Abstract
Development of a mouse model for human immunodeficiency virus type 1 (HIV-1) infection has advanced through the progressive identification of host cell factors required for HIV-1 replication. Murine cells lack HIV-1 receptor molecules, do not support efficient viral gene expression, and lack factors necessary for the assembly and release of virions. Many of these blocks have been described using mouse fibroblast cell lines. Here we identify a postentry block to HIV-1 infection in mouse T-cell lines that has not been detected in mouse fibroblasts. While murine fibroblastic lines are comparable to human T-cell lines in permissivity to HIV-1 transduction, infection of murine T cells is 100-fold less efficient. Virus entry occurs efficiently in murine T cells. However, reduced efficiency of the completion of reverse transcription and nuclear transfer of the viral preintegration complex are observed. Although this block has similarities to the restriction of murine retroviruses by Fv1, there is no correlation of HIV-1 susceptibility with cellular Fv1 genotypes. In addition, the block to HIV-1 infection in murine T-cell lines cannot be saturated by a high virus dose. Further studies of this newly identified block may lend insight into the early events of retroviral replication and reveal new targets for antiretroviral interventions.
Murine cells are refractory to human immunodeficiency virus type 1 (HIV-1) replication at multiple stages of the viral life cycle. While this has allowed a finer inspection of the role of various host factors in HIV-1 replication, it has been an impediment to the development of a genetically modified mouse permissive to HIV-1 infection. A number of host factors have been identified that are indispensable for replication of HIV-1. Necessary factors absent in murine T cells include the human forms of the HIV-1 receptor and coreceptor molecules, CD4 and CCR5, whose murine orthologs do not support HIV-1 entry (10). In contrast, the murine form of CXCR4 can be utilized as a coreceptor by HIV-1 (7, 69). Postentry, human cyclin T1 is necessary for efficient Tat-mediated transactivation of the HIV-1 long terminal repeat (LTR) (22). Additional blocks in later steps of the HIV-1 life cycle in murine cells include excessive splicing of HIV-1 genomic RNA (49) and defects in Rev function (72). Aberrant splicing of HIV-1 mRNA appears to be partially corrected by the human splice inhibitor p32 (80). HIV-1 particle assembly is also impaired in murine cells (6, 53). This restriction can be overcome by fusion of murine cells with human cells, suggesting that murine cells lack a factor (or factors) necessary for HIV-1 Gag assembly and release (52). Substitution of the HIV-1 matrix (MA) region with that of murine leukemia virus (MLV) also circumvents this block, supporting the notion that species-specific cofactors are critical for virion assembly and release (13, 63).
Not only do mouse cells lack some of the factors necessary for HIV-1 replication, they also express factors that actively interfere with retroviral replication. Mouse cells express the factor Fv1 (47, 56, 58), which is present in several allelic forms that specifically restrict infection by certain strains of MLV (48). The two predominant Fv1 forms are Fv1n and Fv1b; Fv1n expression restricts B-tropic MLV infection, whereas Fv1b restricts N-tropic MLV. Laboratory-adapted MLV strains, such as Moloney MLV, proliferate equally well in Fv1n or Fv1b cells, exhibiting N/B-tropism. Cells carrying a nonrestrictive allele, referred to as Fv10, allow infection by both N- and B-tropic MLV (24, 27, 43, 44). Fv1 exerts its antiretroviral effect at a postentry step, after reverse transcription and prior to integration (37, 76). Restriction by Fv1 is not absolute; it can be overcome by a high multiplicity of infection (MOI) (8, 17, 19, 28, 58). Although the Fv1 gene has been identified (5), the restriction mechanism remains elusive (71, 77). N-tropic MLV is also restricted in human cells, and this restriction is due to a factor referred to as Ref1. This restriction is similar to that of Fv1 and can be saturated by challenge with a high MOI by N-tropic MLV (1, 71). In addition to MLV restrictions, human and simian cells contain Lv1 (4, 15, 54), which interferes with the replication of several lentiviruses (including HIV-1, HIV-2, simian immunodeficiency virus, and equine infectious anemia virus) in a virus strain- and cell-specific manner (29). In contrast to Fv1, Lv1 and Ref1 appear to act at a step prior to reverse transcription.
Other factors that broadly interfere with the replication of different virus genera affect retroviruses at distinct stages of infection. ZAP, a zinc-finger protein expressed in rat cells, reduces retroviral gene expression by depleting the cytoplasmic pool of retroviral mRNA (21). The proteins INI1 and PML, constituents of two distinct nuclear protein complexes (SWI/SNF and POD), are exported from the nucleus to the cytoplasm of human cells upon infection by HIV-1 and appear to interfere with HIV-1 replication at a step before integration (73). During virion production in mammalian cells, a cytidine deaminase identified as APOBEC3G (66) is packaged into HIV-1 particles and induces hypermutation of the reverse-transcribed viral genome in a newly infected cell (26, 45, 50, 51, 78). The HIV-1 Vif protein interferes with APOBEC3G incorporation in a species-specific manner. For example, the macaque homolog of APOBEC3G interferes with HIV-1 infectivity but is not counteracted by HIV-1 Vif (51). Thus, both dominant and recessive characteristics of mammalian cell types pose a challenge in the cross-species transmission of retroviruses.
The defects in HIV-1 replication in murine cells have been studied primarily in fibroblastic cell lines transduced with human cofactors. Although fibroblastic lines are easy to manipulate in vitro, HIV-1 does not ordinarily target these types of cells in vivo. We thus examined cell types that were more relevant to HIV-1 infection in vivo and that would serve as HIV-1 target cells within a genetically modified mouse model. Here we show that there is a profound block to HIV-1 replication in murine T-cell lines at an early, postentry replication stage that is not present in murine fibroblastic lines. MLV infection efficiency is also diminished in murine lymphocytic cells, albeit less dramatically. In contrast to Fv1-, Lv1-, and Ref1-mediated blocks to retroviral replication, the murine T-cell restriction of HIV-1 infection does not appear to be saturable by a high virus dose.
MATERIALS AND METHODS
Cell lines.
Human embryonic kidney cells HEK293T (18), mouse fibroblast cell lines NIH 3T3, DBA clone A, 3T3FL, C3H, BLK clone 4, BxN clone 1, and T-lymphoma S1A.TB.4.8.2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, Calif.) supplemented with 10% fetal calf serum (FCS; Atlanta Biologicals, Norcross, Ga.), 2 mM l-glutamine, 50 U of penicillin/ml, and 50 U of streptomycin (Invitrogen)/ml. BALB/3T3 clone A31 cells were grown in DMEM supplemented with 10% newborn calf serum (Invitrogen), 2 mM l-glutamine, and penicillin-streptomycin. SC-1 cells were grown in McCoy's 5A medium with 10% FCS, 2 mM l-glutamine, and penicillin-streptomycin. BLK clone 4 fibroblasts were grown in Iscove's modified Dulbecco's medium with 10% FCS, 2 mM l-glutamine, and penicillin-streptomycin. 2B4 murine T-cell hybridoma and TA3 cells were propagated in RPMI 1640 (Invitrogen) with 10% FCS, 2 mM l-glutamine, and penicillin-streptomycin. T-cell lymphoma TK-1 cells were grown in RPMI 1640 with 10% FCS, 10 mM HEPES, 50 μM β-mercaptoethanol, and 1 mM sodium pyruvate (all from Invitrogen) with 2 mM l-glutamine and penicillin-streptomycin. Rat T-lymphoma C58(NT)D.1.G.OVAR.1 cells, T-lymphoma S49.1 cells, and lymphocytic thymoma R1.1 cells were grown in DMEM with 10% horse serum, 1 mM sodium pyruvate, 2 mM l-glutamine, and penicillin-streptomycin. T-lymphoma LBRM-33 clone 4A2 cells and the B-cell lines WEHI-231, 143-2 M, 22D6, and 1881 were grown in RPMI 1640 with 10% FCS, 50 μM β-mercaptoethanol, 2 mM l-glutamine, and penicillin-streptomycin. T-lymphoma TIMI.4 cells were grown in DMEM with 10% horse serum (Invitrogen), 2 mM l-glutamine, and penicillin-streptomycin. Human T-cell line HuT78/CCR5 (75) was grown in RPMI 1640 with 10% FCS, 1 μg of puromycin/ml, 0.5 mg of G418/ml, 2 mM l-glutamine, and penicillin-streptomycin.
