Abstract
Nodularia spumigena is a bloom-forming cyanobacterium which produces the hepatotoxin nodularin. The complete gene cluster encoding the enzymatic machinery required for the biosynthesis of nodularin in N. spumigena strain NSOR10 was sequenced and characterized. The 48-kb gene cluster consists of nine open reading frames (ORFs), ndaA to ndaI, which are transcribed from a bidirectional regulatory promoter region and encode nonribosomal peptide synthetase modules, polyketide synthase modules, and tailoring enzymes. The ORFs flanking the nda gene cluster in the genome of N. spumigena strain NSOR10 were identified, and one of them was found to encode a protein with homology to previously characterized transposases. Putative transposases are also associated with the structurally related microcystin synthetase (mcy) gene clusters derived from three cyanobacterial strains, indicating a possible mechanism for the distribution of these biosynthetic gene clusters between various cyanobacterial genera. We propose an alternative hypothesis for hepatotoxin evolution in cyanobacteria based on the results of comparative and phylogenetic analyses of the nda and mcy gene clusters. These analyses suggested that nodularin synthetase evolved from a microcystin synthetase progenitor. The identification of the nodularin biosynthetic gene cluster and evolution of hepatotoxicity in cyanobacteria reported in this study may be valuable for future studies on toxic cyanobacterial bloom formation. In addition, an appreciation of the natural evolution of nonribosomal biosynthetic pathways will be vital for future combinatorial engineering and rational design of novel metabolites and pharmaceuticals.
Nodularia species are bloom-forming filamentous cyanobacteria, which are toxic via the production of the cyclic pentapeptide hepatotoxin nodularin. Phylogenetic studies have shown that toxicity is restricted to the bloom-forming strains of N. spumigena isolated from blooms in Australia, New Zealand, and the Baltic Sea (23, 25). Strains of the benthic species N. harveyana and N. sphaerocarpa have been shown to be nontoxic, with the exception of the strain N. harveyana PCC7804, which produces an isoform of nodularin containing a homoarginine (Har) residue at position 5 of nodularin (2, 39).
The consumption of water containing nodularin-producing N. spumigena blooms has led to the death of domestic and native animals by massive liver hemorrhage in Australia, the Baltic Sea, and New Zealand (4, 5, 13, 29). In subacute doses, nodularin is thought to act as a liver tumor initiator and promoter (34). The hepatotoxicity and carcinogenicity of nodularin are associated with the inhibition of eukaryotic protein phosphatase catalytic subunit types 1 and 2A (18).
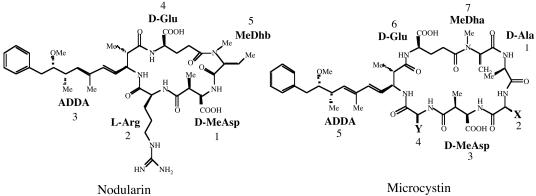
Nodularin is a cyclic pentapeptide consisting of 3-amino-9-methoxy-2,6,8-trimethyl-10-phenyl-4,6-decadienoic acid (Adda), d-glutamic acid (d-Glu), N-methyldehydrobutyrine (MeDhb), d-erythro-β-methylaspartic acid (d-MeAsp), and l-arginine (l-Arg) (Fig. 1) (36). The possible pathway involved in its biosynthesis has been determined previously by labeled-precursor studies (27, 37). The structure of nodularin suggested that the mechanism of biosynthesis is via a nonribosomal mechanism (22). Nonribosomal peptides and polyketides are synthesized by large multienzyme complexes. These large modular proteins catalyze the activation, modification, and condensation of specific amino acid or small chain carboxylic acid substrates. Nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) genes have been associated with the production of nodularin in strains of Nodularia (26).
FIG. 1.
Structure of the cyanobacterial hepatotoxins, nodularin and microcystin. General numbering of residues is indicated. Nodularin is a cyclic pentapeptide (molecular weight, 619). The l-Arg residue of nodularin may be replaced with a homoarginine residue (nodularin-Har) or a valine residue (motuporin). Microcystin is a cyclic heptapeptide with two variable amino acids, indicated by X and Y. The most common isoform is microcystin-LR, where X is l-Leu and Y is l-Arg (molecular weight, 994).
Microcystin-LR is a cyclic heptapeptide similar in structure to nodularin (Fig. 1). Microcystin-LR contains two additional amino acids, d-alanine (d-Ala) and l-leucine (l-Leu), while MeDhb is replaced with N-methyldehydroalanine (MeDha) (36). The gene cluster responsible for microcystin synthesis has been identified in Microcystis aeruginosa strains PCC7806 (50) and K-139 (33), and in Planktothrix agardhii (10). This gene cluster (mcy) is 55 kb in length and is made up of 10 open reading frames (ORFs) (Fig. 2). mcyA to mcyC encode five NRPS modules, mcyD encodes two type I PKS modules, and mcyE and mcyG encode hybrid NRPS-PKS modules. The genes mcyF, mcyH, mcyI, and mcyJ encode a racemase, an ABC transporter, d-3-phosphoglycerate dehydrogenase (d-3-PGDH), and O-methyltransferase (OM), respectively. In addition, the mcy cluster in P. agardhii also contains the gene mcyT, which encodes an additional distinct thioesterase domain (10).
FIG. 2.
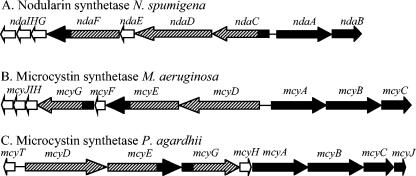
Structures of the gene clusters, nda and mcy, which encode polyketide synthase (hatched), nonribosomal peptide synthetase (black), and tailoring (white) enzymes involved in the biosynthesis of nodularin and microcystin, respectively. A) Structure of the gene cluster, nda, isolated from the nodularin-producing strain N. spumigena NSOR10 (this study). B) Structure of the gene cluster, mcy, isolated from two microcystin-producing strains of Microcystis (32, 33, 50). C) Structure of the mcy gene cluster isolated from the microcystin-producing strain P. agardhii CYA126/8 (10).
While nodularin synthesis is restricted to a single species, the hepatotoxin microcystin is produced by diverse genera of cyanobacteria, including the unicellular Microcystis, the filamentous Planktothrix, and the filamentous, heterocyst-forming Anabaena (3, 10, 28, 41, 50). Characterization of the mcy gene clusters from M. aeruginosa and P. agardhii has revealed that they are structurally distinct (Fig. 2) and are associated with transposases (10, 50). Distribution of the hepatotoxicity of cyanobacteria may occur via transposition of these large gene clusters between genera, as indicated by analysis of the mcy gene clusters and flanking sequences (10, 24, 50).
