ2 was detected with an anti-GST antibody. GST-M9 blots were incubated with Kapβ2 in the absence (lane 4) or presence (lane 5) of Ran-GTP, and bound Kapβ2 was detected with an anti-Kapβ2 antibody. GST-M9 was detected with an anti-GST antibody (lane 3).
FIG. 3.
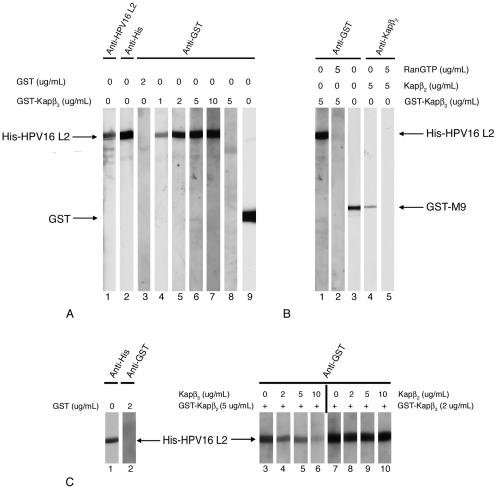
HPV16 L2 interacts specifically with Kapβ3, and Ran-GTP inhibits the interaction. (A) L2 (lanes 1 to 7), BSA (lane 8), and GST (lane 9) were subjected to SDS-PAGE and transferred onto a nitrocellulose membrane. Two L2 blots were detected with anti-HPV16 L2 (lane 1) and anti-His (lane 2) antibodies. Other L2 blots were incubated with increasing concentrations of GST-Kapβ3 (lanes 4 to 7) or with 2 μg of GST per ml (lane 3). The BSA blot was incubated with 5 μg of GST-Kapβ3 per ml (lane 8). Bound GST-Kapβ3 and GST were detected with an anti-GST antibody. (B) L2 blots were incubated with GST-Kapβ3 in the absence (lane 1) or presence (lane 2) of Ran-GTP, and bound GST-Kapβ3 was detected with an anti-GST antibody. GST-M9 blots were incubated with Kapβ2 in the absence (lane 4) or presence (lane 5) of Ran-GTP, and bound Kapβ2 was detected with an anti-Kapβ2 antibody. GST-M9 was detected with an anti-GST antibody (lane 3). (C) Kapβ3 competes with Kapβ2 for binding to HPV16 L2 minor capsid protein. L2 blots were incubated with either 5 μg of GST-Kapβ2 per ml in the absence or presence of increasing concentrations of Kapβ3 (lanes 3 to 6) or 2 μg of GST-Kapβ3 per ml in the absence or presence of increasing concentrations of Kapβ2 (lanes 7 to 10). Bound GST-Kapβ2 and GST-Kapβ3 were detected with an anti-GST antibody.
HPV16 L2 minor capsid protein interacts with both Kapβ2 and Kapβ3 nuclear import receptors, and Ran-GTP inhibits these interactions.
We next investigated if HPV16 L2 can interact directly with Kapβ nuclear import receptors. The L2 blots were incubated with increasing concentrations of GST-Kapβ2, and the bound GST-Kapβ2 was detected with an anti-GST antibody. We discovered that GST-Kapβ2 bound to L2 at concentrations of 5 and 10 μg/ml (Fig. 2A, lanes 6 and 7) and did not at concentrations of 1 and 2 μg/ml (Fig. 2A, lanes 4 and 5). As controls, GST alone did not interact with HPV16 L2 (Fig. 2A, lane 3) and GST-Kapβ2 did not interact with a neutral protein such as bovine serum albumin (BSA) (Fig. 2A, lane 8). These data suggest that HPV16 L2 interacts directly with the Kapβ2 nuclear import receptor. Significantly, Ran-GTP inhibited the interactions between GST-Kapβ2 and L2 (Fig. 2B, compare lanes 1 and 2) and, as a control, between Kapβ2 and the M9 type of NLS of hnRNP A1 (Fig. 2B, lanes 4 and 5). This suggests that the complex of L2 and Kapβ2 can be dissociated by binding of nuclear Ran-GTP to Kapβ2. GST and GST-M9 were detected with an anti-GST antibody (Fig. 2A, lane 9, and B, lane 3).