BALB/3T3 clone A31, BLK clone 4, C58(NT)D.1.G.OVAR.1, LBRM-33 clone 4A2, R1.1, S1A.TB.4.8.2, S49.1, TIMI.4, and TK-1 were obtained from the American Type Culture Collection. 1881, 22D6, 143-2 M, and WEHI-231 were a gift from Naomi Rosenberg (Tufts University, Boston, Mass.). NIH 3T3 cells expressing human CD4 and coreceptors have been described previously (22). The TA3 mouse T cells are a clonal line generated from a thymoma of a p53-deficient C57BL/6 × 129/SV6 mouse (33) by culturing and selection for cells that grew out in culture. TA3 cells expressing human CD4 and coreceptors were produced by transduction of cells with the retroviral vector MX-CD4, followed by cell sorting for human CD4 expression. Cells were then transduced with the MLV vector Babepuro-cycT1, conferring the expression of human cyclin T1, and selected for puromycin resistance. Expression of coreceptors was obtained through transduction with MLV vectors MX-CXCR4 and MX-CCR5, followed by cell sorting for high coreceptor expression.
D10.G4.1 (D10) is a primary murine CD4+ T-cell clone that is specific for hen egg conalbumin peptide 134-146 (40). The primary T cells were stimulated with peptide-pulsed antigen-presenting cells every 2 weeks and expanded in DMEM supplemented with 10% FCS, 200 U of human interleukin-2 (Chiron)/ml, 2 mM l-glutamine, and penicillin-streptomycin.
Virus stocks.
Single-round infectious, pseudotyped HIV-1 stocks were generated by calcium phosphate transfection of HEK293T cells (Transfection MBS kit; Stratagene) with proviral vector constructs pNL-Luc-E−R− (pHIVluc) (14) containing a firefly luciferase reporter gene under transcriptional control of the HIV-1 LTR, pHIVeGFP (74) encoding enhanced green fluorescent protein (eGFP) as reporter gene, or pHIV-HSA (68) encoding the heat-stable antigen (HSA) as reporter gene. pHIV-SVluc contains the firefly luciferase reporter gene under transcriptional control of the simian virus 40 (SV40) early promoter in the nef open reading frame. The vector plasmid was cotransfected with an expression plasmid for CCR5-tropic HIV-1ADA envelope glycoprotein or CXCR4-tropic HIVLAI or the G-protein of vesicular stomatitis virus (VSV-G). For MLV vector production, Phoenix-Eco packaging cells (42) were transfected with the vector plasmid pS003 (16). Virus titers were determined by limiting dilution on GHOST/CCR5/CXCR4 indicator cells (79) for HIVluc, HIV-SVluc, or HIV-HSA, and on NIH 3T3 cells for HIVeGFP and S003 in the presence of 20 μg of Polybrene/ml. If required, virus-containing supernatants from HEK293T cells were additionally concentrated by ultracentrifugation through a 20% sucrose cushion (41). Aliquots of virus-containing supernatants were stored at −80°C until use.
Infection assays.
Adherent target cells were seeded at 5 × 104 cells/well in 12-well tissue culture cluster plates the day before infection. Suspension cells were seeded at 105 cells/well in 24-well plates immediately prior to infection. Infections were performed in a total volume of 500 μl of culture medium containing 10 μg (suspension cells) or 20 μg (adherent cells) of Polybrene (Sigma-Aldrich)/ml. Virus was added in a 10-fold dilution series in duplicate assays. Twenty-four hours after challenge, 500 μl of medium was added, and 48 h later cells were assayed for infection events. To enhance LTR-mediated expression of reporter genes in murine cell lines lacking exogenous human cyclin T1 expression, HIV-1-infected T cells were stimulated using 1 μM ionomycin (Sigma-Aldrich) and 10 ng of phorbol myristate (Sigma-Aldrich)/ml 4 h before harvest. Infection was quantified either by flow cytometry or by luciferase activity present in the cytoplasm. Luciferase activity of lysed cells was measured using the luciferase assay system (Promega, Madison, Wis.) according to the manufacturer's protocol. GFP expression was determined by flow cytometry using a FACSCalibur instrument and CellQuest software (Becton-Dickinson).
For virus saturation experiments, target cells were seeded at 105/well in 24-well plates. A dose calculated to be an MOI of 100 for HIV-HSA/VSV-G was added in the presence of 10 μg of Polybrene/ml for 1 h, before addition of HIVeGFP/VSV-G in 10-fold dilutions. Infection was quantified by flow cytometry 3 days later.
Dual-luciferase assay.
A total of 106 NIH 3T3 cells expressing human cyclin T1 were cotransfected with 4 μg of pHIVluc and 1 μg of pRL-SV40 (Promega), or 107 TA3 cells expressing human cyclin T1 were electroporated (Bio-Rad Genepulser) with 20 μg of pHIVluc and 5 μg of pRL-SV40. Forty-eight hours later, firefly luciferase (pHIVluc) and Renilla luciferase (pRL-SV40) activities were determined using the dual-luciferase assay system (Promega) according to the manufacturer's protocol.
HIVRP entry assay.
HIV reporter particles (HIVRP) pseudotyped with the VSV-G protein were generated by transfecting HEK293T cells with three plasmids: the provirus R8delta-env, with the envelope deleted (5 μg; gift of Chris Aiken); a plasmid encoding VSV-G (pCMV-VSVG; 1 μg; gift of John Kappes); and an expression construct encoding a fusion of β-lactamase to Vpr, pMM310 (5 μg). Cells growing in phenol red-free RPMI 1640 plus 10% FCS and 10 mM HEPES (pH 7.3) were transfected with Fugene 6 (Roche) according to the manufacturer's instructions. Supernatants were harvested 48 h after infection and snap-frozen. Serial twofold dilutions of HIVRP (90 μl/well in the same medium) were incubated with cells (NIH 3T3 at 3 × 104/well; others were at 105/well) in Costar 3603 plates for 3 h at 37°C, and then plates were placed at room temperature. Probenecid (Sigma) and CCF4-AM (Panvera) were added to final concentrations of 2 mM and 5 μM, respectively, and cells were incubated at room temperature overnight to allow substrate cleavage. Blue (460 nm) and green (538 nm) fluorescence emissions in response to excitation at 412 nm were measured in a BMG FluoStar fluorometer.
Determination of copy numbers of late reverse transcription products.
