Abstract
Viral contamination in environmental samples can be underestimated because a single cell line might reproduce only some enteric viruses and some enteric viruses do not exhibit apparent cytopathic effects in cell culture. To overcome this problem, we evaluated a cell culture-PCR assay based on a combination of A549 and Buffalo green monkey kidney (BGMK) cell lines as a tool to monitor infectious adenoviruses and enteroviruses in river water. Water samples were collected 10 times at each of four rivers located in Gyeonggi Province, South Korea, and then cultured on group 1 cells (BGMK cells alone) and group 2 cells (BGMK and A549 cells). Reverse transcription and multiplex PCR were performed, followed by phylogenetic analysis of the amplicons. Thirty (75.0%) of the 40 samples were positive for viruses based on cell culture, and the frequency of positive samples grown on group 2 cells (65.0%) was higher than the frequency of positive samples grown on group 1 cells (50.0%). The number of samples positive for adenoviruses was higher with A549 cells (13 samples) than with BGMK cells (one sample); the numbers of samples positive for enteroviruses were similar with both types of cells. By using phylogenetic analysis, adenoviral amplicons were grouped into subgenera A, C, D, and F, and enteroviral amplicons were grouped into coxsackieviruses B3 and B4 and echoviruses 6, 7, and 30, indicating that A549 and BGMK cells were suitable for recovering a wide range of adenoviral and enteroviral types. The cell culture-PCR assay with a combination of A549 and BGMK cells and molecular identification could be a useful tool for monitoring infectious adenoviruses and enteroviruses in aquatic environments.
Large numbers of human enteric viruses are excreted in human feces and urine, and these viruses have been found in a variety of aquatic environments and shellfish (21, 24, 36, 45). The presence of these viruses in aquatic environments is a public health concern because they can cause illness when even low concentrations are ingested (64). Generally, microbial water quality is assessed by using bacterial indicators such as fecal coliforms, including Escherichia coli. However, bacteria are thought to have limited value as indicators of enteric viruses because these viruses are generally more resistant to water treatment processes than bacteria are (57). Therefore, there is a need to monitor the virological quality of water.
Among the enteric viruses, the adenoviruses and enteroviruses are prevalent in various aquatic environments (12, 22, 36, 51, 53). Enteroviruses consist of more than 70 distinct serotypes of human pathogens and cause illnesses ranging from gastroenteritis to pericarditis and aseptic meningitis (7, 40, 43). Adenoviruses also comprise many serotypes that are classified into six subgenera (subgenera A to F). In particular, members of adenovirus subgenus F (types 40 and 41) have been recognized as important etiological agents of gastroenteritis in children (15), second in importance only to rotaviruses. Subgenera A and C have also been implicated in the etiology of acute diarrhea (9, 29), and other subgenera are frequently associated with respiratory illnesses and epidemic conjunctivitis (25, 39). Both adenoviruses and enteroviruses are listed on the Drinking Water Contaminant Candidate List of the U.S. Environmental Protection Agency as regulated contaminants (63), but little information regarding human health or analytical methods is currently available.
Traditionally, enteric viruses have been detected in environmental samples with a cell culture assay based on the expression of viral cytopathic effects (CPE) in the cultured cell lines. However, with this method the total level of viral contamination can be greatly underestimated because in a single cell line only some of the enteric viruses present may reproduce. Moreover, some human viruses that replicate in cell culture do not cause apparent CPE (12), and additional experiments, which are time-consuming and difficult to interpret, are required to identify the viruses detected.
The limited sensitivity and complicated identification procedure of the cell culture assay for the detection of enteric viruses can be improved by optimizing the combination of appropriate cell lines and by employing molecular techniques. For adenoviruses and enteroviruses, the A549 cell line (a human lung epithelial cell line) and Buffalo green monkey kidney (BGMK) cells might be the optimal combination, as BGMK cells are effective for propagating enteroviruses (12, 41) and A549 cells are susceptible to a broad range of enteric viruses, including adenoviruses and enteroviruses (27-29, 33, 66). Amplification by reverse transcription (RT) and nested multiplex PCR of the viral sequences replicated in a cell culture increases the copy numbers of target nucleic acids and provides an infectivity assay (12, 13). In addition, molecular identification of viral sequences by phylogenetic inference complements the RT-PCR diagnosis and supplies information about the viral types circulating in the community (45), as well as the suitability of the cell lines used for these viruses (33). In this study, we performed the cell culture assay with A549 and BGMK cells, followed by cell culture-RT-multiplex nested PCR and molecular identification. The usefulness of this method for monitoring infectious adenoviruses and enteroviruses in river water was evaluated.
MATERIALS AND METHODS
Virus strains and cell lines.
An adenovirus type 5 strain (ATCC VR-1343) and the adenovirus type 40 Dugan strain (ATCC VR-931) were purchased from the American Type Culture Collection (ATCC), Rockville, Md., and were propagated in A549 cells growing in Dulbecco modified Eagle medium (Gibco-BRL Life Technology, Gaithersburg, Md.) containing 10% (vol/vol) fetal bovine serum (FBS) (HyClone, Logan, Utah). For enteroviruses, the poliovirus type 1 Chat strain (ATCC VR-192) was purchased from the ATCC and propagated in BGMK cells growing in minimal essential medium (Gibco-BRL Life Technology) containing 10% (vol/vol) FBS. Virus stocks were collected from infected A549 or BGMK monolayers by freeze-thaw lysis, low-speed centrifugation (400 × g, 5 min), and filtration through a 0.2-μm-pore-size syringe filter. To confirm the authenticity of the viruses, RT-PCR and sequencing were performed with virus stocks. The stocks were stored at −70°C and used in this study as positive controls.
Water sample collection.
Surface water samples were collected 10 times monthly or semimonthly over a 6-month period (12 May, 21 May, 13 June, 28 June, 17 July, 30 July, 15 August, 3 September, 17 September, and 1 October 2002; from late spring to early autumn) at each of four river tributaries (the Sanbon, Hwajeong, Ansan, and Siheung rivers) located in Gyeonggi Province, South Korea. The water samples were obtained at a depth of 30 cm in the afternoon. All the rivers are typical streams with low flow rates and run through urban areas; the pollution sources of these rivers are untreated domestic and industrial wastewater. Two-liter portions of river water were collected aseptically in sterile polypropylene bottles to avoid contamination. The samples were stored at 4°C and immediately transported to the laboratory for processing. The water temperature was measured in situ by using portable electrode-carrying devices (Checkmate 90; Corning, Inc., Corning, N.Y.). Fecal coliforms were enumerated in triplicate by using the membrane filtration technique (3) and m-FC agar (Difco Laboratories, Detroit, Mich.) for each sample studied. The plates were incubated aerobically at 44.5°C, and blue colonies were counted as fecal coliforms.
Sample processing.