Interestingly, we also discovered that L2 binds with high affinity to the Kapβ3 nuclear import receptor (Fig. 3A, lanes 4 to 7). Again, the GST control did not bind to the L2 protein (Fig. 3A, lane 3) and GST-Kapβ3 did not interact with BSA (Fig. 3A, lane 8). Significantly, Ran-GTP inhibited the interaction between GST-Kapβ3 and L2 (Fig. 3B, compare lanes 1 and 2) and, as a control, between Kapβ2 and GST-M9 (Fig. 3B, compare lanes 4 and 5). This suggests that the L2-Kapβ3 complex can also be dissociated by nuclear Ran-GTP.
In competition assays we found that when the L2 blots were incubated with 5 μg of GST-Kapβ2 per ml in the presence of increasing amounts of Kapβ3, Kapβ3 efficiently competed with Kapβ2 for binding to L2 (Fig. 3C, compare lanes 4 to 6 with lane 3). In contrast, when the L2 blots were incubated with 2 μg of GST-Kapβ3 per ml in the presence of increasing amounts of Kapβ2, Kapβ2 could not compete with Kapβ3 for binding to L2 (Fig. 3C, compare lanes 8 to 10 with lane 7). Again, GST itself did not bind to L2 (Fig. 3C, lane 2). These data suggest that HPV16 L2 has a higher affinity for Kapβ3 than for Kapβ2 and that its two binding sites may overlap.
The binding data suggest that HPV16 L2 may use the Kapα2β1 heterodimers, Kapβ2, or Kapβ3 to enter the nucleus. To investigate the nuclear import pathways for HPV16 L2, we used the standard in vitro nuclear import assay. Digitonin-permeabilized HeLa cells were incubated with wild-type L2 in the presence of either exogenous cytosol or the corresponding recombinant karyopherins. As positive controls for the classical Kapα2β1 pathway, we used the monopartite NLS of HPV16 L1 (17) and for Kapβ2 we used the M9 type of NLS of hnRNP A1 (19). Both L2 and GST-NLSHPV16L1 were imported into the nuclei of digitonin-permeabilized cells in the presence of either cytosol (Fig. 4A, parts B and F) or Kapα2β1 heterodimers plus Ran-GDP (Fig. 4A, parts D and H). In addition, L2 was imported in the presence of either Kapβ2 plus Ran-GDP (Fig. 4B, part C) or Kapβ3 plus Ran-GDP (Fig. 4B, part D). Together the data suggest that HPV16 L2 can enter the nucleus via several pathways mediated by Kapα2β1 heterodimers, Kapβ2, or Kapβ3.
FIG. 4.
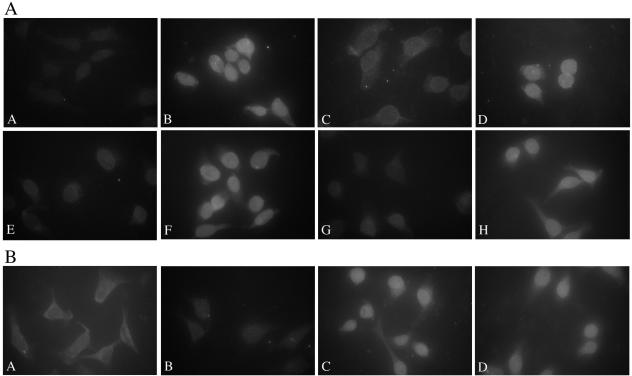
HPV16 L2 can enter the nucleus via Kapα2β1-, Kapβ2-, and Kapβ3-mediated pathways. (A) Digitonin-permeabilized HeLa cells were incubated with either wild-type L2 (panels A to D) or GST-NLSHPV16L1 (panels E to H) in the presence of either transport buffer (panels A and E), cytosol (panels B and F), Kapβ1 plus Ran-GDP (panels C and G), or Kapα2β1 plus Ran-GDP (panels D and H). Note the nuclear import in panels B, D, F, and H. (B) Digitonin-permeabilized HeLa cells were incubated with wild-type L2 in the presence of either transport buffer (panel A), Ran-GDP (panel B), Kapβ2 plus Ran-GDP (panel C), or Kapβ3 plus Ran-GDP (panel D). Note the nuclear import in panels C and D.
HPV16 L2 has two independent NLSs that interact with the Kapα2 adapter and mediate nuclear import via Kapα2β1 heterodimers.