Twenty hours postinfection of T cells with DNase-treated HIVeGFP/VSV-G at an MOI of 10, total cellular DNA was extracted using the Blood DNA kit protocol (QIAGEN). Primers, probes, and PCR conditions for the detection of second-strand transfer products and 2-LTR circle junctions were used as described before (38). Briefly, the forward PCR primer for second-strand transfer product detection (5′-TTT TAG TCA GTG TGG AAA ATC TGT AGC-3′) annealed to sequences within the U5 region, and the reverse PCR primer (5′-TAC TCA CCA GTC GCC GCC-3′) annealed to sequences just downstream of the HIV-1 tRNA primer binding site. For 2-LTR circle junction detection, the forward primer (5′-GCC TGG GAG CTC TCT GGC TAA-3′) was complementary to sequences in the U5 region and the reverse primer (5′-GCC TTG TGT GTG GTA GAT CCA-3′) was annealed to sequences in the U3 region. Fluorescence-labeled specific probes (5′-R-TCG ACG CAG GAC TCG GCT TGC T-Q-3′ for second-strand transfer product detection and 5′-R-AAG TAG TGT GTG CCC GTC TGT TGT GTG ACT C-Q-3′ for 2-LTR circle junction detection, incorporating 6-carboxyfluorescin as 5′ reporter and 6-carboxytetramethyl-rhodamine as 3′ quencher) were designed to bind to sequence amplified by the PCR primers. Taqman probes and primers were used at a final concentration of 200 nM. For quantification of real-time PCR products, duplicate samples of serial dilutions (107 to 101 copies) of plasmid DNA (pHIV-EGFP for second-strand transfer reactions, and a plasmid containing a 2-LTR circle junction [38] for 2-LTR circle junction reactions) were used. Hot start was performed by heating for 2 min at 50°C and 10 min at 95°C. The cycling conditions were 45 cycles of 95°C for 15 s and 60°C for 1 min. Real-time PCR was performed using an Opticon DNA Engine (MJ Research).
RESULTS
HIV-1 infection of murine T cells is restricted.
To determine whether murine T-lymphocytic cells and fibroblastic cell lines are similarly permissive to HIV-1 infection, we challenged the murine thymoma line TA3, generated from a C57BL/6 × 129/SV6 mouse thymoma, and NIH 3T3 cells with several HIV-1 vectors. TA3 cell lines were derived that expressed human cyclin T1, CD4, CCR5, and CXCR4 (Fig. 1A); similar NIH 3T3 lines have been described previously (22). Susceptibility to HIV-1 infection was determined in a single-round infection assay. HIV-1 vector pseudotypes bearing CXCR4-tropic LAI Env or VSV-G infected NIH 3T3 fibroblasts at similar efficiencies (Fig. 1B). In contrast, the murine T cells (TA3.CD4.CXCR4) were less susceptible to HIV-1 infection (Fig. 1B). This infection block was also observed with CCR5-tropic HIV-1 pseudotypes (Fig. 1C); murine T cells (TA3.CD4.CCR5) were challenged with increasing amounts of HIVluc/ADA or HIVluc/VSV-G. Although NIH 3T3.CD4.CCR5 and human HuT78/CCR5 cells were efficiently infected with either virus, the TA3.CD4.CCR5 cells restricted HIV-1 infection in both the CCR5-dependent and the VSV entry pathways (Fig. 1C). At high virus concentrations, the infection block to HIV-1 in the murine T cells increased to over 3 orders of magnitude relative to that in the NIH 3T3 fibroblasts.
FIG. 1.
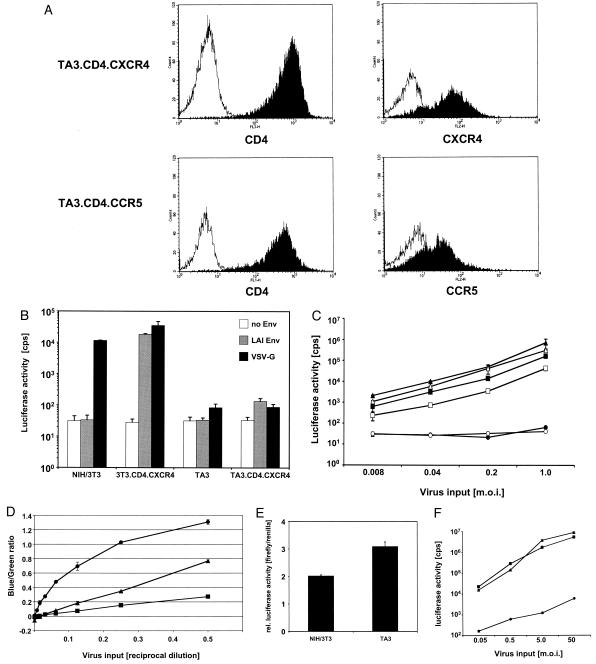
Murine TA3 cells restrict HIV infection. (A) Expression of human CD4, CXCR4, and CCR5 on TA3.CD4.CXCR4 cells (top) and TA3.CD4.CCR5 cells (bottom). The cells were stained with monoclonal antibodies against human CD4, CXCR4, and CCR5. CD4 is shown in the left panels, and coreceptor molecules are in the right panels. Isotype control stainings are represented by the unfilled curves for comparison. (B) Comparison of the transduction efficiency of HIV-1 vectors on murine NIH 3T3 fibroblasts and murine TA3 cells expressing human CD4 and human CXCR4. Predicted MOIs of 0.4 (NIH 3T3) and 2 (TA3) were used in the challenges depicted. Env-negative virus was generated in parallel transfections, and comparable volumes were used in the infections. NIH 3T3, NIH 3T3.CD4.CXCR4, TA3, and TA3.CD4.CXCR4 were challenged with HIVluc without envelope, pseudotyped with LAI Env, or VSV-G. Three days after infection luciferase activity was assayed. Luciferase counts per second (cps) are indicated on the ordinate (logarithmic scale). (C) Restriction is independent of envelope properties. NIH 3T3.CD4.CCR5 (triangles), HuT78/CCR5 (squares), and TA3.CD4.CCR5 (circles) cells were challenged with increasing amounts of HIVluc/ADA (open symbols) and HIVluc/VSV-G (solid symbols). Three days after challenge, infections were assayed by measuring luciferase activity, which is shown as a function of virus input (MOI). Each point represents the average of duplicate samples and is depicted with the standard deviation. (D) Entry of HIV/VSV-G into murine T cells, murine fibroblasts, and human T cells. TA3 (circles), NIH 3T3 (triangles), and HuT78/CCR5 (squares) cells were challenged in a twofold dilution series with HIVRP/VSV-G incorporating β-lactamase (BlaM) fused to Vpr. The cells were then loaded with the BlaM substrate CCF4-AM, which forms a 460 nm (blue) fluorescent product in the presence of BlaM. The 538 nm (green) fluorescent cells are negative for virus penetration. The ratio of blue fluorescence to green fluorescence is proportional to the fraction of cells that have fused with the HIVRP. (E) Comparison of LTR promoter to SV40 promoter activity in murine T cells and fibroblasts. NIH 3T3 and TA3 cells were cotransfected with the vector plasmid pHIVluc (expressing luciferase under transcriptional control of the HIV-1 LTR) and pRL-SV40 (expressing Renillaluciferase under transcriptional control of the SV40 promoter) in a ratio of 4:1, and the activities of firefly luciferase and Renilla luciferase were measured. The y axis shows the ratio of firefly luciferase (cps) over Renilla luciferase (cps) activity in a mean of two transfected wells, each measured in triplicate. (F) Challenge with a vector carrying an internal SV40 promoter. NIH 3T3 (triangles), HuT78/CCR5 (squares), and TA3 (circles) cells were challenged with increasing amounts of HIV-SVluc (carrying the firefly luciferase reporter gene under transcriptional control of the SV40 promoter) pseudotyped with VSV-G. Infections were assayed by measuring luciferase activity, which is shown as a function of virus input (MOI). Each point represents the average of duplicate samples and is depicted with the standard deviation.
Efficient penetration of murine T cells by HIV/VSV-G-pseudotyped virus.