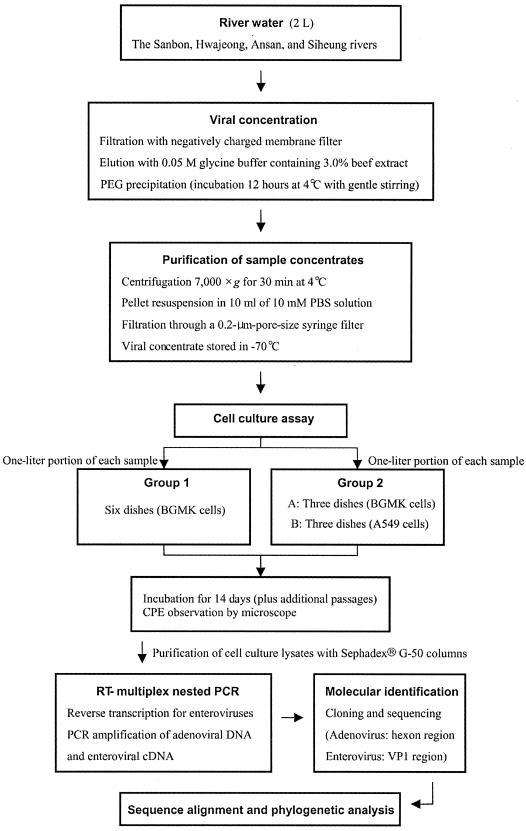
The viruses were concentrated by standard methods (3) (Fig. 1), with a minor modification at the secondary concentration step (38). Briefly, the pH and the MgCl2 level of 2 liters of water were adjusted to pH 5.5 and 0.1 N MgCl2, respectively, by adding 1.0 N HCl and 5 N MgCl2 solutions; each sample was then filtered through a negatively charged 0.45-μm-pore-size membrane filter (cellulose nitrate; diameter, 47 mm; Millipore Corporation, Bedford, Mass.) at a rate not exceeding 130 ml/min, washed with 25 ml of a 0.14 N NaCl solution, and eluted with 7.5 ml of 0.05 M glycine buffer (pH 9.5) containing 3.0% beef extract. The pH values of the eluates were immediately adjusted to neutral with 1.0 N HCl; 13% (wt/vol) polyethylene glycol 8000 (plus 0.2 M NaCl) at pH 7.2 was added to each sample, and the samples were incubated for 12 h at 4°C with gentle stirring. After incubation, the eluates were centrifuged at 7,000 × g for 30 min. The resulting pellets were resuspended in 10 ml of 10 mM phosphate-buffered saline (PBS). The samples were filtered through a 0.2-μm-pore-size filter and stored at −70°C until they were used for cell culture analysis.
FIG. 1.
Procedure for the detection of infectious adenoviruses and enteroviruses in river water. PEG, polyethylene glycol.
Cell culture assay.
Forty water samples were analyzed by using BGMK and A549 cells. The BGMK cell line (group 1), which has been suggested for monitoring enteric viruses in water (3, 42), was used as a control to show the suitability of the combination of the A549 and BGMK cell lines (group 2; the BGMK cell line was designated group 2A, and the A549 cell line was designated group 2B) for detection of infectious adenoviruses and enteroviruses in a cell culture assay (Fig. 1). A 1-liter portion (one-half of the 10-ml concentrate) of each sample was divided equally and inoculated into six individual tissue culture dishes containing confluent monolayers of BGMK cells (group 1). The other 1-liter portion was inoculated equally into three dishes containing confluent monolayers of BGMK cells and three dishes containing confluent monolayers of A549 cells (group 2). All dishes were incubated for 90 min at 37°C with rocking every 15 min. After incubation, the sample inoculum was removed, and the cells were washed twice with 10 mM of PBS. Five milliliters of minimal essential medium containing 2% (vol/vol) FBS was added to each dish containing BGMK cells, and 5 ml of Dulbecco modified Eagle medium containing 2% (vol/vol) FBS was added to each dish containing A549 cells. The adenovirus type 5 strain, the adenovirus type 40 Dugan strain, and the poliovirus type 1 Chat strain were used as positive controls; 10 mM PBS was the negative control. Tissue culture dishes were incubated at 37°C in an atmosphere containing 5% CO2 and were examined for CPE daily for the first 7 days and then every second day for 14 days. When the cell monolayer exhibited CPE, the dish was frozen and thawed three times. All dishes that did not show CPE were frozen and thawed three times on day 14. One hundred microliters of lysate from each culture was subjected to a second passage on a fresh monolayer. Third passages were performed when CPE were observed in second-passage dishes inoculated with lysate from first-passage dishes that had not exhibited CPE.
Detection of adenoviruses and enteroviruses by cell culture-RT-multiplex PCR assay.
Adenovirus- and enterovirus-positive samples were determined by performing RT-multiplex PCR with lysates from cell cultures with and without cytopathic effects. The oligonucleotide primer sequences (Table 1) used for detection of adenoviruses and enteroviruses were identical to those described by Allard et al. (1) and Leparc et al. (37). Prior to PCR amplification, the cell culture lysates were purified as described previously (55), except that Sephadex G-50 columns (Pharmacia Biotech, Uppsala, Sweden) were used. The resulting supernatants were immediately subjected to RT; the target viral genome was heat released and reverse transcribed. The RT-PCR procedure was performed as described by Cho et al. (13), with minor modifications. Briefly, 5 μl of cell culture lysate was added to 5 μl of an RT mixture. RT was carried out at 42°C for 45 min, and then the tubes were heated to 95°C for 5 min to inactivate the enzyme. The mixture was added to 15 μl of a PCR mixture containing (final concentrations) 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 0.1% Triton X-100, each deoxynucleoside triphosphate (dNTP) (Promega Co., Madison, Wis.) at a concentration of 200 μM, 1.25 U of Taq polymerase, 0.25 μM enterovirus primer EV1, 0.1 μM adenovirus primer AV1, 0.1 μM adenovirus primer AV2, and 1.5 mM MgCl2. The PCR protocol included an initial denaturation step at 94°C for 4 min; this was followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min and a final extension at 72°C for 7 min. For nested PCR amplification, 1 μl of each RT-PCR product was added to 25 μl (final volume) of a PCR mixture containing each member of an enterovirus seminested primer pair (EV1 and EV3) at a concentration of 0.25 μM and each member of an adenovirus nested primer pair (AV3 and AV4) at a concentration of 0.1 μM. Thermal cycling was carried out with a Gene Amp 9600 PCR system (Applied Biosystems, Foster City, Calif.). The results were analyzed by electrophoresis with a 2% agarose gel and staining with ethidium bromide.
TABLE 1.
Primers used in the multiplex PCR assays and sequencing
| Virus | Region | Use | Primer | Sequence (5′ → 3′)c | Polarity | Position |
|---|---|---|---|---|---|---|
| Adenovirus | Hexon | Detection by multiplex PCR and typing | AV1 | GCCCGAGTGGTCTTACATGCACATC | Sense | 18858-18883d |
| AV2 | CAGCACGCCGCGGATGTCAAAGT | Antisense | 19136-19158d | |||
| AV3a | GCCACCGAGACGTACTTCAGCCTG | Sense | 18937-18960d | |||
| AV4a | TTGTACGAGTACGCGGTATCCTCGCGGTC | Antisense | 19051-16079d | |||
| Enterovirus | 5′ Nontranslated region | Detection by multiplex PCR | E1 | CAAGCACTTCTGTTTCCCCGG | Sense | 164-184e |
| E2 | ATTGTCACCATAAGCAGCCA | Antisense | 599-578e | |||
| VP1 | Typing | E3b | CTTGCGCGTTACGAC | Antisense | 526-511e | |
| VP1-1S | GGTTYGAYITGGARITIACITTYGT | Sense | 2764-2809f | |||
| VP1-1A | TGIGAYTGRTAYCTIKYKGGRTARTA | Antisense | 3612-3637f | |||
| VP1-2Sa | ARWTWATGTAYRTICCICCIGGIG | Sense | 2874-2898f | |||
| VP1-2Aa | CCIGTKKWRCAIYKRCAYCTIGC | Antisense | 3507-3529f |
Primer used for nested PCR.