It has been shown that HPV6b L2 contains two NLSs, localized near the N terminus and the C terminus, that can independently mediate nuclear import of L2 or of beta-galactosidase (25). A middle region around residues 286 to 306 was proposed to be involved in nuclear retention but alone was unable to mediate nuclear import (25). Dissection of HPV33 L2 domains involved in nuclear import suggested that the corresponding middle and C-terminal regions, but not the N terminus, function in its nuclear import (1), suggesting that there are differences among the L2 proteins of different HPV types. These sequences in the HPV L2 proteins are rich in lysine and arginine residues, a characteristic of many NLSs. We therefore made three GST fusion proteins containing the corresponding N-terminal (1MRHKRSAKRTKR13), middle (296SRRTGIRYSRIGNKQTLRTRSGK318), and C-terminal (454LRKRRKRL462) sequences (the boldface residues are basic amino acids) of HPV16 L2 and called them GST-nNLS, GST-mNLS, and GST-cNLS. We used nuclear import assays in digitonin-permeabilized cells to analyze the ability of each of these potential NLSs to mediate nuclear import of the GST reporter protein. Digitonin-permeabilized cells were incubated separately with GST-nNLS, GST-mNLS, GST-cNLS, or GST itself in the presence of either transport buffer (Fig. 5A to D) or cytosol (Fig. 5E to H). We found that both the nNLS and cNLS were able to mediate nuclear import of the GST reporter protein in the presence of cytosol (Fig. 5E and G), whereas the mNLS was not functional (Fig. 5F). As expected, the GST negative control was not imported into the nuclei (Fig. 5H).
FIG. 5.

Both the nNLS and cNLS of HPV16 L2 can mediate the nuclear import of a GST reporter protein. Digitonin-permeabilized HeLa cells were incubated with either GST-nNLS (panels A and E), GST-mNLS (panels B and F), GST-cNLS (panels C and G), or GST (panels D and H) in the presence of either only transport buffer (panels A to D) or HeLa cytosol (panels E to H). Note the nuclear import in panels E and G.
To map the binding domains of HPV16 L2 for the different karyopherins, we used both deletion mutagenesis and GST-NLS fusion proteins. We obtained three mutant L2 proteins lacking either the N terminus (aa 1 to 9), the middle region (aa 297 to 313), or the C terminus (aa 456 to 461). These were designated L2ΔN, L2ΔM, and L2ΔC, respectively. We also made the L2ΔNΔC double mutant protein and the L2ΔNΔMΔC triple mutant protein.
Blots containing wild-type L2 and the mutant L2 proteins were either probed with anti-His antibody, as a loading control (Fig. 6, lanes 1 to 6), or incubated with Kapα2 (Fig. 6, lanes 7 to 12). Comparison analysis of the interactions of wild-type L2 and the mutant L2 proteins with Kapα2 revealed that deletion of the nNLS of HPV16 L2 strongly reduced Kapα2 binding (Fig. 6, compare lanes 8 and 7). The L2ΔM and L2ΔC mutant proteins bound to Kapα2 as well as did wild-type L2 (Fig. 6, compare lanes 9 and 10 with lane 7). Both the L2 double mutant protein lacking the N and C termini and the L2ΔNΔMΔC triple mutant protein no longer bound to Kapα2 (Fig. 6, lanes 11 and 12). These data suggest that the N terminus contains a high-affinity binding site for Kapα2. Indeed, overlay blot analysis of the interactions of L2 NLSs with Kapα2 showed that the nNLS interacted with Kapα2 with high affinity (Fig. 7A, lanes 2 to 4), similar to the NLS of HPV16 L1, which was used as a positive control (Fig. 7A, lanes 14 to 16). Interestingly, the cNLS also interacted with Kapα2, although with much lower affinity than the nNLS (Fig. 7A, compare lanes 10 to 12 with lanes 2 to 4). The mNLS did not interact specifically with Kapα2 (Fig. 7A, lanes 6 to 8); there was only a low nonspecific interaction at 5 μg of Kapα2 per ml (Fig. 7A, lane 8), at the same level as with GST (Fig. 7A, lane 18). The different GST-NLSs and GST were detected with an anti-GST antibody (Fig. 7A, lanes 1, 5, 9, 13, and 17).
FIG. 6.