The aim of this study was to completely sequence and characterize the gene cluster required for nodularin synthesis and its flanking regions in the toxic strain N. spumigena NSOR10. The previously identified N. spumigena NRPS and PKS genes were used as a basis for this work (26). Following this, the proteins encoded within the putative nodularin synthetase (nda) gene cluster were characterized by comparative analysis with other biosynthetic enzymes, including microcystin synthetase. By using these data along with results of previous labeling studies by Moore et al. (27) and Rinehart et al. (37), a nodularin biosynthetic pathway was proposed. The second aim of this study was to compare and contrast the structures of the nda and mcy gene clusters in order to suggest a mechanism of acquisition and evolution via the deletion or insertion of two NRPS modules. This may have occurred through a natural combinatorial engineering event. A complete understanding of how this event has occurred in evolutionary history can provide invaluable information for the future combinatorial biosynthesis of NRPS and PKS systems. In addition, knowledge of the evolution of these biosynthetic pathways in cyanobacteria will have important implications in understanding the ecological roles of microcystin and nodularin, as well as their association with cyanobacterial bloom formation.
MATERIALS AND METHODS
DNA extraction and amplification.
The Nodularia strains used in this study were obtained from Commonwealth Scientific and Industrial Research Organisation Marine Laboratories (Hobart, Australia) and the Pasteur Culture Collection (PCC). Nodularia cultures were maintained in sterile ASM medium (38) supplemented with 1.5% NaCl. Cultures were grown at 25°C with 12-h light-dark cycling between approximately 16 and 0 μmol of photons · m−2 · s−1. Prior to genomic DNA extraction, Nodularia cultures were filtered through a 3.0-μm-pore-size filter (Millipore, Billerica, Mass.), and cells were washed twice with sterile ASM medium to remove contaminating heterotrophic bacteria. Total genomic DNA was extracted from filtered mid-growth-phase cultures by using a sodium dodecyl sulfate-lysozyme-based method as previously described (31).
Amplification of nodularin-associated NRPS and PKS sequences was performed with NRPS-specific primers NPF (TAT TTT GTG GTG GAG AAG CAC TA) and NPR (GGA ACT ATC TGA TAA TTA GAC) and PKS-specific primers MNKF (GTT CYT CYT CAY TRG TRG CG) and MNKR (CCY AAG AAC AAC WAY TCC ACA) as described previously (26). Nodularia strains were screened for the presence of ORFs flanking the putative nda gene cluster by PCR with primers NPSF3 (CTT ATC GAG GAG GTC GTG AAG) and HLIPR (CAG AAA GTC AGT ATT AGG). Amplification was performed in a 30-μl reaction mixture containing 1× PCR buffer (Fischer Biotech, Perth, Australia), 2.5 mM MgCl2, 130 μM deoxynucleoside triphosphate mix (Fischer Biotech), and 0.5 U of Taq F1 DNA polymerase (Fischer Biotech). PCR was performed with 1 μl of template DNA at a concentration of approximately 100 ng · μl−1. Specific forward and reverse oligonucleotide primers (synthesized by Sigma or GenSet Oligos, Lismore, Australia) were added to a final concentration of 0.3 μM. Thermal cycling was performed in a PCR Sprint temperature cycling system machine (Hybaid Limited, Middlesex, United Kingdom) or a GeneAmp PCR system 2400 thermocycler (Perkin-Elmer Corporation). The initial denaturation step at 94°C for 2 min was followed by 30 cycles of DNA denaturation at 94°C for 5 s, primer annealing for 10 s at the corresponding annealing temperature, and DNA strand extension at 72°C for the appropriate extension time and then by a final extension step at 72°C for 7 min.
Amplification of unknown fragments.
The amplification of the nda gene cluster was performed by using adaptor-mediated, inverse, and “hemidegenerate” PCR methods. Overlapping DNA fragments of up to 5 kb in length were generated by using these approaches, and both DNA strands were sequenced by using primers designed for every 400 to 500 bp.
The adaptor-mediated PCR method was modified from that described by Siebert et al. (43). Short adaptor DNA was ligated to digested genomic DNA, and a specific genomic outward-facing primer was then used with an adaptor primer to amplify a region of the genome. Twenty picomoles of T7 adaptor (Fig. 3) was added to each reaction mixture, containing 1 μg of genomic DNA, 10 U of blunt-ended restriction enzyme, and 5 U of T4 ligase (Promega) in 1× One Phor All buffer (Amersham/Pharmacia), and the one-step digestion-and-ligation reaction mixture was incubated at room temperature overnight.
FIG. 3.
Sequence of the T7 adaptors, utilized for adaptor-mediated PCR. Positions of the primers used for PCR (T7Pr1) and sequencing (T7Pr2) are indicated.
The single-stranded end of the adaptor was blocked with dideoxynucleotides (ddNTP) and subsequent dephosphorylation to prevent extension of the short arm of the adaptor. The blocking reaction was performed in a solution containing 1× PCR buffer (Fischer Biotech), 4 mM MgCl2, and 12.5 μM ddNTP with 1 U of Taq DNA polymerase (Fischer Biotech). Thermal cycling was performed in a PCR Sprint temperature cycling system machine (Hybaid Limited) with an initial step at 70°C for 15 min followed by 10 cycles of DNA denaturation at 95°C for 10 s, DNA reannealing at 40°C for 1 min, and extension of the strand with ddNTP at 70°C for 1 min. Following the PCR cycles, the reaction mixture was incubated with 1 U of shrimp alkaline phosphatase (Boehringer Mannheim, Gottingen, Germany) at 37°C for 20 min, and the enzyme was heat inactivated at 85°C for 5 min.
A PCR mixture containing 1 to 2 μl of adaptor-ligated DNA, 10 pmol of adaptor primer (Fig. 3), and 10 pmol of genome-specific oligonucleotide primers was set up. PCR cycling was performed as described previously, with DNA strand extension at 72°C for 5 min. The primer annealing temperature was decreased by 1°C at each cycle, from 65 to 55°C, followed by primer annealing at 55°C for a further 25 cycles.
Flanking regions were also amplified by using the inverse PCR approach, requiring two specific outward-facing primers as described by Pang and Knecht (35). Genomic DNA was partially digested with between 0.5 and 2 U of the 4-bp cutter Sau3A I, and individual DNA fragments were self-religated at a concentration of 5 ng · μl−1. The ligated DNA was used as a template in PCR with Nodularia-specific primers facing outwards from the known gene sequences.
When possible, a rational hemidegenerate PCR approach was used to take advantage of the modular structure of NRPS and PKS gene clusters. In this case a specific PCR primer was used with a degenerate PCR primer designed to PKS ketosynthase domains DKF (GTG CCG GTN CCR TGN GYY TC) and DKR (GCG ATG GAY CCN CAR CAR MG) or NRPS adenylation domains (A domains) MTF2 (GCN GGY GGY GCN TAY GTN CC) and MTR2 (CCN CGD ATY TTN ACY TG) (26, 30).
Sequencing, analysis, and alignments.
Automated sequencing was performed with the PRISM Big Dye cycle sequencing system and a model 373 sequencer (Applied Biosystems Inc., Foster City, Calif.). Sequence data were analyzed by using the Applied Biosystems Auto-Assembler computer program.