Primer used for seminested PCR.
Degenerate positions: I, deoxyinosine; Y, T or C; S, C or G; R, A or G; M, A or C; W, T or A; K, T or G.
Relative positions of primers in human adenovirus type 2 (accession no. J01917).
Relative positions of primers in coxsackievirus B3 (accession no. M16572).
Relative positions of primers in echovirus 9 strain Barty (accession no. X92886).
Molecular identification of adenoviruses and enteroviruses.
To evaluate the variety of the adenovirus and enterovirus strains detected, the amplified PCR products were sequenced. For adenoviruses, nested PCR products obtained by RT-multiplex nested PCR were used for identification. For enteroviruses, nested PCR products, obtained by nested RT-PCR with degenerate primers (Table 1) that allowed amplification of VP1 regions, were used. Direct PCR amplification with these primers and the concentrates of environmental samples proved to be very inefficient for detection due to the lower sensitivity of the primers compared to the sensitivity of the 5′ nontranslated region primers (21). However, enteroviruses were successfully detected by using a cell culture-PCR assay with degenerate primers and cell culture lysates (data not shown). The PCR procedure with degenerate primers was the procedure described by Casas et al. (11), with some modifications. Five microliters of cell culture lysate was heat released and immediately added to 7 μl of an RT mixture containing 5× reaction buffer, each dNTP at a concentration of 400 μM, 2 mM MgSO4, 100 U of Moloney murine leukemia virus reverse transcriptase, 10 U of RNasin, and 40 pmol of the antisense degenerate primer VP1-1A. RT was carried out at 40°C for 45 min, and then the tubes were heated to 95°C for 5 min to inactivate the enzyme. The mixture was added to 13 μl of a PCR mixture containing (final concentrations) 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 0.1% Triton X-100, each dNTP (Promega Co.) at a concentration of 200 μM, 1.25 U of Taq polymerase, 1.5 mM MgCl2, and 40 pmol of the VP1 antisense and sense primers (VP1-1A and VP1-1S). The following protocol was used: initial denaturation at 94°C for 3 min; 40 cycles of 94°C for 30 s, 50°C for 3 min, and 68°C for 30 s; and final extension at 68°C for 5 min. For the nested PCR amplification, 1 μl of each product of the RT-PCR was added to 25 μl (final volume) of a PCR mixture containing 60 pmol of VP1 antisense and sense nested primers (VP1-2A and VP1-2S). The nested PCR protocol was as follows: initial denaturation at 94°C for 2 min; 30 cycles of 94°C for 30 s, 50°C for 1 min, and 72°C for 30 s; and final extension at 72°C for 5 min. Thermal cycling and gel electrophoresis were carried out as described above.
The amplicons were cut out of the gel and purified. The purified amplicons were ligated into the pGEM-T vector (Promega Co.), and the construct was transformed into E. coli DH5α competent cells. Subsequently, blue-white screening was performed according to the manufacturer's instructions. Plasmid preparations for DNA sequencing were obtained with Wizard mini-preps (Promega Co.). A total of 17 amplicons of adenoviruses and 24 amplicons of enteroviruses were sequenced with the ABI BigDye terminator by using ABI 3730 automated sequencers (Applied Biosystems) and the T7 primer. The sequences were compared with those available in the EMBL/GenBank databases by using the PubMed NCBI BLAST program.
Quality control of the amplification method.
To avoid false-positive results due to contamination with DNA amplified in previous PCR assays, separate areas and apparatus were used for sample preparation, reagent preparation, and amplified samples. Each cabinet was equipped with an independent batch of reagents, micropipettes, pipette tips, and sterile reagent tubes. Virus-seeded positive controls (0.1 50% tissue culture infective dose [TCID50] of adenovirus and 0.1 TCID50 of enterovirus), as well as negative controls, were included in all PCR assays to ensure the propriety of the PCR assay. Negative controls were added to all PCR assays to ensure that there was no carryover contamination.
Sequence alignment and phylogenetic analysis.
Reference strains from a previously described database (2, 11, 14, 35, 46, 47, 62) were included in the analysis. For the most conserved and consistent alignment, deduced amino acid sequences from enterovirus DNA sequences were preliminarily aligned with Clustal X (version 1.8) (60), with the multiple alignments set to a gap opening penalty equal to 75 and a gap extension penalty equal to 6.66, as described by Palacios et al. (50). For adenoviruses, DNA sequences were aligned as described above, with the multiple alignments set to default. Regions of ambiguous alignment were excluded, and all gaps were treated as missing data. Human rhinovirus type 89 (18) and bovine adenovirus 4 strain THT/62 (16) were used as outgroups. Phylogenetic relationships were inferred with the PROTPARS program (as a character state method) and the PRODIST program (as a distance method) for enteroviruses and with the DNAPARS and the DNADIST programs for adenoviruses, followed by NEIGHBOUR from the PHYLIP package (version 3.57) (20). The phylogenetic trees demonstrated the most robust relationships observed and were determined by using the Kimura two-parameter distance and neighbor joining for adenoviruses and the Dayhoff PAM matrix and neighbor joining for enteroviruses. The phylogenetic tree obtained was plotted by using TREEVIEW (version 1.6.1) (49). The statistical significance of phylogenies was estimated by using the SEQBOOT program by bootstrap analysis with 1,000 pseudoreplicate data sets; only groups with bootstrap values higher than 50% were considered significant.
Statistical analysis.
Statistical analyses were performed with the SAS system (release v.6.12) and EXCEL (Microsoft Office 2000) by using a Pentium III computer. The possible differences in levels of fecal coliforms from sampling sites and viral frequencies from cell culture groups were analyzed by using Student's t test at a 5% significance level. The possible relationships between levels of fecal coliforms and viral frequency and between the results for adenoviruses and the results for enteroviruses at the study sites were evaluated by Pearson correlation analysis.
Nucleotide sequence accession numbers.
The original DNA sequences obtained in this study have been deposited in the GenBank database under accession numbers AY280872 to AY280912.
RESULTS
Water temperature and levels of fecal coliforms.
At each sampling site, we measured the water temperature and the level of fecal coliforms (Table 2). The mean water temperatures were similar at the sites studied; they ranged from 22.7°C at the Siheung River to 23.4°C at the Hwajeong River. A seasonal difference in water temperature was not observed at the sites studied. The water at four of the sites studied was constantly affected by sewage effluent; the mean values for fecal coliforms ranged from 1.78 log10 CFU/100 ml at the Siheung River to 2.44 log10 CFU/100 ml at the Sanbon River. The levels of fecal coliforms fluctuated between 2 and 3 log10 CFU/100 ml at the Sanbon River, the Hwajeong River, and the Ansan River and between 1 and 2 log10 CFU/100 ml at the Siheung River (data not shown). However, the seasonal pattern during the sampling period was not determined. The mean level of fecal coliforms was lower at the Siheung River than at the other rivers (P < 0.05, as determined by Student's t test).
TABLE 2.
Water temperatures and levels of fecal coliforms at sampling sites
| Sampling site | Temp (°C)a | Fecal coliforms (log10 CFU/100 ml)a |
|---|---|---|
| Sanbon River | 23.3 ± 1.2 | 2.44 ± 2.01 |
| Hwajcong River | 23.3 ± 0.7 | 2.32 ± 1.84 |
| Ansan River | 22.9 ± 1.3 | 2.29 ± 1.74 |
| Siheung River | 22.7 ± 1.3 | 1.78 ± 1.64 |
Means ± standard deviations.