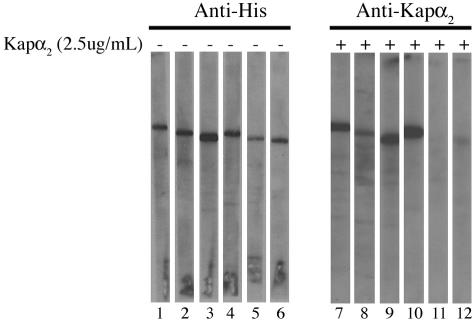
Deletion of both the N and C termini of HPV16 L2 abolishes interaction with the Kapα2 adapter. Blots containing wild-type L2 protein (lanes 1 and 7), L2ΔN mutant protein (lanes 2 and 8), L2ΔM mutant protein (lanes 3 and 9), L2ΔC mutant protein (lanes 4 and 10), L2ΔNΔC double mutant protein (lanes 5 and 10), and L2ΔNΔMΔC triple mutant protein (lanes 6 and 12) were either probed with an anti-His antibody (lanes 1 to 6) or incubated with 2.5 μg of Kapα2 per ml, and bound Kapα2 was detected with an anti-Kapα2 antibody (lanes 7 to 12).
FIG. 7.
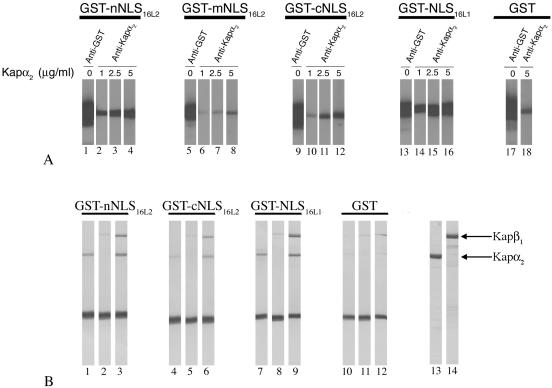
Both the nNLS and cNLS of HPV16 L2 interact with the Kapα2 adapter and form a complex with Kapα2β1 heterodimers. (A) Blots containing GST-nNLS16L2 (lanes 2 to 4), GST-mNLS16L2 (lanes 6 to 8), GST-cNLS16L2 (lanes 10 to 12), GST-NLS16L1 (positive control; lanes 14 to 16), or GST (negative control; lane 18) were incubated with increasing amounts of Kapα2, and bound Kapα2 was detected with an anti-Kapα2 antibody. The different GST-NLS proteins and GST were detected with an anti-GST antibody (lanes 1, 5, 9, 13, and 17, respectively). (B) GST-nNLS16L2 (lanes 1 to 3), GST-cNLS16L2 (lanes 4 to 6), GST-NLSHPV16L1 (lanes 7 to 9), and GST (lanes 10 to 12) immobilized on glutathione-Sepharose (2 μg/10 μl of beads) were incubated with either Kapα2 (lanes 1, 4, 7, and 10), Kapβ1 (lanes 2, 5, 8, and 11), or Kapβ1 plus Kapα2 (lanes 3, 6, 9, and 12). Bound proteins were eluted with sample buffer and analyzed by SDS-PAGE and Coomassie blue staining. The input of Kapα2 and Kapβ1 is shown in lanes 13 and 14.
In agreement with the overlay blot data, in-solution binding assays revealed that both the nNLS and the cNLS bound to Kapα2 (Fig. 7B, lanes 1 and 4) and formed a complex with the Kapα2β1 heterodimer (Fig. 7B, lanes 3 and 6) and that the nNLS had a higher binding affinity than the cNLS. Neither the nNLS nor the cNLS interacted directly with the Kapβ1 import receptor (Fig. 7B, lanes 2 and 5). As expected, the NLS of HPV16 L1, used as a positive control, interacted with Kapα2 and formed a complex with the Kapα2β1 heterodimer (Fig. 7B, lanes 7 to 9), whereas no interactions were observed with GST itself (Fig. 7B, lanes 10 to 12).
We next investigated if the nNLS and the cNLS could mediate nuclear import of a GST reporter protein via the classical Kapα2β1 pathway. Digitonin-permeabilized HeLa cells were incubated with the GST-nNLS, the GST-cNLS, GST (negative control), or the GST-NLSHPV16L1 (as a positive control) in the presence of either only transport buffer or the corresponding Kaps (Fig. 8). The nNLS of HPV16 L2 mediated nuclear import via a Kapα2β1 pathway as efficiently as the NLS of HPV16 L1 (Fig. 8, compare panels C and I). In agreement with the binding data, the cNLS of HPV16 L2 was less efficient in mediating nuclear import via Kapα2β1 heterodimers (Fig. 8F). The mNLS was unable to mediate nuclear import of the GST reporter via the Kapα2β1 pathway (data not shown).
FIG. 8.