ORFs were identified and translated and homology was inferred by using programs accessed through BioManager at the Australian National Genome Information Service, Sydney, Australia, and through the Basic Local Alignment Search Tool at the National Center for Biotechnology Information (1).
Protein sequences were aligned by using the program Pileup from the Genetics Computer Group (GCG) (12), accessed through BioManager at the Australian National Genome Information Service, and the ClustalX multiple-sequence alignment tool (version 1.8) (48). Divergence between amino acid sequences was calculated by using a PAM-Dayhoff matrix (42). Gene sequences used in this study were aligned by using the programs Pileup from GCG and the Clustal W multiple-sequence alignment tool (49). Manual alignment of the output was also performed. Evolutionary relationships and genetic distances were determined by the alignment of 1,236 bp from within the 16S rRNA gene sequence and 1,623 bp from within the ABC transporter gene sequence. Genetic distances (D) between strains were calculated by using the following formula, described by Jukes and Cantor (20): D = −3/4[ln(1 − 4/3d)], where d is the level of sequence dissimilarity. Phylogenetic trees were constructed with the ClustalX program, by the neighbor-joining method of Saito and Nei (40). Statistical confidence values were determined by performing 1,000 bootstrap trials (11).
Nucleotide sequence accession number.
The nucleotide sequence of the nodularin biosynthetic gene cluster described in this paper has been submitted to GenBank under accession number AY210783.
RESULTS
Description of the nodularin synthetase (nda) gene cluster and its flanking regions.
The NRPS and PKS gene fragments previously associated with nodularin synthesis were identified in N. spumigena NSOR10 (26), and regions flanking these NRPS and PKS fragments were amplified in order to completely sequence the 47-kb nda gene cluster. Knowledge of the modular structure of NRPS and PKS genes allowed a logical approach to the design of a hemidegenerate PCR by using N. spumigena NSOR10 genome-specific primers in a PCR with previously described NRPS and PKS degenerate primers (26, 30). This approach, along with both adaptor-mediated PCR, which requires a single specific primer and an adaptor primer (43), and inverse PCR, which requires two specific outward-facing primers (35), was successful in generating a series of overlapping fragments of unknown sequence up to 5 kb in size. The fragments generated by PCR were designed with an overlap of at least 100 bp, which were then bidirectionally sequenced by design of primers every 400 to 500 bp, generating 55 kb of sequence including the 47 kb nda gene cluster and its flanking regions.
Sequence analysis indicates that nda includes two large operons, upstream and downstream of a bidirectional promoter (Fig. 2). Downstream of the promoter, three NRPS modules are encoded within the two ORFs ndaA and ndaB. Transcribed in the opposite direction, upstream of the promoter, are three ORFs, ndaCDF, which encode NRPS and PKS modules, along with four other ORFs, ndaE and ndaGHI, which encode tailoring enzymes.
A 4.5-kb region downstream of the nda gene cluster was sequenced. Five ORFs were identified; the first three, ORF1 to -3, were transcribed in the same direction as ndaAB, while ORF4 and -5 were transcribed on the opposing strand. Upstream of ndaI, a 2.5-kb region was sequenced and characterized. The three ORFs, ORF6 to -8 were found to be transcribed in the opposite direction from ndaI.
Putative functions of enzymes encoded by the nda gene cluster.
The protein sequences predicted from analysis of the nda gene cluster were compared to those of homologous proteins in order to propose functions and catalytic activities (Table 1). The ORFs ndaCDEF encode enzymes that are responsible for the biosynthesis of Adda, based on protein homology to microcystin biosynthetic enzymes, while ndaFGHAB encode the enzymes responsible for peptide synthesis, cyclization, and transport.
TABLE 1.
Deduced functions of encoded proteins in the nda gene cluster of N. spumigena NSOR10
| Protein | Amino acids | Proposed function and domain similarity detecteda |
|---|---|---|
| NdaA | 2,607 | NRPS |
| Module 1 | AThr, NM, PCP | |
| Module 2 | C, Ad-MeAsp, PCP | |
| NdaB | 1,299 | |
| Module 1 | C, AArg, PCP, TE | |
| NdaC | 2,640 | NRPS/PKS |
| Module 1 | APhenylacetate, PCP | |
| Module 2 | KS, ATAcetate, CM, KR, ACP, | |
| NdaD | 3,872 | PKS |
| Module 1 | KS, ATAcetate, CM, DH, KR, ACP | |
| Module 2 | KS, ATAcetate, DH, KR, ACP, | |
| NdaE | 927 | OM |
| NdaF | 3,475 | PKS/NRPS |
| Module 1 | KS, ATAcetate, CM, ACP, AMT | |
| Module 2 | C, Ad-Glu, PCP, C, | |
| NdaG | 235 | Racemase |
| NdaH | 341 | D-3-PGDH |
| NdaI | 601 | ABC transporter |
The domains of the NRPS and PKS modules are listed from left to right in the N-C-terminal order in which they occur. NM, N-methyltransferase; PCP, peptidyl carrier protein; TE, thioesterase; KS, ketosynthase; AT, acyltransferase; CM, C-methyltransferase; KR, ketoreductase; ACP, acyl carrier protein; DH, dehydratase; AMT, aminotransferase. The A domain and AT domain specificities are given as subscripts.
The first ORF upstream of the promoter encodes the putative hybrid NRPS-PKS megasynthase NdaC, with 70% identity to McyG of microcystin synthetase. The A domain of the NdaC NRPS module was analyzed, and the residues lining the substrate-binding pocket were compared to other characterized A domains in order to predict the substrate amino acid (Table 2). Based on the identity to the binding pocket of McyG, NdaC is predicted to activate the starter unit as phenylalanine or phenylacetate. Interestingly, the Asp235 residue, which is found in most binding pockets to date, is replaced by a Val residue in the A domains of both NdaC and McyG. The PKS module at the C terminus of NdaC contains the same order of catalytic domains as the McyG PKS module, which is required for polyketide extension of the phenylacetate unit. Analysis of this region in the acyltransferase domains of nodularin synthetase indicated that these PKS modules are responsible for the recruitment of the malonyl group from malonyl coenzyme A and its subsequent modification (16, 19).
TABLE 2.