Frequency of viral occurrence in river water determined by cell culture assay.
A total of 40 water samples were analyzed for infectious viruses by cell culture assays. Overall, 30 (75.0%) of the 40 samples, representing 20 (50.0%) of the 40 samples in group 1 and 26 (65.0%) of the 40 samples in group 2, exhibited CPE (Table 3).
TABLE 3.
River water results for detection of culturable viruses by the cell culture assay
| Sampling site | No. of positive samples/no. of samples tested (no. of positive subsamples/no. of subsamples tested)
|
|||
|---|---|---|---|---|
| Group 1 (BGMK) | Group 2 (BGMK + A549)
|
|||
| Group 2A (BGMK) | Group 2B (A549) | Totala | ||
| Sanbon River | 8/10 (23/60) | 8/10 (15/30) | 9/10 (22/30) | 10/10 (37/60) |
| Hwajeong River | 5/10 (10/60) | 5/10 (8/30) | 7/10 (18/30) | 8/10 (26/60) |
| Ansan River | 3/10 (4/60) | 3/10 (4/30) | 2/10 (2/30) | 4/10 (6/60) |
| Siheung River | 4/10 (5/60) | 0/10 (0/30) | 4/10 (8/30) | 4/10 (8/60) |
| Total | 20/40 (42/240) | 16/40 (27/120) | 22/40 (50/120) | 26/40 (77/240) |
Total number of samples that exhibited CPE in both group 2A and group 2B.
The viral frequency in the Sanbon River (60 [50.0%] of 120 subsamples) was the highest frequency, followed by the viral frequencies in the Hwajeong River (36 [30.0%] of 120 subsamples), the Siheung River (13 [10.8%] of 120 subsamples), and the Ansan River (10 [8.3%] of 120 subsamples) (Table 3). However, a seasonal pattern of viral frequency at each site studied was not observed. The viral frequencies in the water samples for group 2 (77 [32.1%] of 240 subsamples) were higher than the viral frequencies in the water samples for group 1 (42 [17.5%] of 240 subsamples) for all of the subsamples tested (P < 0.05, as determined by Student's t test). In addition, the viral frequencies detected were higher when A549 cells were used (group 2B) than when BGMK cells were used (groups 1 and 2A; P < 0.05, as determined by Student's t test), and the difference in viral frequency between group 1 and group 2A was not significant. The viral frequency was positively correlated for groups 1 and 2 and for group 2A (BGMK cells) and group 2B (A549 cells) (P < 0.05, as determined by Pearson correlation analysis).
The viral frequency at each site studied was not related to the level of fecal coliforms; the viral frequencies at the Siheung River (13 [10.8%] of 120 subsamples) and the Ansan River (10 [8.3%] of 120 subsamples) were similar, while the fecal coliform level was lower at the Siheung River than at the other rivers (Table 2).
Detection of adenoviruses and enteroviruses by cell culture-RT-multiplex nested PCR assay.
Forty samples were analyzed by the cell culture-RT-multiplex nested PCR assay for the detection of adenoviruses and enteroviruses. Overall, 31 (72.5%) of the 40 samples, representing 19 (47.5%) of the 40 samples in group 1 and 26 (65.0%) of the 40 samples in group 2, were positive for adenoviruses and/or enteroviruses (Table 4).
TABLE 4.
Detection of adenoviruses and enteroviruses in river water samples by cell culture-RT-multiplex PCR assay
| Sampling site | Virus detection by cell culture-RT-multiplex PCR assay
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Group 1 (BGMK)
|
Group 2 (BGMK + A549)
|
||||||||
| Group 2A (BGMK)
|
Group 2B (A549)
|
Total (groups 2A and 2B)
|
|||||||
| Adenovirus | Enterovirus | Othersa | Adenovirus | Enterovirus | Adenovirus | Enterovirus | Adenovirus | Enterovirus | |
| Sanbon River | 1/10 (2/60)b,c | 8/10 (22/60) | 1/10 (1/60) | 0/10 (0/30) | 8/10 (15/30) | 6/10 (10/30) | 5/10 (12/30) | 6/10 (10/60) | 8/10 (27/60)f |
| Hwajeong River | 0/10 (0/60) | 4/10 (8/60) | 2/10 (2/60) | 0/10 (0/30) | 6/10 (8/30) | 3/10 (4/30)e | 7/10 (15/30)e | 3/10 (4/60) | 8/10 (23/60)f |
| Ansan River | 0/10 (0/60) | 3/10 (3/60) | 1/10 (1/60) | 0/10 (0/30) | 3/10 (4/30) | 2/10 (2/30) | 0/10 (0/30) | 2/10 (2/60) | 3/10 (4/60) |
| Siheung River | 0/10 (0/60) | 4/10 (4/60)d | 2/10 (2/60) | 0/10 (0/30) | 0/10 (0/30) | 2/10 (3/30) | 3/10 (5/30) | 2/10 (3/60) | 3/10 (5/60) |
| Total | 1/40 (2/240) | 19/40 (37/240) | 6/40 (6/60) | 0/40 (0/120) | 17/40 (27/120) | 13/40 (19/120) | 15/40 (32/120) | 13/40 (19/240) | 22/40 (59/240) |
Samples exhibited CPE, but the cell culture-RT-multiplex PCR results were negative.
No. of positive samples/no. of samples tested. (No. of positive subsamples/no. of subsamples tested).
Two subsamples obtained on 13 June 2002 did not exhibit CPE but were positive for adenoviruses as determined by a nested PCR.
One subsample obtained on 28 June 2002 that did not exhibit CPE was positive for enteroviruses as determined by a nested PCR.
One subsample obtained on 13 June 2002 was coinfected with both adenoviruses and enteroviruses.
Total number of positive samples detected in either group 2A or group 2B.
We found that 37 (15.4%) of the 240 subsamples in group 1 and 59 (24.6%) of the 240 subsamples in group 2 (27 [21.7%] of the 120 subsamples in group 2A and 32 [26.7%] of the 120 subsamples in group 2B) were positive for enteroviruses. Three subsamples that did not exhibit CPE were subsequently determined to be positive by the cell culture-RT-multiplex nested PCR assay. One subsample taken from the Siheung River on 28 June that did not exhibit CPE was positive for enteroviruses as determined by the cell culture-RT-multiplex nested PCR assay (Table 4). Two group 1 subsamples taken from the Sanbon River on 1 October did not exhibit CPE but were positive for adenoviruses as determined by a nested PCR. Nineteen subsamples (15.8%) from 13 samples (32.5%) of group 2B exhibited CPE and were also successfully amplified by a nested PCR. In addition, one group 2B subsample taken from the Hwajeong River on 13 June was coinfected with both adenoviruses and enteroviruses. Six group 1 subsamples (2.1%) exhibited CPE but were negative for both viruses as determined by the cell culture-RT-multiplex nested-PCR assay.
The data obtained were analyzed further to determine whether there was any possible correlation between the occurrence patterns of adenoviruses and enteroviruses in the groups. The occurrence patterns of enteroviruses for groups 1 and 2 and for group 2A (BGMK cells) and group 2B (A549 cells) were positively correlated according to a Pearson correlation analysis (P < 0.05). However, the occurrence patterns for adenoviruses and enteroviruses did not show statistically significant correlations in the groups used.