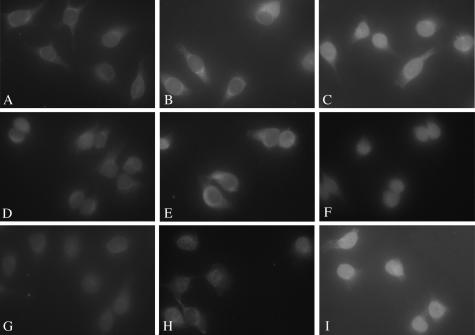
The nNLS of HPV16 L2 efficiently mediates the nuclear import of a GST reporter via the Kapα2β1 pathway. Digitonin-permeabilized cells were incubated with either GST-nNLS (panels A to C), GST-cNLS (panels D to F), or GST-NLSHPV16L1 (panels G to I) in the presence of either transport buffer (panels A, D, and G), Kapβ1 plus Ran-GDP (panels B, E, and H), or Kapα2 plus Kapβ1 plus Ran-GDP (panels C, F, and I). Nuclear import of the GST-NLSs was detected with an anti-GST antibody.
The nNLS of HPV16 L2 is essential for the interaction with and import via either Kapβ2 or Kapβ3.
To determine the domain of L2 required for Kapβ2 interaction, blots containing wild-type L2 and the mutant L2 proteins were either probed with anti-His antibody, as a loading control (Fig. 9A, lanes 1 to 6), or incubated with Kapβ2 (Fig. 9A, lanes 7 to 12). This analysis revealed that deletion of the N terminus of L2 strongly reduced Kapβ2 binding (Fig. 9A, compare lanes 8 and 7). Deletion of the C terminus of L2 caused a small reduction in Kapβ2 binding (Fig. 9A, compare lanes 10 and 7), whereas Kapβ2 binding to L2ΔM was not affected (Fig. 9A, lane 9). The double mutant protein lacking the N and C termini and the L2ΔNΔMΔC triple mutant protein no longer interacted with Kapβ2 (Fig. 9A, lanes 11 and 12). A similar analysis was performed to determine the domain of L2 required for Kapβ3 interaction. The blots containing wild-type L2 and the mutant L2 proteins were either probed with anti-His antibody (Fig. 9B, lanes 1 to 6) or incubated with Kapβ3 (Fig. 9B, lanes 7 to 12). This analysis revealed that deletion of the nNLS also strongly reduced Kapβ3 binding (Fig. 9B, compare lanes 8 and 7). Deletion of the mNLS or cNLS of L2 had no significant effect on Kapβ3 binding (Fig. 9B, lanes 9 and 10). Both the L2ΔNΔC double mutant protein and the L2ΔNΔMΔC triple mutant protein no longer bound Kapβ3 (Fig. 9B, lanes 11 and 12).
FIG. 9.
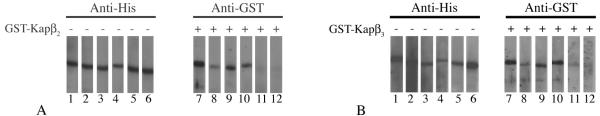
The N terminus of HPV16 L2 is essential for the interaction with Kapβ2 and Kapβ3 nuclear import receptors. (A) Blots containing wild-type L2 protein (lanes 1 and 7), L2ΔN mutant protein (lanes 2 and 8), L2ΔM mutant protein (lanes 3 and 9), L2ΔC mutant protein (lanes 4 and 10), L2ΔNΔC double mutant protein (lanes 5 and 10), and L2ΔNΔMΔC triple mutant protein (lanes 6 and 12) were either probed with anti-His antibody (lanes 1 to 6) or incubated with 10 μg of GST-Kapβ2 per ml, and bound GST-Kapβ2 was detected with an anti-GST antibody (lanes 7 to 12). (B) Blots containing wild-type L2 protein (lanes 1 and 7), L2ΔN mutant protein (lanes 2 and 8), L2ΔM mutant protein (lanes 3 and 9), L2ΔC mutant protein (lanes 4 and 10), L2ΔNΔC double mutant protein (lanes 5 and 10), and L2ΔNΔMΔC triple mutant protein (lanes 6 and 12) were either probed with an anti-His antibody (lanes 1 to 6) or incubated with 10 μg of GST-Kapβ3 per ml, and bound GST-Kapβ3 was detected with an anti-GST antibody (lanes 7 to 12).