Analysis of the residues of the A domain-binding pockets that confer substrate specificity in nodularin synthetase and related A domains
| A domain | Organism | Substrate | Binding pocket | Reference |
|---|---|---|---|---|
| NdaC | N. spumigena | Phenylacetate | V G I W V A A S | This study |
| McyG | M. aeruginosa | Phenylacetate | V G I W V A A S | 50 |
| BarE | L. majuscula | Trichloro-isovalerate | V G I I V G G T | 8 |
| NdaFA1 | N. spumigena | d-Glu | D P R H S G V V | This study |
| McyEA1 | M. aeruginosa | d-Glu | D P R H S G V V | 50 |
| McyEA1 | P. agardhii | d-Glu | D P R H S G V V | 10 |
| NdaAA1 | N. spumigena | l-Thr | D F W N I G M V | This study |
| FxbCA2 | M. smegmatis | l-Thr | D F W N I G M V | 7 |
| NdaAA2 | N. spumigena | d-MeAsp | D A R H V G I F | This study |
| McyB2 | M. aeruginosa | d-MeAsp | D A R T V G I F | 50 |
| McyB2 | P. agardhii | d-MeAsp | D P R H L S I F | 10 |
| NdaBA1 | N. spumigena | l-Arg | D V W N F G F V | This study |
| McyCA1 | M. aeruginosa | l-Arg | D V W T I G A V | 50 |
| McyCA1 | P. agardhii | l-Arg | D P W V F G L V | 10 |
| FenAA2 | B. subtilis | l-Glu | D A W H F G S V | 7 |
| EntFA1 | E. coli | l-Ser | D V W H F S L V | 7 |
| FxbCA2 | M. smegmatis | l-Thr | D F W N I G M V | 7 |
| NdaBA1 | N. harveyana | l-Har | D V W S F G F V | This study |
The NdaD protein, encoded directly upstream of ndaC, is 67% identical to McyD of M. aeruginosa. This protein is made up of two PKS modules that may catalyze two further polyketide extension steps in a manner identical to that of McyD. The ndaE ORF encodes a small protein with 81% identity to the OM, McyJ, of M. aeruginosa. The NdaE protein sequence contains conserved motifs characteristic of methyltransferases, including the glycine-rich motif I, which is required for S-adenosyl-l-methionine binding (21). Upstream, the ndaF ORF encodes a hybrid PKS-NRPS enzyme complex. The protein sequence is 74% identical to the sequence of McyE, and both proteins have the same order of catalytic domains. The PKS module of NdaF catalyzes the final round of polyketide extension and the biosynthesis of Adda completed via the aminotransferase domain.
The NdaF NRPS module contains the same catalytic domains as the NRPS module of McyE. The specificity-conferring recognition sequences of the A domains in NdaF and McyE from M. aeruginosa PCC7806 and P. agardhii CYA126/8 are all identical and are probably responsible for the activation of d-Glu. Epimerization of l-Glu may be catalyzed by NdaG, which is encoded by ndaG, directly upstream of ndaF. This 26,538-Da protein shows 77% identity to McyF, which may be a Glu racemase (32), although the structures of the active sites of NdaG and McyF appear to be distinct from those of other bacterial l-Glu racemases (14). More recent studies have indicated that McyF may also be an l-Asp epimerase (44).
NdaH is 71 and 44% identical to McyI and a putative d-3-PGDH identified in the hyperthermophilic archaeon Methanopyrus kandleri, respectively (46). In Escherichia coli, d-3-PGDH enzymes are responsible for the first step in the pathway for Ser biosynthesis, via the oxidation of 3-phosphoglycerate to phosphohydroxypyruvate (15). The role of the d-3-PGDH homologs in microcystin biosynthesis is unknown; however, it has been proposed that McyI catalyzes the formation of MeDha from activated MeSer or the biosynthesis of Ser (50). NdaH is likely to catalyze the conversion of MeThr to MeDhb, requiring NADH as a cofactor. Clear differences in the specificities of McyI for MeSer and NdaH for MeThr could not be detected through their alignment; however, a region C terminal of the putative NADH binding site was distinct between the two proteins, indicating that this may be a region of substrate specificity (data not shown).
Located upstream of ndaH and transcribed in the same direction, ndaI encodes a 69,010-Da protein which is 73% identical to McyH, a putative ABC transporter, and 69% identical to NosG of nostopeptolide synthetase from Nostoc sp. strain GSV224, also a putative ABC transporter associated with a nonribosomal biosynthetic gene cluster (17).
The first ORF downstream of the promoter region ndaA encodes two NRPS modules. The first NRPS module (NdaA-M1) is 50% identical to the first NRPS module of McyA. The specificity-conferring recognition sequence of the first A domain of NdaA (NdaA-A1) was identical to the Thr-specific consensus sequence (Table 2) (47). The NdaA-A1 domain is likely to activate Thr as the precursor of MeDhb. The second NRPS module is 67% identical to the second NRPS module of McyB of M. aeruginosa PCC7806. The specificity-conferring region of the second A domain of NdaA (NdaA-A2) is most similar to the D-MeAsp activation domain of McyB-A2, although it shows little homology to other A domain active sites. Downstream of ndaA, ndaB encodes a protein with 71% similarity to the NRPS module McyC, which catalyzes the final peptide extension reaction, cyclization, and release of the peptide. Analysis of the recognition sequence of NdaB revealed some similarity to the McyC A domain of P. agardhii; however, there was little homology to any other A domains. The structure of nodularin suggests that this A domain activates l-Arg. A recently characterized isoform of nodularin contains a Har residue at position 5 of nodularin and is produced by N. harveyana PCC7804 (2, 39). Comparison of the specificity-conferring regions of these strains identified that the sequence reflected the change in specificity, with the Asn of the NSOR10 active site replaced with a Ser residue. The longer Har structure more appropriately fits within the structure of the binding pocket due to the replacement of Asn with Ser.
ORFs flanking nodularin synthetase (nda) genes.
The ORFs flanking the nda gene cluster were sequenced and analyzed. Downstream of ndaB, ORF1 encodes a small, 19,729-Da protein with similarity to a group of transposases. The ORF1 protein lacks 160 amino acids of the N-terminal region that are present in other homologous transposases, and it is unknown whether the ORF1 transposase would be functional in this form. Interestingly, the mcy gene cluster also has an ORF, uma4, downstream of the NRPS gene mcyC in M. aeruginosa PCC7806 and downstream of the OM gene mcyJ in P. agardhii CYA126/8, both of which have homology to transposases. While they are associated with similar gene clusters, the transposases identified in Microcystis, Planktothrix, and Nodularia are members of three distinct families of transposases (10, 50).
ORF2 encodes a 63-amino-acid protein which is homologous to a family of high-light-inducible chlorophyll-binding proteins, Hli, identified in cyanobacteria. ORF3 to -5 encode a β-carotene ketolase homolog, a hypothetical protein, and a heat shock transcriptional repressor protein homolog, respectively. Upstream of ndaI, ORF6 encodes a protein with similarity to 3-oxoacyl-[acyl-carrier-protein] synthase proteins, which are involved in bacterial fatty acid biosynthesis. ORF7 and ORF8 encode two hypothetical proteins.