Molecular identification of adenoviruses and enteroviruses by sequence analysis.
For adenoviruses, nested PCR products were used for molecular identification. In the case of enteroviruses, PCR products generated from the VP1 gene were used for molecular identification instead of products from the 5′ nontranslated region; the VP1 region was the most suitable target, due to the high correlation between serotypes and sequences and the availability of a large database of enteroviral sequences (47). Seventeen amplicons (one amplicon from group 1 and 16 amplicons from group 2A) of 21 subsamples that were positive for adenoviruses and 24 amplicons (9 amplicons from group 1, 6 amplicons from group 2A, and 9 amplicons from group 2B) of 96 subsamples that were positive for enteroviruses were sequenced and initially compared to sequences available in the GenBank databases by using the BLAST program. All of the amplicons sequenced belonged to human adenoviruses or enteroviruses.
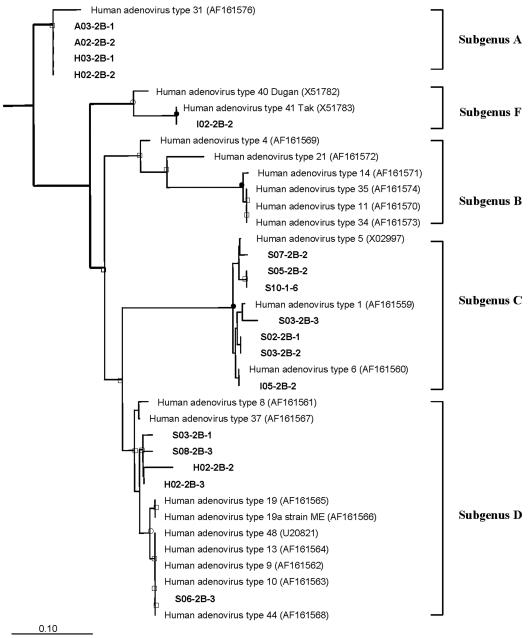
To obtain accurate molecular identification of the amplicons sequenced, a phylogenetic inference analysis was performed (6, 50). For adenoviruses, 90-bp nucleotide sequences from which the primer sequences were excised were used for the analysis. Because all the phylogenetic methods generated the same tree topology, the phylogenetic tree was determined by using the Kimura two-parameter distance and neighbor joining, and bootstrap values were obtained (Fig. 2). Five subgenera (subgenera A, B, C, D, and F) supported by bootstrap values were clustered in a pattern similar to the patterns obtained in previously published studies (2, 6). The reference strains of subgenus E were omitted in order to simplify the tree. Six amplicons were grouped with reference strains with bootstrap values that were highly significant (90%; one amplicon grouped with enteric adenovirus type 40 Dugan) and moderately significant (50%; four amplicons grouped with human adenovirus type 31 and one amplicon grouped with human adenovirus type 44). Seven amplicons were grouped with reference stains without significant bootstrap values (<50%; three amplicons grouped with human adenovirus type 1, three amplicons grouped with human adenovirus type 5, and one amplicon grouped with human adenovirus type 6), but these amplicons were assigned to subgenus C with a highly significant value (>90%). The remaining four amplicons were not grouped with reference strains but could be assigned to subgenus D with a moderately significant value (>50%).
FIG. 2.
Phylogenetic tree based on nucleotide sequences of 17 adenoviral amplicons. Subgenera A, B, C, D, and F of the adenoviruses are indicated. The reference strains of subgroup E were omitted to simplify the tree. Bovine adenovirus 4 strain THT/62 was used as the outgroup. Nodes supported by bootstrap values of >90% are indicated by solid circles; open circles indicate >70% support, and open squares indicate >50% support. The scale bar corresponds to 0.10 substitution per nucleotide position. Amplicon designations are in boldface type. A, H, I, and S indicate the Ansan River, the Hwajeong River, the Siheung River, and the Sanbon River, respectively, and these letters are followed by sampling dates (01, 12 May 2002; 02, 21 May 2002; 03, 13 June 2002; 04, 28 June 2002; 05, 17 July 2002; 06, 30 July 2002; 07, 15 August 2002; 08, 3 September 2002; 09, 17 September 2002; 10, 1 October 2002); 1, 2A, and 2B indicate the cell line groups and are followed by the subsample number. The numbers in parentheses are accession numbers. See Materials and Methods for details.
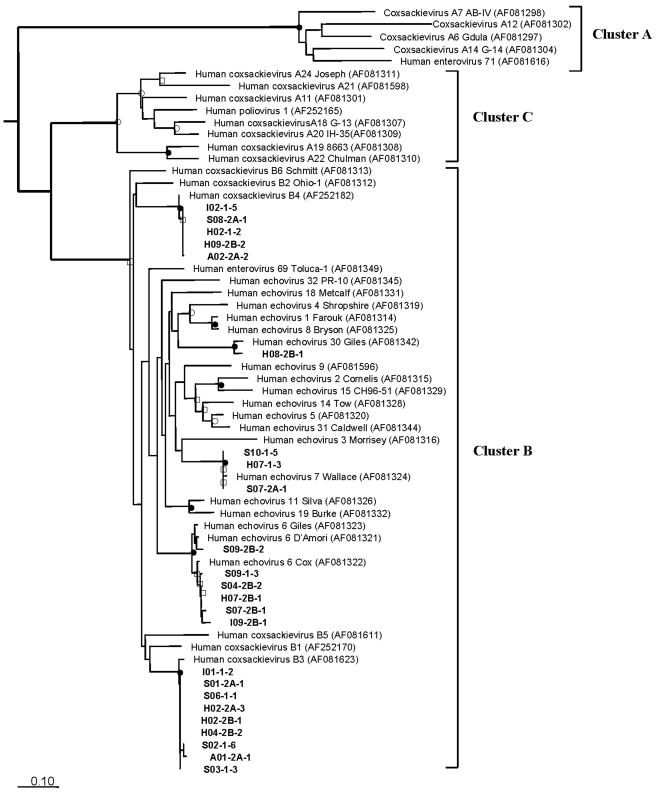
Alternatively, the deduced 101 amino acid sequences of enteroviruses were used for analysis. Because all the phylogenetic methods generated the same tree topology, the phylogenetic tree was determined by using the Dayhoff PAM matrix and neighbor joining, and bootstrap values were obtained (Fig. 3). Three separate clusters (clusters A to C) supported by bootstrap values (>50%) clustered in a pattern similar to the patterns reported previously (44, 50, 54). The reference strains of cluster D were omitted in order to simplify the tree. All 24 amplicons were grouped with reference strains with highly significant bootstrap values (>90%; nine amplicons grouped with human coxsackievirus B3, six amplicons grouped with human echovirus 6 strains, five amplicons grouped with human coxsackievirus B4, three amplicons grouped with human echovirus 7 Wallace, and one amplicon grouped with human echovirus 30 Giles).
FIG. 3.
Phylogenetic tree based on deduced amino acid sequences of 24 enteroviral amplicons. Clusters A, B, and C of the enteroviruses are indicated. The reference strains of cluster D were omitted to simplify the tree. Human rhinovirus 89 was used as the outgroup. The scale bar corresponds to 0.10 substitution per amino acid position. The bootstrap values and amplicon designations are described in the legend to Fig. 2. The numbers in parentheses are accession numbers. See Materials and Methods for details.