In agreement with the binding data, deletion of the nNLS strongly reduced the nuclear import of L2ΔN via either Kapβ2- or Kapβ3-mediated pathways (Fig. 10, compare panels E and F with panels B and C). Deletion of the mNLS had no effect on nuclear import of L2, as L2ΔM was imported into the nucleus via either pathway as well as wild-type L2 (data not shown). Unfortunately, the results obtained with the L2ΔC mutant protein were not informative as the protein aggregated in the cytoplasmic area (data not shown). The L2ΔNΔC double mutant protein was no longer imported into the nucleus via either the Kapβ2 or the Kapβ3 pathway (data not shown). Together these data suggest that the nNLS of HPV16 L2, rich in arginine and lysine residues, is essential for the interaction with Kapβ2 and Kapβ3 and consequently for Kapβ2- and Kapβ3-mediated nuclear import. However, GST-nNLS alone did not efficiently interact with either Kapβ2 or Kapβ3 and consequently was not imported via Kapβ2- and Kapβ3-mediated pathways (data not shown). This suggests that the nNLS is necessary, but not sufficient, for the interaction with Kapβ2 and Kapβ3 and that additional residues may be required.
FIG. 10.

The nNLS is essential for Kapβ2- or Kapβ3-mediated nuclear import of HPV16 L2. Digitonin-permeabilized HeLa cells were incubated with either wild-type L2 protein (panels A to C) or L2ΔN mutant protein (panels D to F) in the presence of either transport buffer (panels A and D), Kapβ2 plus Ran-GDP (panels B and E), or Kapβ3 plus Ran-GDP (panels C and F). Note the nuclear import in panels B and C.
DISCUSSION
The L2 minor capsid proteins enter the nucleus twice during the viral life cycle: in the initial phase after virion disassembly and in the productive phase when newly synthesized L2, together with L1, encapsidates the viral DNA into new virions. Expression and nuclear import of L2 precede those of L1 during the productive phase of viral infection in natural lesions (9a).
In this study we investigated the interactions of the HPV16 L2 minor capsid protein with the host nuclear import receptors and analyzed its NLSs and nuclear import pathways. We discovered that L2 forms a complex with Kapα2β1 heterodimers via interaction with the Kapα2 adapter and also interacts with high affinity with the Kapβ2 and Kapβ3 nuclear import receptors. Moreover, binding of Ran-GTP to either Kapβ2 or Kapβ3 inhibits their interaction with L2, suggesting that the Kapβ/L2 complexes are import competent. In in vitro nuclear import assays with digitonin-permeabilized cells, wild-type L2 was imported into the nuclei in the presence of exogenous cytosol containing the karyopherins. In agreement with the binding data, L2 could enter the nucleus via several pathways mediated by either Kapα2β1 heterodimers or Kapβ2 or Kapβ3 receptors. We have previously established that HPV16 L1 major capsid protein interacts with the Kapα2 adapter and enters the nucleus via a classical Kapα2β1-mediated pathway (17). Interestingly, although HPV16 L1 also interacts with lower affinities with Kapβ2 and Kapβ3, it does not exploit these interactions to enter the nucleus via two additional pathways (17). The significance of the interactions between HPV16 L1 and Kapβ2 and Kapβ3 receptors during the productive phase of infection remains to be determined. Thus, the L1 and L2 capsid proteins of high-risk HPV16 evolved both common and distinct strategies to enter the nuclei of host cells.
Preliminary studies on the L2 proteins of other HPV types suggest that those of high-risk HPV18 and low-risk HPV11 also interact with Kapβ2 and Kapβ3 import receptors and form a complex with Kapα2β1 heterodimers (M. S. Darshan, E. Harding, and J. Moroianu, unpublished data). In addition, HPV11 L2 interacts with low affinity directly with Kapβ1 import receptor. In contrast, the L2 protein of BPV1 interacts only with Kapα2β1 heterodimers and not with either Kapβ1, Kapβ2, or Kapβ3 (7a). This suggests that the HPV L2 minor capsid proteins evolved multiple interactions with the host nuclear import receptors in comparison with the BPV1 L2 protein.
When the L2 proteins of either BPV1 or different HPV types are expressed in BPHE-1, HuTK−, or COS-7 cells they accumulate in the nucleus in specific nuclear domains called PML oncogenic domains or ND10 (1, 6, 18, 20). For HPV33 L2, the domain involved in targeting L2 to ND10 was mapped in the C-terminal half of L2 (1). In our study, when digitonin-permeabilized HeLa cells were incubated with HPV16 L2 in the presence of either cytosol or recombinant karyopherins, the L2 imported into the nucleus was diffusely distributed throughout the nucleoplasm and did not accumulate at ND10. It is not clear if this is due to the fact that the cells are permeabilized with digitonin (and consequently a targeting factor is missing) or because ND10 localization is too slow to occur within the 30-min time of the nuclear import assay.