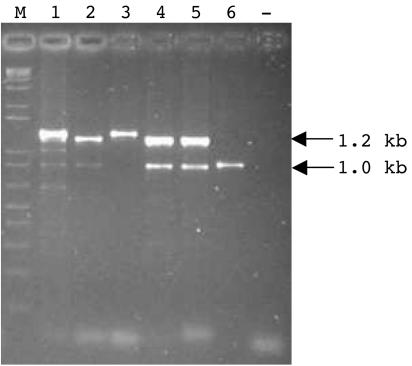
In order to determine whether ORF1 and ORF2 were associated with nodularin production, the toxic N. spumigena strains BY1, NSGL02A10, HEM, and L575 and the toxic N. harveyana strain PCC7804 were screened by specific PCR for the distribution of the ORFs downstream of ndaAB. This specific PCR amplified the expected 1.2-kb fragment from ndaB to ORF2 in the toxigenic N. spumigena strains NSOR10, BY1, NSGL02A10, HEM, and L575 (Fig. 4). ORF1 and ORF2 were not identified in nontoxic Nodularia strains (data not shown). An additional 1-kb band was amplified from the N. spumigena strains HEM and L575. N. harveyana strain PCC7804 did not produce the expected 1.2-kb band; however, it did yield a smaller, 1-kb fragment. Sequencing of this region found that ORF1 was absent from this strain.
FIG. 4.
Amplification of the open reading frames downstream of the nodularin synthetase (nda) gene cluster from toxic Nodularia strains. PCR with primers NPSF3 and HLIPR generated 1.2- and/or 1.0-kb fragments. Lanes 1 to 6 correspond to PCR-amplified genes from the toxic strains N. spumigena NSOR10 (lane 1), N. spumigena BY1 (lane 2), N. spumigena NSGL02A10 (lane 3), N. spumigena HEM (lane 4), N. spumigena L575 (lane 5), and N. harveyana PCC7804 (lane 6). —, no-DNA control. Lane M contains 150 ng of 1-kb DNA ladder marker (Invitrogen).
Characterization of the C domain of NdaA may indicate mechanism of nodularin evolution.
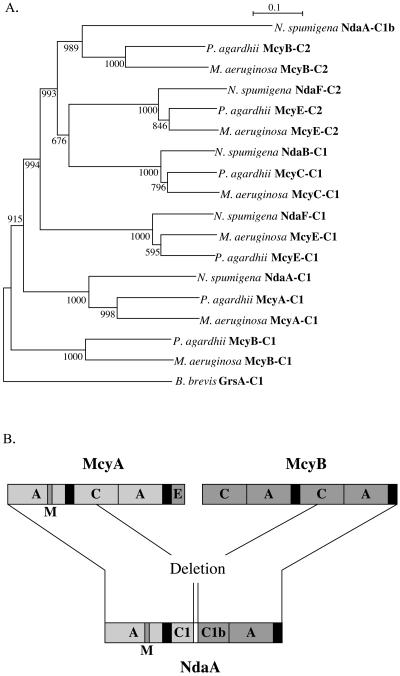
A duplication of conserved regions C4, C5, and C6 was detected in the condensation domain (C domain) of the NRPS module of NdaA (NdaA-C). To analyze this duplication, the protein sequence of NdaA-C was divided into two sections, NdaA-C1 and the duplicated region (NdaA-C1b). These protein sequences were then aligned with the other C domains of nodularin synthetase and microcystin synthetase from M. aeruginosa. The alignment shows a high degree of homology between McyA-C and NdaA-C1 (54% identical over 290 amino acids), while C4 to C7 of the duplication region NdaA-C1b and McyB-C2 are highly homologous (62% identical over 167 amino acids). The unusual structure of the NdaA-C domain may have evolved from the fusion of the McyA-C domain and the McyB-C2 domain following deletion of two NRPS modules (Fig. 5). Analysis of the conserved motifs C4 to C7 of both sections of NdaA-C1 indicated that C4 and C5 of NdaA-C1b are highly divergent, while C7 of NdaA-C1 is not present. Alignment of the C domains indicated that the NdaA-C1 C6 motif and the NdaA-C1b C4 motif were separated by a putative linker region that had no homology to other C domains.
FIG. 5.
Characterization of the NdaA condensation domain. A) Phylogenetic analysis of the amino acid sequences of the condensation (C) domains from nodularin synthetase of N. spumigena NSOR10, microcystin synthetases of M. aeruginosa PCC7806 and P. agardhii CYA126/8, and gramicidin S synthetase (GrsA) of Bacillus brevis ATCC 9999. The sequence alignment and phylogenetic tree were created by using Pileup and ClustalX from GCG (40, 48). The scale bar represents one substitution per 10 amino acids. Significant bootstrap values calculated from 1,000 bootstrap trees are indicated at the nodes (11). B) Diagram of the proposed deletion of the second and first NRPS modules of McyA and McyB, respectively. A, adenylation; C, condensation; E, epimerization; M, N-methyltransferase. PCP domains are shown in black; the NdaA condensation domain linker region is shown in white.
In order to confirm the origin of NdaA-C1 and NdaA-C1b, the protein sequences of these domains were aligned with the C domain protein sequences from nodularin synthetase of N. spumigena NSOR10 and microcystin synthetase of M. aeruginosa PCC7806 and P. agardhii CYA126/8 (10, 50). The resulting phylogenetic tree showed that each of the nodularin C domains was most similar to its catalytic equivalent C domain in microcystin (Fig. 5). As expected, a McyB-C1 homologue was not identified in nodularin synthetase, since the corresponding amino acid activated by this module in microcystin is absent from nodularin. NdaA-C1 was shown to be most similar to the C domain of McyA (McyA-C1), while the NdaA-C1b region was most closely related to McyB-C1 of both M. aeruginosa and P. agardhii. The results also showed that the protein sequence of McyA-C1 from M. aeruginosa was more similar to that of McyA-C1 from P. agardhii than to that of NdaA-C1 from N. spumigena. This phylogenetic relationship was seen for each of the C domains analyzed.
Phylogenetic comparisons between nodularin and microcystin synthetases.
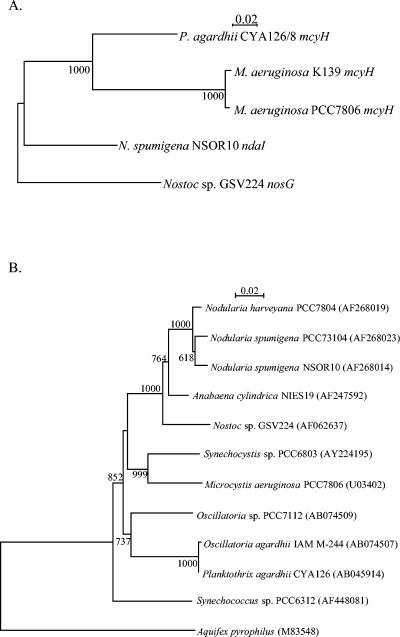
In order to further infer genetic relationships between the mcy and nda gene clusters, phylogenetic analysis of the genes that encode the putative ABC transporters, mcyH and ndaI, was performed, with the nosG ABC transporter from Nostoc sp. strain GSV224 used as an outgroup. The ABC transporter gene was chosen to analyze the relationship between these clusters because it is associated with each of the nda and mcy clusters characterized to date. The results of the phylogenetic analysis indicated that the mcyH genes of M. aeruginosa and P. agardhii are more similar to each other than to the ndaI sequence of N. spumigena (Fig. 6). These species represent distinct orders within the cyanobacteria. The evolutionary distances between these genera were also determined by 16S ribosomal DNA (rDNA) nucleotide sequence analysis (Fig. 6). Phylogenetic analysis of the 16S rRNA gene indicated that the evolutionary distances between the species N. spumigena, P. agardhii, and M. aeruginosa were similar (Fig. 6).