Previous studies had indicated that the HIV-1 receptor molecules in the murine TA3 cells supported infection by MLV pseudotyped with HIV-1 Env (J. G. Baumann, G. E. Ahn, and V. N. KewalRamani, unpublished data). We next assayed the efficiency of VSV-G-mediated viral entry in TA3 cells, NIH 3T3 cells, and human T cells (HuT78/CCR5) by using HIVRP. These HIVRP were VSV-G-pseudotyped HIV-1 particles that contained the enzyme β-lactamase (BlaM) as a fusion to the HIV-1 Vpr protein in their core. Viral entry is measured by monitoring the delivery of BlaM into the cytoplasm of a target cell by adding the cell-permeant fluorescent BlaM substrate CCF4-AM (81). Cells displaying blue fluorescence have fused with the HIVRP, whereas cells displaying green fluorescence have not fused with particles (70). HIVRP fusion depends on an active envelope glycoprotein (e.g., HIV gp120:gp41 or VSV-G) but does not require any of the subsequent steps in the virus life cycle. Using this system, we found that fusion with HIVRP was more efficient in murine T cells than murine fibroblasts or human T cells (Fig. 1D). These data demonstrated that the block to HIV-1 transduction in murine T cells occurs after virus entry.
Differences in susceptibility are independent of HIV-1-mediated gene expression.
To determine whether HIV-1 LTR-mediated expression of the luciferase reporter gene was responsible for the reduced detection of HIV-1 in mouse T cells, we cotransfected murine fibroblasts and T cells (both stably expressing human cyclin T1) with plasmids encoding firefly and Renilla luciferase genes under transcriptional control of different enhancer-promoter elements. The vector plasmid pHIVluc expressed Tat and firefly luciferase under transcriptional control of the Tat-dependent HIV-1 LTR promoter, whereas Renilla luciferase expression was dependent on the SV40 promoter of pRL-SV40. Inefficient HIV-1 LTR activity in murine T cells would result in a reduced amount of firefly luciferase activity relative to Renilla luciferase compared to similarly transfected mouse fibroblastic cells. As shown in Fig. 1E, no significant differences in relative promoter activity were observed. Mouse T cells and fibroblasts were similarly permissive to HIV-1-mediated gene expression, suggesting other differences were responsible for the inefficient infection of the mouse T cells. Supporting this conclusion, murine T cells infected with another HIV vector (pHIV-SVluc) expressing the firefly luciferase reporter gene under control of an internal SV40 promoter also showed a strong restriction compared to NIH 3T3 and Hut78/CCR5 cells (Fig. 1F).
Restriction of HIV-1 infection in murine T cells is independent of Fv1 genotypes.
We next tested an HIV-1 vector encoding an eGFP cassette, HIVeGFP, pseudotyped with VSV-G. In contrast to challenges with HIVluc pseudotypes, the number of infected cells could be directly quantified by assaying GFP-positive cells, and the utilization of VSV-G pseudotypes allowed us to examine a broad number of cell types.
Challenges of HuT78/CCR5 cells versus murine TA3 cells with an increasing HIVeGFP/VSV-G inoculum indicated that the murine T cells were approximately 100-fold less sensitive to infection over a range of virus concentrations (Fig. 2A). Similar results were obtained when comparing NIH 3T3 infection efficiency to that of TA3 cells. To determine if the differences in infection rates simply reflected an unusual defect in TA3 cells, we examined additional murine cell lines, selecting those with similar or different genetic backgrounds (Table 1).
FIG. 2.
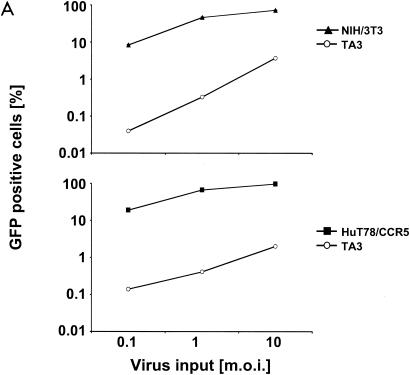
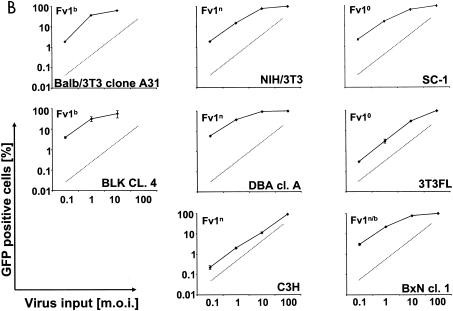
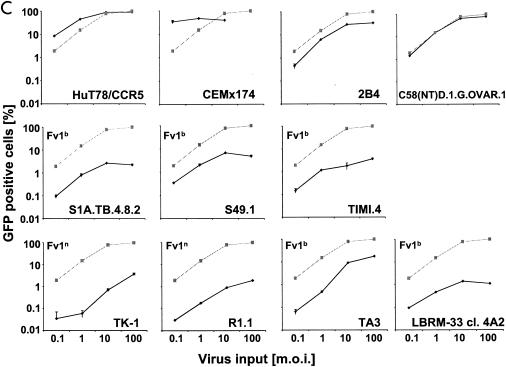
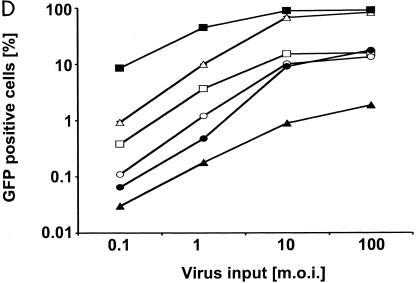
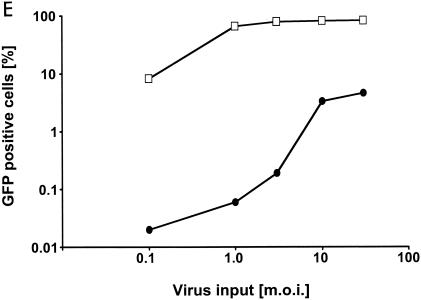
Infection of murine fibroblasts, T cells, and B cells with HIVeGFP/VSV-G. Ten-fold serial dilutions of HIVeGFP pseudotyped with VSV-G were used to infect lymphocytic and fibroblast lines (105 cells each). Three days after challenge, GFP expression in infected cells was measured using flow cytometry. Infections (percent GFP-positive cells) are shown as a function of virus input (MOI) on a logarithmic scale. (A) Comparison of human T cells (HuT78/CCR5), mouse fibroblasts (NIH 3T3), and mouse T cells (TA3) for susceptibility to HIV-1. (B) Infection of fibroblasts from mouse strains expressing different Fv1 alleles. A fixed reference curve (grey line) is included in all plots. (C) Challenge of mouse T-cell lines, two human lymphocytic cell lines (HuT78/CCR5 and CEMx174), the mouse T-cell hybridoma 2B4, and rat T cells C58(NT)D.1.G.OVAR.1 (top row, from left to right). Infection of NIH 3T3 fibroblasts is included as a light grey reference curve in all plots. (D) Challenge of mouse B-cell lines. Mouse B-lymphocytic cell lines WEHI-231 (open triangles), 22D6 (open circles), 143-2 M (open squares), and 1881 (solid triangles) were challenged with HIVeGFP/VSV-G and compared to HuT78/CCR5 (solid squares) and TA3 (solid circles) cells. (E) Infection of primary mouse CD4+ T cells (D10; solid circles) and human HuT78/CCR5 cells (open squares) with HIVeGFP/VSV-G at various MOIs.
TABLE 1.