It has been recently reported that in cells that express L2, Hsc70 chaperone is relocalized from the cytoplasm to ND10 (9b). Moreover, L2 was found in a complex with Hsc70 and this interaction was required for nuclear translocation of L2 in intact cells; in the absence of Hsc70, L2 was retained in the cytoplasm most likely via interaction with another cytoplasmic protein(s) (9b). In our system nuclear import of HPV16 L2 in digitonin-permeabilized cells in the presence of recombinant karyopherins does not require exogenous Hsp70. This suggests that the cytoplasmic protein(s) involved in L2 retention in the cytoplasm is washed out in digitonin-permeabilized cells.
By deletion mutagenesis and with GST fusion proteins, we determined that HPV16 L2 contains two NLSs rich in basic amino acids in the N terminus (nNLS) and in the C terminus (cNLS) that can function in its nuclear import. The mNLS region was unable to mediate nuclear import of a GST reporter in digitonin-permeabilized cells, and deletion of the mNLS had no effect on L2 nuclear import. These data reveal that HPV16 L2 is similar to HPV6b L2 in that both have two functional NLSs in the N and C termini (25; this study). It is possible that the mNLS of HPV16 L2 has a role in nuclear retention, as suggested for the mNLS of HPV6b L2 (25).
Both the nNLS and the cNLS interacted with the Kapα2 adapter and mediated nuclear import of a GST reporter protein via Kapα2β1 heterodimers. The nNLS had higher affinity for the Kapα2 adapter and consequently was more efficient in mediating nuclear import via a classical Kapα2β1 pathway. The nNLS, which is rich in arginines and lysines, was also essential for the interaction with Kapβ2 and Kapβ3, and consequently deletion of the nNLS abolished Kapβ2- or Kapβ3-mediated nuclear import. Interestingly, the nNLS alone was not sufficient to interact efficiently with either Kapβ2 or Kapβ3 and was unable to mediate nuclear import of a GST fusion protein via Kapβ2 and Kapβ3. This suggests that the nNLS is only part of the binding domain for Kapβ2 or Kapβ3 and that additional residues are required for the interaction with Kapβ2 and Kapβ3. It is difficult to speculate about whether the cNLS contributes to the Kapβ2 and Kapβ3 binding site. Future determination of the tertiary structure of L2 combined with point mutation studies will give insight into potential additional residues involved in Kapβ2 and Kapβ3 interaction.
It is unclear why HPV16 L2 has more than one NLS, interacts with at least three nuclear import receptors, and can enter the nucleus via several pathways. It is possible that HPV16 L2 uses a different NLS and nuclear import pathway(s) in the initial phase of viral infection after virion disassembly versus in the productive phase taking place in differentiated epithelial cells. Future studies in this area may help in elucidating the trafficking pathways in the different stages of viral infection.
Acknowledgments
We thank G. Blobel, Y. M. Chook, S. Adam, G. Dreyfuss, K. Weis, A. Lamond, and N. Yaseen for the generous gifts of expression vectors. We thank R. B. S. Roden for the kind gift of the HPV16 L2 expression plasmid and for the anti-HPV16 L2 polyclonal antibody. We thank James Daley and Sophie Forte for technical assistance in preparation of recombinant transport factors. We thank A. Annunziato, C. Hoffman, and Y. M. Chook for helpful discussions and C. Hoffman and R. B. S. Roden for critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health (1-RO1 CA94898-01) to J.M.