FIG. 6.
Evolution of the hepatotoxicity in cyanobacterial genera by using phylogenetic characterization of the ABC transporter and 16S rRNA genes. All sequences were obtained from GenBank, and the accession numbers are given in parentheses. The sequence alignment and phylogenetic tree were created by using Pileup and ClustalX from GCG (40, 48). The scale bar represents two substitutions per 100 nucleotides. Significant bootstrap values calculated from 1,000 bootstrap trees are indicated at the nodes (11). A) Phylogenetic analysis of the putative ABC transporter gene. The ndaI gene from N. spumigena NSOR10 was aligned with the mcyH sequences from M. aeruginosa strains PCC7806 and K-139 and from P. agardhii CYA126/8 and with the putative ABC transporter nosG sequence from Nostoc sp. strain GSV224. B) Phylogenetic analysis of the 16S rRNA gene.
DISCUSSION
Comparative analysis of the nodularin and microcystin biosynthetic pathways.
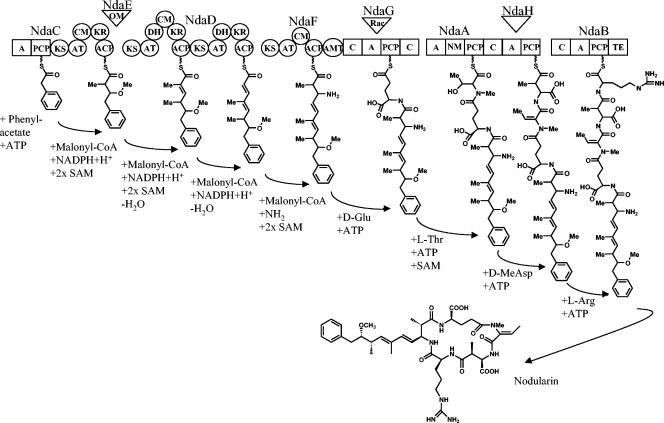
Previously, labeling studies have shown that nodularin is biosynthesized from a mixed polyketide-peptide pathway (27, 37). This study has identified the complete hybrid NRPS-PKS gene cluster putatively involved in the biosynthesis of nodularin. Previous knockout studies have shown that the mcy gene cluster is required for microcystin biosynthesis in M. aeruginosa and P. agardhii (10, 50). The high degree of sequence homology between the mcy and nda clusters clearly indicates that the nda cluster is responsible for nodularin biosynthesis. Using results of the previous labeling studies (27, 37) and the genetic analysis of mcy (50) it has been possible to predict the enzymes responsible for each reaction involved in nodularin biosynthesis (Fig. 7).
FIG. 7.
Model of the nodularin biosynthetic pathway catalyzed by proteins encoded by ndaC to -F. The squares represent the NRPS adenylation (A), peptidyl carrier protein (PCP), and condensation (C) domains. The circles represent the PKS, ketosynthase (KS), acyltransferase (AT), C-methyltransferase (CM), dehydratase (DH), ketoreductase (KR), and acyl carrier protein (ACP) domains. Condensation reactions are indicated in red. The O-methyltransferase (OM) NdaE catalyzes the transfer of the methyl group to the C9 hydroxyl. The racemase NdaG catalyzes the epimerization of the Glu residue. The aminotransferase (AMT) domain catalyzes the transfer of the amine group to Adda. Peptide condensation reactions are catalyzed by NdaA and NdaB. The putative dehydrogenase, NdaH, catalyzes the formation of the MeDhb residue following peptide bond formation. The thioesterase (TE) domain catalyzes the cyclization and release of the peptide.
In this study, a combination of PCR methods was employed to amplify and sequence the nda gene cluster. Fragments of the nda gene cluster were amplified by using adaptor-mediated PCR, inverse PCR, and a hemidegenerate PCR approach, which utilized the modular structure of the NRPS and PKS genes to amplify DNA by using a specific primer and a degenerate primer. There were no specific problems associated with this method of amplification and sequencing of the gene cluster, and the quality of the sequence data generated was quality comparable to that for other gene clusters sequenced from cosmid libraries. These techniques may be useful for future sequencing of large NRPS-PKS gene clusters in such cases where construction of a suitable, complete cosmid library is problematic, for example, from unculturable organisms present in environmental samples.
The structure of the cyanobacterial toxin nodularin is similar to that of the toxin microcystin (Fig. 1). It is therefore interesting to note the differences in the structures of the nda gene cluster sequenced in this study and the mcy gene clusters sequenced from M. aeruginosa strains and P. agardhii CYA126/8 (10, 33, 50). The nda genes are located upstream and downstream of a bidirectional promoter, similar to the case for the mcy cluster of M. aeruginosa; however, the order of proteins encoded by the nda gene cluster is more colinear, generally representing the order of each catalytic step required for the biosynthesis of nodularin (Fig. 2). The mcy gene cluster encodes an additional two NRPS modules, responsible for the activation of the amino acids d-Ala and l-Leu, that are not present in the structure of nodularin. Four NRPS modules of nodularin synthetase were compared to the modules of microcystin synthetase. The active sites of the d-Glu and d-MeAsp A domains are identical in both nodularin and microcystin synthetases. The active site of NdaA clearly activates and methylates l-Thr as a precursor of MeDhb and has little homology to the active site of McyA, which activates and methylates l-Ser. Conversion of MeThr to MeDhb probably then occurs via the NdaH enzyme via a mechanism similar to the conversion of MeSer to MeDha.
Interestingly, the substrate-binding pockets of the terminal activation domains of nodularin and microcystin synthetases show only 50% identity. This A domain in nodularin is likely to be specific for the activation of l-Arg. Many isoforms of microcystin have been shown to have variations at this position (6, 37, 45). Perhaps the relaxed specificity of this A domain in microcystin synthetase is reflected in the sequence of the active site of the A domain. P. agardhii CYA126/8 also produces the l-Arg isoform; however, the substrate-binding pocket is distinct from those of both NdaB and McyC of M. aeruginosa (Table 2). Further expression and activation studies are required to determine the degree of relaxed specificity of the McyC and NdaB A domains. The A domain of NdaB of N. harveyana PCC7804, which produces the nodularin-Har isoform (2, 39), was analyzed, and the active site was compared with that of NdaB of N. spumigena NSOR10. Indeed, the sequence reflects the change in specificity with the replacement of Asn with Ser in the binding pocket of N. harveyana PCC7804, clearly allowing a more compatible fit for the longer Har residue.
Evolution of hepatotoxin biosynthesis in cyanobacteria.