Summary of murine cell lines tested for HIV-1 susceptibilitya
| Cell line | Strain | Reference | Fv1b | Restrictionc |
|---|---|---|---|---|
| Fibroblasts | ||||
| BALB/3T3 clone A31 | BALB/c | 2 | b | + |
| BLK clone 4 | C57BL/6 | 57 | b | + |
| BxN clone 1 | BALB/c × NIH | 64 | n/b | + |
| 3T3FL | BALB/c | 24 | Null | ++ |
| DBA clone A | DBA/2 | 3 | n | + |
| C3H | C3H | 65 | n | ++ |
| NIH 3T3 | NIH/Swiss | 34 | n | + |
| SC-1 | (Feral mouse) | 27 | Null | + |
| B cells | ||||
| 1881 | BALB/c | 62 | b | +++ |
| 22D6 | BALB/c | 62 | b | +++ |
| 143-2M | BALB/c | 60 | b | ++ |
| WEHI-231 | BALB/c × NZB | 25 | nr/bd | + |
| T cells | ||||
| 2B4 | B10.A/AKR | 30 | n/b | ++ |
| C58(NT)D.1.G.OVAR.1 | WFu (rat) | 32 | NAe | + |
| LBRM-33 clone 4A2 | B10.BR | 23 | b | +++ |
| R1.1 | C58.J | 61 | n | +++ |
| S1A.TB.4.8.2 | BALB/c | 31 | b | +++ |
| S49.1 | BALB/c | 31 | b | +++ |
| TA3 | C57BL/6 × 129/SV6 | This study | nr/bd | +++ |
| TIMI.4 | C57BL/6 | 61 | b | +++ |
| TK-1 | AKR/Cum | 12 | n | +++ |
aGenetic backgrounds, Fv1 alleles, and levels of restriction to HIV-1 for fibroblasts, B cells, and T cells are listed.
Restriction to HIV-1, scored as follows: +, <10-fold; ++, 10- to 100-fold; +++, >100-fold (compared to HuT78/CCR5 human T cells).
Fv1nr was described in reference 39.
NA, not applicable.
The genetic background of the murine cell lines initially tested differed in a way that provided a potential model for restriction: NIH 3T3 cells are derived from NIH Swiss mice, the prototype Fv1n mouse (47), whereas TA3 cells are derived from C57BL/6 mice (55), which exhibit a strong Fv1b phenotype. Although Fv1 has been classically defined in its restriction of murine retroviruses, Fv1 is weakly related to MLV isolates and instead has greater homology to human endogenous retrovirus type L (5). In addition, because Fv1 is unique to the mouse genome, Fv1 effects on infection by nonmurine retroviruses have not been described. To determine if the HIV-1 restriction observed in the TA3 cells was a general phenomenon related to mouse T-cell physiology or if the underlying Fv1 genotype was responsible, we tested several murine fibroblast and T-cell lines with different Fv1 backgrounds for their susceptibility to HIV-1 infection. Eight fibroblastic cell lines were challenged with 10-fold serial dilutions of HIVeGFP/VSV-G (Fig. 2B). Predicted MOIs in this experiment and subsequent experiments were based on prior titration of virus stocks on NIH 3T3 cells and calculated as a ratio of NIH 3T3 infectious units of HIVeGFP/VSV-G applied per target cell number. Most of the cell lines were infected at frequencies of 1 to 5% at an MOI of 0.1, with only the C3H and 3T3FL cells having less than 1% of cells infected. C3H cells are derived from an Fv1n mouse, and 3T3FL cells are of an Fv10 background (24). However, all of the cell lines could be infected at levels exceeding 90% by using a higher MOI irrespective of their genetic background or specific Fv1 genotype. As an example, the fibroblast line BLK clone 4 encodes Fv1b and was similar to the Fv1n NIH 3T3 cells in susceptibility to HIV-1 infection. These data suggested that the Fv1 genotype does not affect infection by HIV-derived vectors in murine fibroblast cell lines.
We also tested seven murine T-lymphocytic cell lines for susceptibility to HIV-1 infection (Fig. 2C). The cell lines were challenged at predicted MOIs ranging from 0.1 to 100. For comparison, we included two human lymphocytic cell lines, the rat T-lymphoma cell line C58(NT)D.1.G.OVAR.1 and the 2B4 mouse T-cell hybridoma line. The human T cells were infected efficiently at low MOIs, whereas the rat T cells and the 2B4 T-hybridoma cells were slightly less susceptible to infection when the predicted MOI was less than or equal to 1. However, infection levels greater than 35% (2B4) and 80% (rat) were achieved with higher virus MOIs. A different outcome was seen with the mouse T-cell lines. All seven cell lines were poorly infected; infection rates ranged from 0.08% (R1.1) to 0.3% (S49.1) at a low MOI. The maximum rate was 1.6% (R1.1) to 11% (TA3) at a high MOI. As with the fibroblasts, the restriction in T cells did not coincide with the Fv1 status of the mice. Murine T cells were generally restricted, while the fibroblasts were not, even when some of the murine T cells were of the same genetic background as the murine fibroblasts.
We extended the infection studies to a different lymphocytic cell lineage. Four murine B-cell lines were challenged with the same vector (Fig. 2D). Three of these cell lines (143-2 M, 22D6, and 1881) showed a strong restriction, similar to what was observed with the mouse T-cell lines. The efficiency of infection ranged from 0.03 to 0.4% at an MOI of 0.1, reaching 2 to 16% at the highest MOI. WEHI-231 cells, however, displayed a more moderate restriction, with an infection rate of 0.9% at a low MOI but reaching over 80% at a high MOI.
We also asked whether primary murine CD4+ T cells restrict HIV-1 infection. Mouse D10 cells, a primary ovalbumin-specific murine CD4+ T-cell clone, were challenged with increasing concentrations of HIVeGFP/VSV-G (Fig. 2E). D10 cells, like the other mouse T-cell lines, were significantly less susceptible to transduction by HIV-1 than the human T-cell line.
TA3 cell restriction is nonsaturable with high virus input.
Fv1, Lv1, and Ref1 restrictions can be overcome with saturating amounts of challenge virus, which presumably sequesters a factor present in limiting amounts. We asked whether treatment with a GFP-negative HIV-1 vector at an MOI of 100 would permit a higher level of infection by the HIVeGFP reporter virus. As shown in Fig. 3, this prechallenge with GFP-negative HIV-1 did not increase infection by HIVeGFP/VSV-G. In fact, diminished infection was observed after prechallenge. These data indicated that these cells either lacked a factor necessary for efficient transduction or, if interfering factors were present, they were not saturated by pretreatment at an MOI of 100.
FIG. 3.
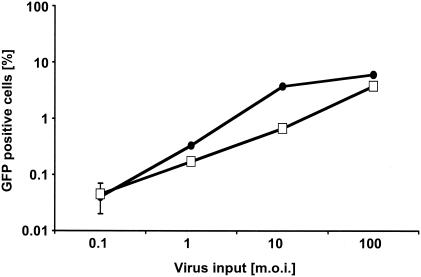
Infection of TA3 cells after pretreatment with a second HIV-1 vector. TA3 cells were inoculated with serial dilutions of HIVeGFP/VSV-G. In one infection set, the cells were pretreated with HIV-HSA/VSV-G (open squares) at a predicted MOI of 100. Parallel infections were performed without pretreatment (solid circles). HIVeGFP/VSV-G virus input is shown on a logarithmic scale on the abscissa, and the percentage of infected cells as measured by GFP expression in flow cytometry is represented on the ordinate.
Mouse T cells restrict HIV-1 infection at and after reverse transcription.