The results of the phylogenetic analysis demonstrated that none of the amplicons was significantly different from the other amplicons, irrespective of the sampling site and time. However, differences in viral types between cell lines were observed in this study. Three amplicons (S10-1-1, H07-1-1, and S07-2A-1) that grouped with echovirus 7 were recovered from only BGMK cells, whereas one amplicon (H08-2B-1) that grouped with echovirus 30 was recovered from only A549 cells.
DISCUSSION
The cell culture-PCR method described here provided reliable information about the presence of infectious adenoviruses and enteroviruses in river water. This method included a cell culture assay and a cell culture-RT-multiplex nested PCR assay to eliminate false-positive results and to provide high levels of sensitivity and specificity (13); this allowed us to overcome the technical limitations of isolating the viruses in cell culture assays and to use sequence analysis to molecularly identify viral amplicons recovered from water samples.
Although the cell culture-PCR method is a useful method for detection of enteric viruses in water samples, the viral level can be underestimated because in a single cell only some of the enteric viruses present may reproduce. To reduce false-negative results resulting from the limited susceptibility of a single cell line for enteric viruses, several investigators have combined appropriate cell lines for expanding the range of detectable viruses. Taylor et al. (59) reported cell culture assays based on PLC/PRF/5 and FRhK-4 cells, and PLC/PRF/5 and CaCo-2 cells enhanced the detection of human hepatitis A viruses and human astroviruses, respectively. Grabow et al. (26) also reported similar findings for poliovirus. Overall, these findings indicate that a combination of different cell lines could be more useful than single cell line for monitoring enteric viruses in water samples.
We used A549 cells together with BGMK cells for the cell culture assay. A549 cells are susceptible to a broad range of viruses, including adenoviruses and enteroviruses (27-29, 33, 66), and showed good integrity of cell monolayers during the 2-week incubation period (33). Although the BGMK cell line was shown in previous studies (12, 36) to be suitable for enteroviruses and to be able to propagate some enteric viruses, it is difficult to produce CPE in BGMK cells with adenoviruses, and BGMK cells seem to select for enteroviruses, which replicate more rapidly than adenoviruses (36). We obtained a higher viral frequency when we used A549 cells than we used BGMK cells in this study (Table 3). While the enteroviral frequencies in A549 and BGMK cells were similar, the adenoviral frequency was much higher with A549 cells (13 samples) than with BGMK cells (one sample) (Table 4). It seemed that the adenoviral frequency obtained with BGMK cells could be influenced by interference from enteroviruses, and thus adenoviruses in aquatic environments could be very inefficiently propagated in BGMK cells. These hypotheses are supported by the results of previous studies of adenoviral receptors (4, 5, 8, 10). The binding of adenovirus to target cells is mediated by its fiber (17), and the cellular receptor for adenovirus subgenera A, C, E, and F and for adenovirus type 9 and type 19p of subgenus D has been identified recently as the coxsackievirus-adenovirus receptor (CAR) (8). It has been noted that CAR is differentially regulated during development, that the expression of CAR appears to be dependent on both tissue type and development stage, and that the expression in various tissues and cell lines corresponds closely to the known tropisms of the viruses (10). These findings explain the increased susceptibility to adenoviruses and coxsackievirus B during the early neonatal period and the differential susceptibility to adenoviruses among cell lines. Therefore, it seems that CAR in BGMK cells has a higher affinity for coxsackievirus B than for adenoviruses, and thus adenoviruses are inefficiently recovered in BGMK cells. In this study, subgenus D adenoviruses were recovered from five river water samples by using A549; however, they were not detected with several other cell lines, including BGMK cells, as has been noted in previous studies (13, 33, 35, 50). Several subgenus D adenoviruses are known to use sialic acid as a receptor instead of CAR (5). Because A549 cells express sialic acid as well as CAR (4), they can propagate a broad range of adenoviruses, including subgenus D adenoviruses. Consequently, our study demonstrated that the A549 cell line was suitable for recovering human adenoviruses from environmental samples and could be useful for detecting culturable enteric viruses, including adenoviruses and enteroviruses, at least in polluted river water.
On the basis of the results of the molecular identification of the recovered adenoviruses, we concluded that a broad range of adenoviruses belonging to subgenera A, C, D, and F were present in river water (Fig. 2). One reason for this may be the input of a variety of adenoviruses excreted from patients. In addition, adenoviruses can survive and persist longer than other enteric viruses in different environments, and they are more resistant to UV radiation than other viruses are in water (19, 23, 58, 61).
The following two approaches were used for suitable molecular identification of enteroviruses: (i) cell culture and PCR amplification of the VP1 region with degenerate primers and (ii) phylogenetic inference from comparisons of deduced amino acid sequences of recovered enteroviruses and reference strains (11, 50). Twenty (83.3%) of 24 amplicons sequenced were grouped as coxsackievirus B3 and B4 and echovirus 6 (Fig. 3). These viruses are similar to the causative agent of enteroviral diseases in Korea (32) and have also been found in aquatic environments and in shellfish around the world (21, 34, 52). Interestingly, three amplicons grouped with echovirus 7 were recovered from only BGMK cells, and one amplicon grouped with echovirus 30 was recovered from only A549 cells (Fig. 3). However, these results may not reflect a difference in cell tropism for these viruses, as they use the same cellular receptor, decay-accelerating factor (65). Although it is currently accepted that BGMK cells are effective for the propagation of enteroviruses, especially coxsackievirus B (12, 41, 48), many types of enteroviruses, especially coxsackievirus A, are still unable to propagate in BGMK cells, and the levels of enteroviruses in some environments analyzed with BGMK cells could be underestimated. A large number of enteroviral types, including the types that do not grow in BGMK cells (33), could propagate in A549 cells, and thus the use of both A549 and BGMK cells could reduce false-negative results. However, there is still a problem due to the lack of sensitivity of these cells to a large number of coxsackievirus A viruses. Therefore, it is necessary to look for improved cell lines and better combinations of appropriate cell lines for enteroviral detection.
The occurrence patterns of adenoviruses and enteroviruses did not show statistically significant correlations in this study. A difference in the occurrence patterns of adenoviruses and enteroviruses has also been reported in previous studies (34, 36). One reason for this phenomenon is the difference in the input frequency between adenoviruses and enteroviruses excreted from patients in the community (57). In addition, this phenomenon could be affected by the different survival rates of adenoviruses and enteroviruses and by interference with each other (23, 36).
In previous studies, it was documented that the cell culture-PCR method was more sensitive than the cell culture assay alone for the detection of enteric viruses, including adenoviruses and enteroviruses that did not produce CPE (12, 59). However, only three subsamples (0.6%) that did not exhibit CPE were determined to be virus positive by the cell culture-PCR method in this study. The small increase in the sensitivity in this study is not presumed to be due to the misuse of the method, as the two positive controls (0.1 TCID50 of adenovirus and 0.1 TCID50 of enterovirus) included with each PCR procedure were successfully amplified by this method. Rather, because the sampling sites in this study were directly affected by domestic sewage and because the viruses were exposed to inactivating agents, such as sunlight, for a relatively short period, it seems that most of the viruses in this study were very stable, thus easily replicating and exhibiting CPE on the cell lines.
On the other hand, we also found that five samples that exhibited CPE on BGMK cells were negative for both adenoviruses and enteroviruses according to the cell culture-PCR assay. Chapron et al. (12) reported similar results and suggested that their results could be explained by the presence of reoviruses. This explanation is supported by the findings that reoviruses are capable of replicating in BGMK cells and are present in environmental samples (30).