While nodularin production is restricted to Nodularia, the production of microcystin is not consistently present in distinct evolutionary clades of cyanobacterial species, in particular M. aeruginosa (51). It has recently been proposed that the transfer of large NRPS genes clusters is responsible for the presence of the highly homologous gene clusters in different strains of Bacillus subtilis (52). This indicates that the transfer of large fragments of DNA is more common than previously thought. The transposases associated with the mcy gene clusters in M. aeruginosa and P. agardhii and the nda gene cluster in N. spumigena may have mediated transfer of these gene clusters between cyanobacterial genera. Previous studies of mcy in Microcystis and Planktothrix proposed a recent insertion of the second and first NRPS modules of mcyA and mcyB, respectively, into the nda gene cluster (10, 24). This indicated that mcy evolved from the nda gene cluster. Characterization of the nda gene cluster from N. spumigena NSOR10 in this study has provided new data which may suggest an alternative hypothesis for hepatotoxin evolution in cyanobacteria.
A duplicated region, separated by a linker region with little homology to other proteins, was identified within the NdaA C domain. We hypothesized that the duplication within the NdaA C domain and the putative linker region are remnants of a deletion event which had occurred during evolution (Fig. 5). To confirm this hypothesis, sequence alignments and phylogenetic analysis found that NdaA-C1 was most similar to the C domain of McyA (McyA-C1), while the NdaA-C1b region was most closely related to McyB-C1 of both M. aeruginosa and P. agardhii. Thus, we propose that the structure of nodularin synthetase has evolved from microcystin synthetase via the deletion of the second and first NRPS modules of McyA and McyB, respectively. This differs from previous hypotheses that microcystin synthetase evolved from nodularin synthetase (10, 24).
To further characterize evolution of hepatotoxin biosynthesis in cyanobacteria, phylogenetic analysis of ABC transporter genes, ndaI and mcyH, was performed, and the results were compared with 16S rDNA phylogeny. Comparative analysis of the branch lengths observed in the 16S rDNA and ABC transporter gene phylogenies may suggest that the nda lineage diverged from the mcy lineage well before the transfer of the mcy gene cluster between P. agardhii CYA126/8 and M. aeruginosa PCC7806. This result supports previous hypotheses that the mcy gene cluster was transferred between strains via a transposase (24, 51); however, additional phylogenetic analysis using a larger group of sequences is required to strengthen this theory.
From these results, we propose that hepatotoxins in cyanobacteria most likely originated as microcystin-LR, the most common isoform of microcystin. The original biosynthetic gene cluster may have been similar in structure to the nda gene cluster from N. spumigena NSOR10. An MeDhb microcystin-LR isoform, the most likely direct nda ancestor, may have been inserted within the genome of Nodularia via transposition. During or following insertion of this gene cluster into the genome of Nodularia, a deletion event may have occurred, resulting in the loss of the two NRPS modules. We propose that the wide distribution of this gene cluster across the cyanobacteria and the 60 isoforms of microcystin that have been reported to date (9, 37, 45) may be attributed to transposition of the gene cluster followed by further genetic shuffling, mutagenesis, deletion, and recombination events. This is exemplified by the distinct differences present in the structures of the mcy gene clusters of M. aeruginosa and P. agardhii. Further characterization of the gene clusters, their associated transposases, and the extent of recombination and shuffling in these strains is required to more accurately describe the evolution of hepatotoxigenicity in cyanobacterial genera.
Unlike microcystin biosynthesis, nodularin biosynthesis is restricted to strains of N. spumigena and one strain of N. harveyana. Since nodularin biosynthesis has been identified in all strains of N. spumigena, it is likely that the transposition event occurred early in the delineation of the species N. spumigena. The toxic strain N. harveyana PCC7804 is phylogenetically distinct from the toxic species N. spumigena, and the toxigenicity of this strain cannot be easily explained (25). Nodularin biosynthesis in this strain may have resulted from a separate transposition-acquisition event, leading to convergent phenotypes. A putative transposase gene could not be identified downstream of ndaB in N. harveyana PCC7804; thus, the mechanism of transfer of this gene cluster is unclear.
This study reports for the first time the complete sequence and characterization of the nda gene cluster in the bloom-forming cyanobacterium N. spumigena. The sequence of the nda gene cluster will be useful for future genetic and biochemical studies into the biosynthesis of nodularin and its regulation, which will provide a strong foundation for understanding biosynthesis of toxins in blooms of N. spumigena and related cyanobacterial species. A new hypothesis describing the evolution of nodularin synthetase from microcystin synthetase has been proposed from analyses reported in this study. These results may provide important insight into mechanisms of natural evolution of nonribosomal biosynthetic clusters in cyanobacteria and how these mechanisms may be utilized for the directed evolution of such clusters for the rational design of novel metabolites.
Acknowledgments
The Australian Research Council (ARC) financially supported this work. M.C.M. is jointly funded by the ARC and the CRC for Water Quality and Treatment. B.A.N. is a fellow of the ARC.
We thank Elke Dittman for invaluable advice throughout this project and Bradley S. Moore for helpful discussions regarding the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beattie, K. A., K. Kaya, and G. A. Codd. 2000. The cyanobacterium Nodularia PCC7804, of freshwater origin, produces [l-Har2]nodularin. Phytochemistry 54:57-61. [DOI] [PubMed] [Google Scholar]
- 3.Beattie, K. A., K. Kaya, T. Sano, and G. A. Codd. 1998. Three dehydrobutyrine-containing microcystins from Nostoc. Phytochemistry 47:1289-1292. [DOI] [PubMed] [Google Scholar]
- 4.Carmichael, W. W. 1994. The toxins of cyanobacteria. Sci. Am. 270:74-86. [DOI] [PubMed] [Google Scholar]
- 5.Carmichael, W. W., J. T. Eschedor, G. M. Patterson, and R. E. Moore. 1988. Toxicity and partial structure of a hepatotoxic peptide produced by the cyanobacterium Nodularia spumigena Mertens emend. L575 from New Zealand. Appl. Environ. Microbiol. 54:2257-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castenholz, R. W. 2001. Phylum BX. Cyanobacteria. Oxygenic photosynthetic bacteria, p. 473-599. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 7.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Z., P. Flatt, W. Gerwick, V. Nguyen, C. Willis, and D. Sherman. 2002. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene 296:235. [DOI] [PubMed] [Google Scholar]
- 9.Chorus, I., and J. Bartram (ed.). 1999. Toxic cyanobacteria in water. A guide to their public health consequences, monitoring and management. E & FN Spon, London, United Kingdom.