REFERENCES
- 1.Becker, K. A., L. Florin, C. Sapp, and M. Sapp. 2003. Dissection of human papillomavirus type 33 L2 domains involved in nuclear domains (ND) 10 homing and reorganization. Virology 314:161-167. [DOI] [PubMed] [Google Scholar]
- 2.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 3.Chi, N. C., E. J. Adam, G. D. Visser, and S. A. Adam. 1996. RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import. J. Cell Biol. 135:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chook, Y. M., and G. Blobel. 1999. Structure of the nuclear transport complex karyopherin-β2-Ran x GppNHp. Nature 399:230-237. [DOI] [PubMed] [Google Scholar]
- 5.Coutavas, E., M. Ren, J. D. Oppenheim, P. D'Eustachio, and M. G. Rush. 1993. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature 366:585-587. [DOI] [PubMed] [Google Scholar]
- 6.Day, P. M., R. B. S. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doorbar, J., and P. H. Gallimore. 1987. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus 1a. J. Virol. 61:2793-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Fay, A. K., W. Yutzy, R. B. S. Roden, and J. Moroianu. The positively charged termini of L2 minor capsid protein required for bovine papillomavirus infection function separately in nuclear import and DNA binding. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 8.Floer, M., and G. Blobel. 1996. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J. Biol. Chem. 271:5313-5316. [DOI] [PubMed] [Google Scholar]
- 9a.Florin, L., C. Sapp, R. E. Streeck, and M. Sapp. 2002. Assembly and translocation of papillomavirus capsid proteins. J. Virol. 76:10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b.Florin, L., K. A. Becker, C. Sapp, C. Lambert, H. Sirma, M. Muller, R. E. Streeck, and M. Sapp. 2004. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J. Virol. 78:5546-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fried, H., and U. Kutay. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagensee, M. E., N. H. Olson, T. S. Baker, and D. A. Galloway. 1994. Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J. Virol. 68:4503-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirnbauer, R., J. Taub, H. Greenstone, R. B. S. Roden, M. Durst, L. Gissmann, D. R. Lowy, and J. T. Schiller. 1993. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 67:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merle, E., R. C. Rose, L. LeRoux, and J. Moroianu. 1999. Nuclear import of HPV11 L1 capsid protein is mediated by karyopherin α2β1 heterodimers. J. Cell. Biochem. 74:628-637. [PubMed] [Google Scholar]
- 15.Moroianu, J. 1999. Nuclear import and export pathways. J. Cell. Biochem. 74(Suppl.):76-83. [DOI] [PubMed] [Google Scholar]
- 16.Nelson, L. M., R. C. Rose, L. LeRoux, C. Lane, K. Bruya, and J. Moroianu. 2000. Nuclear import and DNA binding of human papillomavirus type 45 L1 capsid protein. J. Cell. Biochem. 79:225-238. [PubMed] [Google Scholar]
- 17.Nelson, L. M., R. C. Rose, and J. Moroianu. 2002. Nuclear import strategies of high risk HPV16 L1 major capsid protein. J. Biol. Chem. 277:23958-23964. [DOI] [PubMed] [Google Scholar]
- 18.Okun, M. M., P. M. Day, H. L. Greenstone, F. P. Booy, D. R. Lowy, J. T. Schiller, and R. B. S> Roden. 2001. L1 interaction domains of papillomavirus l2 necessary for viral genome encapsidation. J. Virol. 75:4332-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 20.Roden, R. B. S., P. M. Day, B. K. Bronzo, W. H. Yutzy IV, Y. Yang, D. R. Lowy, and J. T. Schiller. 2001. Positively charged termini of the L2 minor capsid protein are necessary for papillomavirus infection. J. Virol. 75:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roden, R. B. S., H. L. Greenstone, R. Kirnbauer, F. P. Booy, J. Jessie, D. R. Lowy, and J. T. Schiller. 1996. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 70:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roden, R. B. S., R. Kirnbauer, A. B. Jenson, D. R. Lowy, and J. T. Schiller. 1994. Interaction of papillomaviruses with the cell surface. J. Virol. 68:7260-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roden, R. B. S., W. H. Yutzy IV, R. Fallon, S. Inglis, D. R. Lowy, and J. T. Schiller. 2000. Minor capsid protein of human genital papillomaviruses contains subdominant, cross-neutralizing epitopes. Virology 270:254-257. [DOI] [PubMed] [Google Scholar]
- 24.Stauffer, Y., K. Raj, K. Masternak, and P. Beard. 1998. Infectious human papillomavirus type 18 pseudovirions. J. Mol. Biol. 283:529-536. [DOI] [PubMed] [Google Scholar]
- 25.Sun, X. Y., I. Frazer, M. Muller, L. Gissmann, and J. Zhou. 1995. Sequences required for the nuclear targeting and accumulation of human papillomavirus type 6B L2 protein. Virology 213:321-327. [DOI] [PubMed] [Google Scholar]
- 26.Unckell, F., R. E. Streeck, and M. Sapp. 1997. Generation and neutralization of pseudovirions of human papillomavirus type 33. J. Virol. 71:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 28.Yaseen, N. R., and G. Blobel. 1997. Cloning and characterization of human karyopherin β3. Proc. Natl. Acad. Sci. USA 94:4451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]