Several investigations have pointed out that bacterial indicators are not related to the presence of enteric viruses (3, 53, 59). Our results also showed that the level of fecal coliforms, which is one of standard bacterial indicators, was not related to the viral frequency at each site studied, confirming that bacterial indicators have limited value as a parameter for the virological quality of water. Moreover, the frequencies of viral occurrence in river water were very high, indicating that the river water at the study sites was chronically exposed to viral contamination. Previous studies have also reported contaminating levels of adenoviruses and enteroviruses in a variety of aquatic environments in Korea, including tap water (13, 36). Several types of the viruses detected in this study and previous studies are similar to the causative agents of viral diseases in Korea (31, 32). Because drinking water may be exposed to unusual contamination by river water leaking into distribution systems or household wells, the presence of infectious adenoviruses and enteroviruses in river tributaries may be potentially harmful to public health. Therefore, it is necessary to monitor these viruses periodically in the aquatic environments of Korea.
Understanding the origin of fecal pollution and predicting the presence of fecal pollution in water are important for maintenance of the microbial quality and safety for human public health. The methods of microbial source tracking and the concept that the origin of fecal pollution can be traced by microbiological, genotypic, phenotypic, and chemical methods have been developed to predict and identify sources of fecal pollution in the environment (reviewed in reference 56). One significant result of this study was the extension of the information about infectious adenoviruses and enteroviruses generated with the specific cell lines and primers into molecular identification of recovered viral amplicons. Therefore, our method is potentially useful for investigators attempting to monitor infectious enteric viruses for microbial source tracking.
Overall, the monitoring of infectious adenoviruses and enteroviruses in aquatic environments is useful for the assessment of public health risks associated with these viruses. The cell culture-PCR assay based on a combination of the A549 and BGMK cell lines and the molecular identification established in this study provide a more efficient way of detecting infectious adenoviruses and enteroviruses and thus could be useful for monitoring these viruses in aquatic environments.
Acknowledgments
This work was supported by grant R01-2002-000-00304-0 from the Basic Research Program of the Korea Science & Engineering Foundation. C.L. and E.H. were supported by a BK21 Research Fellowship from the Ministry of Education and Human Resources Development.
REFERENCES
- 1.Allard, A., B. Albinsson, and G. Wadell. 1992. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J. Med. Virol. 37:149-157. [DOI] [PubMed] [Google Scholar]
- 2.Allard, A., B. Albinsson, and G. Wadell. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 4.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 5.Arnberg, N., A. H. Kidd, K. Edlund, F. Olfat, and G. Wadell. 2000. Initial interactions of subgenus D adenoviruses with A549 cellular receptors: sialic acid versus αv integrins. J. Virol. 74:7691-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey, A., and V. Mautner. 1994. Phylogenetic relationships among adenovirus serotypes. Virology 205:438-452. [DOI] [PubMed] [Google Scholar]
- 7.Berlin, L. E., M. L. Rorabaugh, F. Heldrich, K. Roberts, T. Doran, and J. F. Modlin. 1993. Aseptic meningitis in infants <2 yr of age: diagnosis and etiology. J. Infect. Dis. 168:888-892. [DOI] [PubMed] [Google Scholar]
- 8.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Cromwell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 9.Brown, M., J. D. Grysduk, E. Fortsas, and M. Petric. 1996. Structural features unique to enteric adenoviruses. Arch. Virol. Suppl. 12:301-307. [DOI] [PubMed] [Google Scholar]
- 10.Carson, S. D. 2001. Receptor for the group B coxsackieviruses and adenoviruses: CAR. Rev. Med. Virol. 11:219-226. [DOI] [PubMed] [Google Scholar]
- 11.Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. [PubMed] [Google Scholar]
- 12.Chapron, C. D., N. A. Ballester, J. H. Fontaine, C. N. Frades, and A. B. Margolin. 2000. Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the Information Collection Rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 66:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, H.-B., S.-H. Lee, J.-C. Cho, and S.-J. Kim. 2000. Detection of adenoviruses and enteroviruses in tap water and river water by reverse transcription multiplex PCR. Can. J. Microbiol. 46:417-424. [PubMed] [Google Scholar]
- 14.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz, J. R., P. Caceres, F. Cano, J. Flores, A. Bartlett, and B. Torun. 1990. Adenovirus types 40 and 41 and rotaviruses associated with diarrhea in children form Guatemala. J. Clin. Microbiol. 28:1780-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dan, A., Z. Ruzsics, W. C. Russell, M. Benko, and B. Harrach. 1998. Analysis of the hexon gene sequence of bovine adenovirus type 4 provides further support for a new adenovirus genus (Atadenovirus). J. Gen. Virol. 79:1453-1460. [DOI] [PubMed] [Google Scholar]
- 17.Defer, C., M.-T. Belin, M.-L. Caillet-Boudin, and P. Boulanger. 1990. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J. Virol. 64:3661-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duechler, M., T. Skern, W. Sommergruber, C. Neubauer, P. Gruendler, I. Fogy, D. Blaas, and E. Kuechler. 1987. Evolutionary relationships within the human rhinovirus genus: comparison of serotypes 89, 2, and 14. Proc. Natl. Acad. Sci. USA 84:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enriquez, C. E., C. J. Hurst, and C. P. Gerba. 1995. Survival of the enteric adenovirus 40 and 41 in tap, sea, and wastewater. Water Res. 29:2548-2553. [Google Scholar]
- 20.Feltenstein J. 1995. PHYLIP: phylogeny inference package, version 3.57c. University of Washington, Seattle.
- 21.Formiga-Cruz, M., G. Tofiño-Quesada, S. Bofill-Mas, D. N. Lees, K. Henshilwood, A. K. Allard, A.-C. Conden-Hansson, B. E. Hernroth, A. Vantarakis, A. Tsibouxi, M. Papapetropoulou, M. D. Furones, and R. Girones. 2002. Distribution of human virus contamination in shellfish from different growing areas in Greece, Spain, Sweden, and the United Kingdom. Appl. Environ. Microbiol. 68:5990-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gantzer, C., A. Maul, J. M. Audic, and L. Schwartzbrod. 1998. Detection of infectious enterovirus genomes, somatic coliphages, and Bacteroides fragilis phages in treated wastewater. Appl. Environ. Microbiol. 64:4307-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerba, C. P., D. M. Gramos, and N. Nwachuku. 2002. Comparative inactivation of enteroviruses and adenovirus 2 by UV light. Appl. Environ. Microbiol. 68:5167-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerba, C. P., and J. B. Rose. 1990. Viruses in source and drinking water, p. 380-396. In G. A. McFeters (ed.), Drinking water microbiology. Springer-Verlag, New York, N.Y.
- 25.Gordon, Y. J., K. Aoki, and P. R. kinchington. 1996. Adenovirus keratoconjunctivitis, p. 877-894. In J. S. Pepose (ed.), Ocular infections and immunity. Mosby, St. Louis, Mo.