To determine if reverse transcription was completed in murine T cells, we measured the amount of viral DNA synthesized in cells infected by HIVeGFP/VSV-G using real-time PCR. Primers and probes were used that detected the second strand transfer (i.e., forward primer within the U5 region, reverse primer downstream of the primer binding site), thereby measuring a late reverse transcription product. The murine T cells exhibited a moderate reduction (approximately threefold overall) in late reverse transcription products compared to that in NIH 3T3 fibroblasts (Fig. 4A). To test whether HIV-1 preintegration complexes efficiently entered the nuclei of mouse T cells, we next performed real-time PCR measurements for 2-LTR circle junction formation. 2-LTR circle junctions are the result of end ligations of the full-length, double-stranded viral genome by nuclear ligases (35, 46). Although they represent a dead-end by-product of fully reverse-transcribed retroviral DNA, they can be used as a surrogate marker for nuclear entry (9). Monitoring of 2-LTR circle junctions revealed 7- to 17-fold-reduced amounts of circularized viral genomes in murine T cells compared to NIH 3T3 fibroblasts (Fig. 4B). These data indicated the presence of a cumulative infection block of HIV-1 reverse transcription and nuclear migration of the HIV-1 preintegration complex in murine T cells.
FIG. 4.
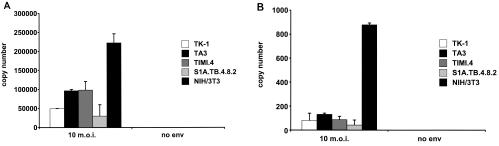
HIV-1 reverse transcription is less efficient in murine T cells. TK-1, TA3, TIMI.4, S1A.TB.4.8.2, and NIH 3T3 cells were challenged with an MOI of 10 using HIVeGFP/VSV-G and HIVeGFP (without Env). Real-time PCR was used to quantify reverse transcription products. Copy numbers of late reverse transcription (second-strand transfer) products (A) and 2-LTR circle junctions (B) are indicated on the y axes.
Murine T cells are less susceptible to Moloney MLV infection.
To determine if the reduced infection susceptibility in mouse T cells was restricted to HIV-1 or extended to other retroviruses, we compared infection of mouse T-lymphocytic lines and fibroblastic cell lines with Moloney MLV vectors expressing a GFP reporter gene (Fig. 5). MLV infection in the murine T cells was approximately 5- to 10-fold less efficient relative to that in the NIH 3T3 cells. Although the infection block was less, the relative susceptibility of cells to Moloney MLV infection correlated to their susceptibility to HIV-1 infection.
FIG. 5.
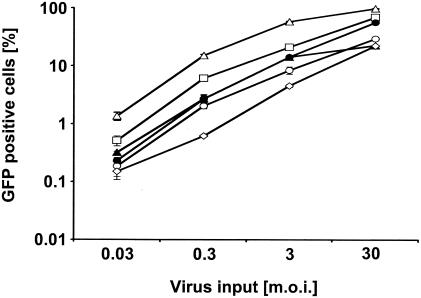
MLVeGFP infection of murine fibroblasts and T cells. Tenfold dilutions of ecotropic MoMLV vector pseudotypes encoding a GFP cassette were used to infect NIH 3T3 (open triangles), 2B4 (open squares), S49.1 (solid triangles), TA3 (solid circles), S1A.TB.4.8.2 (open circles), and TIMI.4 (open diamonds) cells. GFP expression was assayed using flow cytometry 3 days postinfection. Infections (percent GFP; y axis) are shown as a function of virus input (MOI; x axis).
DISCUSSION
Our results indicate that there are significant differences in the susceptibility of different murine cell types to HIV-1 infection. Murine T cells are 100- to 1,000-fold less susceptible to HIV-1 infection than murine fibroblasts at a postentry step. Challenge of TA3 mouse T cells reveals a strong restriction to HIV-1 infection relative to that in NIH 3T3 fibroblasts or human T cells. The restriction is irrespective of the entry pathway, as similar data were observed using HIV-1 particles pseudotyped with different HIV-1 Env proteins (LAI, ADA) or VSV-G. Strikingly, this murine T-cell block to lentiviral infection occurred at and after reverse transcription and was not saturable at high virus concentrations.
The restriction to HIV-1 infection in murine cells is closely linked to cell tissue type. We observed that primary murine T cells, seven of seven mouse T-cell lines, and three of four mouse B-cell lines derived from different inbred strains were highly restrictive to transduction by HIV-1, whereas eight of eight mouse fibroblast lines were largely permissive to HIV-1 transduction (summarized in Table 1). Mouse T cells were also less susceptible than fibroblasts to MLV vector transduction; however, the magnitude of this restriction was significantly less than what was observed with HIV-1 vectors. Moloney MLV packaging constructs, which are N/B-tropic, were used in preparation of the vectors and would not be restricted by Fv1n or Fv1b. Although HIV-1 and MLV were restricted at an early step in murine T cells, we do not know whether the underlying basis for this reduced susceptibility is the same for both viruses. Given the tumor-causing potential of retroviral infection in murine B and T lymphocytes (11, 20, 59), the acquisition of additional restriction mechanisms to retroviral infection could provide a selective advantage to the host. Curiously, such an antiviral defense would be epigenetically regulated, as it appears to be a function of cell type, suggesting that such mechanisms may latently exist in other murine cell types.
Examination of HIV-1 DNA by real-time PCR revealed significantly reduced amounts of late reverse transcription products in the murine T cells compared to NIH 3T3 cells. In addition, the nuclear transfer of the preintegration complexes was reduced even further, up to 17-fold compared to that in NIH 3T3 cells. Both staging assays were performed at high MOIs to maximize the likelihood of detection of viral DNA in the murine T cells. The reduced amount of reverse transcription products was even more striking when taking into account the higher virus penetration rate measured for murine T cells than for fibroblasts (Fig. 1D). This discrepancy would suggest an even stronger restriction in the murine T cells at the level of reverse transcription than our assay suggests. Although our analyses did not distinguish whether an additional HIV-1 infection block(s) preceded integration, we observed a similar restriction in the transduction of HIV-1 vectors encoding a luciferase marker expressed by an internal SV40 promoter. In addition, transfection analyses of the NIH 3T3 and TA3 cells used in this study revealed comparable HIV-1 LTR-mediated gene expression in both cell types. Collectively, our data suggest that the HIV-1 infection block in the murine T cells occurs postentry and reflects a progressive inefficiency manifested to the point of integration.
The cumulative nature of the murine T-cell block bears some similarity to the one imposed by Fv1. In contrast to Fv1 restriction of MLV, prechallenge of TA3 cells with a large inoculum of HIV-1 (MOI, 100) was unable to saturate a putative restrictive factor and enable more efficient infection by a second challenge virus. In fact, prechallenge with HIV-1 actually reduced the infection rate of the second challenge virus, suggesting possible saturation of a necessary factor (Fig. 3). Whereas challenge of the various murine T-cell lines with HIV-1 at low to intermediate MOI yielded an infection rate that resembled one-hit kinetics (Fig. 2C), challenge from intermediate to high MOIs resulted in an infection rate that quickly hit a plateau, with <10% infection of the cell populations. These data indicate that the restriction might be due to the lack of necessary factors. Notably, the T-cell hybridoma 2B4 showed a reduced restriction pattern. The increased susceptibility of 2B4 cells would support a model that the restriction in mouse T cells is due to the absence of a necessary factor, and the factor is contributed by the fusion partner in the T-cell hybridization.