- 26.Grabow, W. O. K., K. L. Botma, J. C. de Villiers, C. G. Clay, and B. Erasmus. 1999. An assessment of cell culture and PCR procedures for the detection of polio viruses in water. Bull. W. H. O. 77:973-980. [PMC free article] [PubMed] [Google Scholar]
- 27.Greening, G. E., J. Hewitt, and G. D. Lewis. 2002. Evaluation of integrated cell culture-PCR for virological analysis of environmental samples. J. Appl. Microbiol. 93:745-750. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto, S., N. Sakakibara, H. Kumai, M. Nakai, S. Sakuma, S. Chiba, and K. Fujinaga. 1991. Fastidious human adenovirus type 40 can propagate efficiently and produce plaques on a human cell line, A549, derived from lung carcinoma. J. Virol. 65:2429-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horwitz, M. S. 1996. Adenoviruses, p. 2149-2171. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 30.Irving, L. G., and F. A. Smith. 1981. One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 41:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, D. S. 1999. The epidemiology of enteroviruses in Korea. Pediatr. Infect. 6:1-3. [Google Scholar]
- 32.Kim, K. H., J. M. Yang, S. I. Joo, Y. G. Cho, R. I. Glass, and Y. J. Cho. 1990. Importance of rotavirus and adenovirus types 40 and 41 in acute gastroenteritis in Korean children. J. Clin. Microbiol. 28:2279-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kok, T. W., T. Pryor, and L. Payne. 1998. Comparison of rhabdomyosarcoma, buffalo green monkey kidney epithelial, A549 (human lung epithelial) cells and human embryonic lung fibroblasts for isolation of enteroviruses from clinical samples. J. Clin. Virol. 11:61-65. [DOI] [PubMed] [Google Scholar]
- 34.Krikelis, V., N. Spyrou, P. Markoulatos, and C. Serie. 1985. Seasonal distribution of enteroviruses and adenoviruses in domestic sewage. Can. J. Microbiol. 31:24-25. [DOI] [PubMed] [Google Scholar]
- 35.Kruijer, W., F. M. van Schaik, and J. S. Sussenbach. 1980. Nucleotide sequence analysis of a region of adenovirus 5 DNA encoding a hitherto unidentified gene. Nucleic Acids Res. 8:6033-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, S.-H., and S.-J. Kim. 2002. Detection of infectious enteroviruses and adenoviruses in tap water in urban areas in Korea. Water Res. 36:248-256. [DOI] [PubMed] [Google Scholar]
- 37.Leparc, I., M. Aymard, and F. Fuchs. 1994. Acute, chronic and persistent enterovirus and poliovirus infection: detection of viral genome by semi-nested PCR amplification in culture-negative samples. Mol. Cell. Probes 8:487-495. [DOI] [PubMed] [Google Scholar]
- 38.Lewis, G. D., and T. G. Metcalf. 1988. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 54:1983-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNeil, K. M., R. M. Hendrix, J. L. Lindner, R. R. Benton, S. C. Monteith, M. A. Tuchoscherer, G. C. Gray, and J. C. Gaydos. 1999. Large, persistent epidemic of adenovirus type 4-associated acute respiratory disease in U.S. army trainees. Emerg. Infect. Dis. 5:798-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 655-712. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 41.Menegus, M., and G. E. Hollick. 1982. Increased efficiency of group B coxsackievirus isolation from clinical specimens by use of BGM cells. J. Clin. Microbiol. 15:945-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Messer, J. W., G. S. Fout, F. W. Schaefer III, D. R. Dahling, and R. E. Stetler. 1995. ICR microbial laboratory manual. U.S. Environmental Protection Agency, Cincinnati, Ohio.
- 43.Minor, P. D., P. Morgan-Capner, and G. C. Schild. 1994. Enteroviruses, p. 417-439. In A. J. Zuckerman, J. E. Banatvala, and J. R. Pattison (ed.), Principles and practice of clinical virology, 3rd ed. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 44.Muir, P., U. Kammerer, K. Korn, M. N. Mulders, T. Poyry, B. Weissbrichi, R. Kandolf, G. M. Cleator, and A. M. van Loon for The European Union Concerted Action on Virus Meningitis and Encephalitis. 1998. Molecular typing of enteroviruses: current status and future requirements. Clin. Microbiol. Rev. 11:202-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muscillo, M., G. La Rosa, C. Marianelli, S. Zaniratti, M. R. Capobianchi, L. Cantiani, and A. Carducci. 2001. A new RT-PCR method for the identification of reoviruses in seawater samples. Water Res. 35:548-556. [DOI] [PubMed] [Google Scholar]
- 46.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otero, J. R., L. Folgueira, G. Trallero, C. Prieto, S. Maldonaldo, M. J. Babiano, and I. Martinez-Alonso. 2001. A-549 is a suitable cell line for primary isolation of coxsackie B viruses. J. Med. Virol. 65:534-536. [PubMed] [Google Scholar]
- 49.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 23:357-358. [DOI] [PubMed] [Google Scholar]
- 50.Palacios, G., I. Casas, A. Tenorio, and C. Freire. 2002. Molecular identification of enterovirus by analyzing a partial VP1 genome region with different methods. J. Clin. Microbiol. 40:182-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papapetropoulou, M., and A. C. Vanatarakis. 1998. Detection of adenovirus outbreak at a municipal swimming pool by nested PCR amplification. J. Infect. 36:101-103. [DOI] [PubMed] [Google Scholar]
- 52.Payment, P., R. Ayache, and M. Trudel. 1983. A survey of enteric viruses in domestic sewage. Can. J. Microbiol. 29:111-119. [DOI] [PubMed] [Google Scholar]
- 53.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pöyry, T., L. Kinnunen, T. Hyypiä, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 55.Reynolds, K. A., C. P. Gerba, and I. L. Pepper. 1996. Detection of infectious enteroviruses by an integrated cell culture-PCR procedure. Appl. Environ. Microbiol. 62:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sellwood, J., J. Dadswell, and J. Slade. 1981. Viruses in sewage as an indicator of their presence in the community. J. Hyg. 86:217-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sobsey, M. D. 1989. Inactivation of health-related microorganisms in water by disinfection process. Water Sci. Technol. 21:179-195. [Google Scholar]
- 59.Taylor, M. B., N. Cox, M. A. Very, and W. O. K. Grabow. 2001. The occurrence of hepatitis A and astroviruses in selected river and dam waters in South Africa. Water Res. 35:2653-2660. [DOI] [PubMed] [Google Scholar]
- 60.Thompson, J. D., T. J. Gibson, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, K. Riley, and C. P. Gerba. 2003. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl. Environ. Microbiol. 69:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toogood, C. I., R. Murali, R. M. Burnett, and R. T. Hay. 1989. The adenovirus type 40 hexon: sequence, predicted structure and relationship to other adenovirus hexons. J. Gen. Virol. 70:3203-3214. [DOI] [PubMed] [Google Scholar]
- 63.U.S. Environmental Protection Agency. 1998. Announcement of the drinking water contaminant candidate list: notice. Fed. Regist. 63:10273-10287. [Google Scholar]
- 64.Ward, R. L., and E. W. Akin. 1984. Minimum infective dose of animal viruses. Crit. Rev. Environ. Contam. 14:297-310. [Google Scholar]
- 65.Ward, T., R. M. Powell, P. A. Pipkin, D. J. Evans, P. D. Minor, and J. W. Almond. 1998. Role for β2-microglobulin in echovirus infection of rhabdomyosarcoma cells. J. Virol. 72:5360-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Witt, D. J., and E. B. Bousquet. 1988. Comparison of enteric adenovirus infection in various human cell lines. J. Virol. Methods 20:295-308. [DOI] [PubMed] [Google Scholar]