This line of reasoning does not completely rule out the possibility of high intracellular amounts of a restricting factor or the presence of several independent factors interfering with the preintegration complex. In either scenario, inhibition via a large virus inoculum might be difficult to accomplish. Recently it has been reported that Trim5α, a component of cytoplasmic bodies, is responsible for a strong restriction of HIV-1 infection present in rhesus macaque cells (67). Restriction of HIV-1 by rhesus Trim 5α is mediated at or before reverse transcription. Similar to Fv1-mediated restriction, the antiviral activity of Trim5α is specific and is dependent on the capsid of incoming viral particles. It is currently unclear if Trim5α is identical to or different from Lv1 and/or Ref1. However, given that there are now at least two members of the Trim family (Trim5α and PML [73]) with antiretroviral activity, it is conceivable that a murine ortholog of Trim5 might play a role in HIV-1 restriction in murine T cells. Several mouse-encoded Trim genes bear significant homologies to rhesus Trim5. It will be interesting to determine whether these are differentially expressed in mouse T cells and fibroblasts.
The results presented here have implications in the development of a genetically modified mouse model for HIV-1 infection, given that CD4+ T cells comprise the largest population of infected cells during in vivo infection. Mice have not been successfully colonized by lentiviruses, and the reduced susceptibility of mouse lymphocytes to HIV-1 at an early replication stage suggests that these cells may provide unique barriers to retroviral zoonoses. Ultimately, identifying the underlying restriction mechanisms present in murine T cells will enhance our understanding of cellular factors that modulate viral infection and may provide guidance in the development of new antiretroviral therapeutics and strategies.
Acknowledgments
We thank John Coffin, Eric Freed, and Steve Hughes for manuscript comments and Paul Bieniasz for discussion of results prior to submission. We thank John Julias and S. Hughes for advice on real-time PCR, Naomi Rosenberg for the gift of murine B-cell lines, Michael Emerman for the gift of pLAI-Env, and Thomas Martin for help with cell sorting.
Funding for this research was provided by the NCI Center for Cancer Research. Funding was also provided by NIH grants AI49131 (D.U.) and AI33856 and AI36606 (D.R.L.).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Aagaard, L., J. G. Mikkelsen, S. Warming, M. Duch, and F. S. Pedersen. 2002. Fv1-like restriction of N-tropic replication-competent murine leukaemia viruses in mCAT-1-expressing human cells. J. Gen. Virol. 83:439-442. [DOI] [PubMed] [Google Scholar]
- 2.Aaronson, S. A., and G. J. Todaro. 1968. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J. Cell Physiol. 72:141-148. [DOI] [PubMed] [Google Scholar]
- 3.Benjers, B. M., R. H. Bassin, A. Rein, B. I. Gerwin, and G. Duran-Troise. 1979. Mechanism of restriction of murine leukemia viruses varies between different strains of Fv-1n mice. Int. J. Cancer 24:600-607. [DOI] [PubMed] [Google Scholar]
- 4.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74:9868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieniasz, P. D., R. A. Fridell, K. Anthony, and B. R. Cullen. 1997. Murine CXCR-4 is a functional coreceptor for T-cell-tropic and dual-tropic strains of human immunodeficiency virus type 1. J. Virol. 71:7097-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boone, L. R., C. L. Innes, P. L. Glover, and E. Linney. 1989. Development and characterization of an Fv-1-sensitive retrovirus-packaging system: single-hit titration kinetics observed in restrictive cells. J. Virol. 63:2592-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, P. O. 1997. Integration, p. 161-203. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Browning, J., J. W. Horner, M. Pettoello-Mantovani, C. Raker, S. Yurasov, R. A. DePinho, and H. Goldstein. 1997. Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc. Natl. Acad. Sci. USA 94:14637-14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bundy, L. M., M. Ru, B. F. Zheng, L. Cheng, P. K. Pattengale, J. L. Portis, and H. Fan. 1995. Biological characterization and molecular cloning of murine C-type retroviruses derived from the TSZ complex from mainland China. Virology 212:367-382. [DOI] [PubMed] [Google Scholar]
- 12.Butcher, E. C., R. G. Scollay, and I. L. Weissman. 1980. Organ specificity of lymphocyte migration: mediation by highly selective lymphocyte interaction with organ-specific determinants on high endothelial venules. Eur. J. Immunol. 10:556-561. [DOI] [PubMed] [Google Scholar]
- 13.Chen, B. K., I. Rousso, S. Shim, and P. S. Kim. 2001. Efficient assembly of an HIV-1/MLV Gag-chimeric virus in murine cells. Proc. Natl. Acad. Sci. USA 98:15239-15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 15.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Göttlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dardalhon, V., N. Noraz, K. Pollok, C. Rebouissou, M. Boyer, A. Q. Bakker, H. Spits, and N. Taylor. 1999. Green fluorescent protein as a selectable marker of fibronectin-facilitated retroviral gene transfer in primary human T lymphocytes. Hum. Gene Ther. 10:5-14. [DOI] [PubMed] [Google Scholar]
- 17.Declève, A., O. Niwa, E. Gelmann, and H. S. Kaplan. 1975. Replication kinetics of N- and B-tropic murine leukemia viruses on permissive and nonpermissive cells in vitro. Virology 65:320-332. [DOI] [PubMed] [Google Scholar]
- 18.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duran-Troise, G., R. H. Bassin, A. Rein, and B. I. Gerwin. 1977. Loss of Fv-1 restriction in Balb/3T3 cells following infection with a single N tropic murine leukemia virus particle. Cell 10:479-488. [DOI] [PubMed] [Google Scholar]
- 20.Fan, H., B. Brightman, B. Davis, and Q. Li. 1991. Leukemogenesis by Moloney murine leukemia virus, p. 155-174. In H. Fan (ed.), Viruses that affect the immune system. American Society for Microbiology, Washington, D.C.
- 21.Gao, G., X. Guo, and S. P. Goff. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703-1706. [DOI] [PubMed] [Google Scholar]
- 22.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillis, S., M. Scheid, and J. Watson. 1980. Biochemical and biologic characterization of lymphocyte regulatory molecules. III. The isolation and phenotypic characterization of interleukin-2 producing T cell lymphomas. J. Immunol. 125:2570-2578. [PubMed] [Google Scholar]
- 24.Gisselbrecht, S., R. H. Bassin, B. I. Gerwin, and A. Rein. 1974. Dual susceptibility of a 3T3 mouse cell line to infection by N- and B-tropic murine leukemia virus: apparent lack of expression of the FV-1 gene. Int. J. Cancer 14:106-113. [DOI] [PubMed] [Google Scholar]
- 25.Gutman, G. A., N. L. Warner, and A. W. Harris. 1981. Immunoglobulin production by murine B-lymphoma cells. Clin. Immunol. Immunopathol. 18:230-244. [DOI] [PubMed] [Google Scholar]
- 26.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 27.Hartley, J. W., and W. P. Rowe. 1975. Clonal cells lines from a feral mouse embryo which lack host-range restrictions for murine leukemia viruses. Virology 65:128-134. [DOI] [PubMed] [Google Scholar]
- 28.Hartley, J. W., W. P. Rowe, and R. J. Huebner. 1970. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J. Virol. 5:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hedrick, S. M., L. A. Matis, T. T. Hecht, L. E. Samelson, D. L. Longo, E. Heber-Katz, and R. H. Schwartz. 1982. The fine specificity of antigen and Ia determinant recognition by T cell hybridoma clones specific for pigeon cytochrome c. Cell 30:141-152. [DOI] [PubMed] [Google Scholar]
- 31.Horibata, K., and A. W. Harris. 1970. Mouse myelomas and lymphomas in culture. Exp. Cell Res. 60:61-77. [DOI] [PubMed] [Google Scholar]
- 32.Hyman, R., and I. Trowbridge. 1981. Two complementation classes of T200 (Ly-5) glycoprotein-negative mutants. Immunogenetics 12:511-523. [DOI] [PubMed] [Google Scholar]
- 33.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 34.Jainchill, J. L., S. A. Aaronson, and G. J. Todaro. 1969. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J. Virol. 4:549-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeanson, L., and J. F. Mouscadet. 2002. Ku represses the HIV-1 transcription: identification of a putative Ku binding site homologous to the mouse mammary tumor virus NRE1 sequence in the HIV-1 long terminal repeat. J. Biol. Chem. 277:4918-4924. [DOI] [PubMed] [Google Scholar]
- 36.Jolicoeur, P. 1979. The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr. Top. Microbiol. Immunol. 86:67-122. [DOI] [PubMed] [Google Scholar]
- 37.Jolicoeur, P., and D. Baltimore. 1976. Effect of Fv-1 gene product on proviral DNA formation and integration in cells infected with murine leukemia viruses. Proc. Natl. Acad. Sci. USA 73:2236-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung, Y. T., and C. A. Kozak. 2000. A single amino acid change in the murine leukemia virus capsid gene responsible for the Fv1nr phenotype. J. Virol. 74:5385-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaye, J., S. Porcelli, J. Tite, B. Jones, and C. A. Janeway, Jr. 1983. Both a monoclonal antibody and antisera specific for determinants unique to individual cloned helper T cell lines can substitute for antigen and antigen-presenting cells in the activation of T cells. J. Exp. Med. 158:836-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.KewalRamani, V. N., C. S. Park, P. A. Gallombardo, and M. Emerman. 1996. Protein stability influences human immunodeficiency virus type 2 Vpr virion incorporation and cell cycle effect. Virology 218:326-334. [DOI] [PubMed] [Google Scholar]
- 42.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 43.Kozak, C. A. 1985. Analysis of wild-derived mice for Fv-1 and Fv-2 murine leukemia virus restriction loci: a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J. Virol. 55:281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lander, M. R., and S. K. Chattopadhyay. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J. Virol. 52:695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 46.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lilly, F. 1967. Susceptibility to two strains of Friend leukemia virus in mice. Science 155:461-462. [DOI] [PubMed] [Google Scholar]
- 48.Lilly, F., and T. Pincus. 1973. Genetic control of murine viral leukemogenesis. Adv. Cancer Res. 17:231-277. [Google Scholar]
- 49.Malim, M. H., D. F. McCarn, L. S. Tiley, and B. R. Cullen. 1991. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 65:4248-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 51.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Münk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 52.Mariani, R., B. A. Rasala, G. Rutter, K. Wiegers, S. M. Brandt, H. G. Kräusslich, and N. R. Landau. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J. Virol. 75:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Kräusslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Münk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Odaka, T. 1969. Inheritance of susceptibility to Friend mouse leukemia virus. V. Introduction of a gene responsible for susceptibility in the genetic complement of resistant mice. J. Virol. 3:543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Odaka, T., and T. Yamamoto. 1965. Inheritance of susceptibility to Friend mouse leukemia virus. II. Spleen foci method applied to test the susceptibility of crossbred progeny between a sensitive and a resistant strain. Jpn. J. Exp. Med. 35:311-314. [PubMed] [Google Scholar]
- 57.Patek, P. Q., J. L. Collins, and M. Cohn. 1978. Transformed cell lines susceptible or resistant to in vivo surveillance against tumorigenesis. Nature 276:510-511. [DOI] [PubMed] [Google Scholar]
- 58.Pincus, T., J. W. Hartley, and W. P. Rowe. 1971. A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J. Exp. Med. 133:1219-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poirier, Y., and P. Jolicoeur. 1989. Distinct helper virus requirements for Abelson murine leukemia virus-induced pre-B- and T-cell lymphomas. J. Virol. 63:2088-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radfar, A., I. Unnikrishnan, H. W. Lee, R. A. DePinho, and N. Rosenberg. 1998. p19(Arf) induces p53-dependent apoptosis during Abelson virus-mediated pre-B cell transformation. Proc. Natl. Acad. Sci. USA 95:13194-13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ralph, P. 1973. Retention of lymphocyte characteristics by myelomas and theta + -lymphomas: sensitivity to cortisol and phytohemagglutinin. J. Immunol. 110:1470-1475. [PubMed] [Google Scholar]
- 62.Ramakrishnan, L., Q. Wu, A. Yue, M. D. Cooper, and N. Rosenberg. 1990. BP-1/6C3 expression defines a differentiation stage of transformed pre-B cells and is not related to malignant potential. J. Immunol. 145:1603-1608. [PubMed] [Google Scholar]
- 63.Reed, M., R. Mariani, L. Sheppard, K. Pekrun, N. R. Landau, and N. W. Soong. 2002. Chimeric human immunodeficiency virus type 1 containing murine leukemia virus matrix assembles in murine cells. J. Virol. 76:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rein, A., S. V. Kashmiri, R. H. Bassin, B. L. Gerwin, and G. Duran-Troise. 1976. Phenotypic mixing between N- and B-tropic murine leukemia viruses: infectious particles with dual sensitivity to Fv-1 restriction. Cell 7:373-379. [DOI] [PubMed] [Google Scholar]
- 65.Reznikoff, C. A., D. W. Brankow, and C. Heidelberger. 1973. Establishment and characterization of a cloned line of C3H mouse embryo. Cancer Res. 33:3231-3238. [PubMed] [Google Scholar]
- 66.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 67.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 68.Sundrud, M. S., S. M. Grill, D. Ni, K. Nagata, S. S. Alkan, A. Subramaniam, and D. Unutmaz. 2003. Genetic reprogramming of primary human T cells reveals functional plasticity in Th cell differentiation. J. Immunol. 171:3542-3549. [DOI] [PubMed] [Google Scholar]
- 69.Tachibana, K., T. Nakajima, A. Sato, K. Igarashi, H. Shida, H. Iizasa, N. Yoshida, O. Yoshie, T. Kishimoto, and T. Nagasawa. 1997. CXCR4/fusin is not a species-specific barrier in murine cells for HIV-1 entry. J. Exp. Med. 185:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tobiume, M., J. E. Lineberger, C. A. Lundquist, M. D. Miller, and C. Aiken. 2003. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J. Virol. 77:10645-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trono, D., and D. Baltimore. 1990. A human cell factor is essential for HIV-1 Rev action. EMBO J. 9:4155-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turelli, P., V. Doucas, E. Craig, B. Mangeat, N. Klages, R. Evans, G. Kalpana, and D. Trono. 2001. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell 7:1245-1254. [DOI] [PubMed] [Google Scholar]
- 74.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu, L., T. D. Martin, R. Vazeux, D. Unutmaz, and V. N. KewalRamani. 2002. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J. Virol. 76:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, W. K., J. O. Kiggans, D. M. Yang, C. Y. Ou, R. W. Tennant, A. Brown, and R. H. Bassin. 1980. Synthesis and circularization of N- and B-tropic retroviral DNA Fv-1 permissive and restrictive mouse cells. Proc. Natl. Acad. Sci. USA 77:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yap, M. W., and J. P. Stoye. 2003. Intracellular localisation of Fv1. Virology 307:76-89. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, Y. J., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. KewalRamani, and J. P. Moore. 1998. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng, Y. H., H. F. Yu, and B. M. Peterlin. 2003. Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell Biol. 5:611-618. [DOI] [PubMed] [Google Scholar]
- 81.Zlokarnik, G., P. A. Negulescu, T. E. Knapp, L. Mere, N. Burres, L. Feng, M. Whitney, K. Roemer, and R. Y. Tsien. 1998. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279:84-88. [DOI] [PubMed] [Google Scholar]