- 10.Christiansen, G., J. Fastner, M. Erhard, T. Borner, and E. Dittmann. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 185:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:166-170. [DOI] [PubMed] [Google Scholar]
- 12.Feng, D. F., and R. F. Doolittle. 1987. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol. 25:351-360. [DOI] [PubMed] [Google Scholar]
- 13.Francis, G. 1878. Poisonous Australian lake. Nature 18:11-12. [Google Scholar]
- 14.Glavas, S., and M. E. Tanner. 2001. Active site residues of glutamate racemase. Biochemistry 40:6199-6204. [DOI] [PubMed] [Google Scholar]
- 15.Grant, G. A., S. J. Kim, X. L. Xu, and Z. Hu. 1999. The contribution of adjacent subunits to the active sites of d-3-phosphoglycerate dehydrogenase. J. Biol. Chem. 274:5357-5361. [DOI] [PubMed] [Google Scholar]
- 16.Haydock, S. F., J. F. Aparicio, I. Molnar, T. Schwecke, L. E. Khaw, A. Konig, A. F. Marsden, I. S. Galloway, J. Staunton, and P. F. Leadlay. 1995. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 374:246-248. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann, D., J. M. Hevel, R. E. Moore, and B. S. Moore. 2003. Sequence analysis and biochemical characterization of the nostopeptolide A biosynthetic gene cluster from Nostoc sp. GSV224. Gene 311:171-180. [DOI] [PubMed] [Google Scholar]
- 18.Honkanen, R. E., M. Dukelow, J. Zwiller, R. E. Moore, B. S. Khatra, and A. L. Boynton. 1991. Cyanobacterial nodularin is a potent inhibitor of type 1 and type 2A protein phosphatases. Mol. Pharmacol. 40:577-583. [PubMed] [Google Scholar]
- 19.Ikeda, H., T. Nonomiya, M. Usami, T. Ohta, and S. Omura. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 96:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York, N.Y.
- 21.Kagan, R. M., and S. Clarke. 1994. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 310:417-427. [DOI] [PubMed] [Google Scholar]
- 22.Kleinkauf, H., and H. Von Dohren. 1996. A nonribosomal system of peptide biosynthesis. Eur. J. Biochem. 236:335-351. [DOI] [PubMed] [Google Scholar]
- 23.Lehtimaki, J., C. Lyra, S. Suomalainen, P. Sundman, L. Rouhiainen, L. Paulin, M. Salkinoja-Salonen, and K. Sivonen. 2000. Characterization of Nodularia strains, cyanobacteria from brackish waters, by genotypic and phenotypic methods. Int. J. Syst. Evol. Microbiol. 50:1043-1053. [DOI] [PubMed] [Google Scholar]
- 24.Mikalsen, B., G. Boison, O. M. Skulberg, J. Fastner, W. Davies, T. M. Gabrielsen, K. Rudi, and K. S. Jakobsen. 2003. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 185:2774-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moffitt, M. C., S. I. Blackburn, and B. A. Neilan. 2001. rRNA sequences reflect the ecophysiology and define the toxic cyanobacteria of the genus Nodularia. Int. J. Syst. Evol. Microbiol. 51:505-512. [DOI] [PubMed] [Google Scholar]
- 26.Moffitt, M. C., and B. A. Neilan. 2001. On the presence of peptide synthetase and polyketide synthase genes in the cyanobacterial genus Nodularia. FEMS Microbiol. Lett. 196:207-214. [DOI] [PubMed] [Google Scholar]
- 27.Moore, R. E., J. L. Chen, B. S. Moore, G. M. L. Patterson, and W. W. Carmichael. 1991. Biosynthesis of microcystin-LR. Origin of the carbons in the Adda and Masp units. J. Am. Chem. Soc. 113:5083. [Google Scholar]
- 28.Namikoshi, M., M. Yuan, K. Sivonen, W. W. Carmichael, K. L. Rinehart, L. Rouhiainen, F. Sun, S. Brittain, and A. Otsuki. 1998. Seven new microcystins possessing two l-glutamic acid units, isolated from Anabaena sp. strain 186. Chem. Res. Toxicol. 11:143-149. [DOI] [PubMed] [Google Scholar]
- 29.Nehring, S. 1993. Mortality of dogs associated with a mass development of Nodularia spumigena (Cyanophyceae) in a brackish lake at the German North Sea coast. J. Plankton Res. 15:867-872. [Google Scholar]
- 30.Neilan, B. A., E. Dittmann, L. Rouhiainen, R. A. Bass, V. Schaub, K. Sivonen, and T. Borner. 1999. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 181:4089-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neilan, B. A., D. Jacobs, and A. E. Goodman. 1995. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 61:3875-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishizawa, T., M. Asayama, and M. Shirai. 2001. Cyclic heptapeptide microcystin biosynthesis requires the glutamate racemase gene. Microbiology 147:1235-1241. [DOI] [PubMed] [Google Scholar]
- 33.Nishizawa, T., A. Ueda, M. Asayama, K. Fujii, K. Harada, K. Ochi, and M. Shirai. 2000. Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J. Biochem (Tokyo) 127:779-789. [DOI] [PubMed] [Google Scholar]
- 34.Ohta, T., E. Sueoka, N. Iida, A. Komori, M. Suganuma, R. Nishiwaki, M. Tatematsu, S. J. Kim, W. W. Carmichael, and H. Fujiki. 1994. Nodularin, a potent inhibitor of protein phosphatases 1 and 2A, is a new environmental carcinogen in male F344 rat liver. Cancer Res. 54:6402-6406. [PubMed] [Google Scholar]
- 35.Pang, K. M., and D. A. Knecht. 1997. Partial inverse PCR: a technique for cloning flanking sequences. BioTechniques 22:1046-1048. [DOI] [PubMed] [Google Scholar]
- 36.Rinehart, K. L., K. Harada, M. Namikoshi, C. Chen, C. A. Harvis, M. H. G. Munro, J. W. Blunt, P. E. Mulligan, V. R. Beasley, A. M. Dahlem, and W. W. Carmicheal. 1988. Nodularin, microcystin, and the configuration of Adda. J. Am. Chem. Soc. 110:8557-8558. [Google Scholar]
- 37.Rinehart, K. L., M. Namikoshi, and B. W. Choi. 1994. Structure and biosynthesis of toxins from blue-green algae (cyanobacteria). J. Appl. Phycol. 6:159-176. [Google Scholar]
- 38.Rippka, R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol. 167:3-28. [DOI] [PubMed] [Google Scholar]
- 39.Saito, K., A. Konno, H. Ishii, H. Saito, F. Nishida, T. Abe, and C. Chen. 2001. Nodularin-Har: a new nodularin from Nodularia. J. Nat. Prod. 64:139-141. [DOI] [PubMed] [Google Scholar]
- 40.Saito, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 41.Sano, T., and K. Kaya. 1998. Two new (E)-2-amino-2-butenoic acid (Dhb)-containing microcystins isolated from Oscillatoria agardhii. Tetrahedron 54:463-470. [Google Scholar]
- 42.Schwartz, R. M., and M. O. Dayhoff. 1978. Matrices for detecting distant relationships, p. 353-358. In M. O. Dayhoff (ed.), Atlas of protein sequence and structure, vol. 5, suppl. 3. National Biomedical Research Foundation, Washington, D.C.
- 43.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sielaff, H., E. Dittmann, N. Tandeau De Marsac, C. Bouchier, H. Von Dohren, T. Borner, and T. Schwecke. 2003. The mcyF gene of the microcystin biosynthetic gene cluster from Microcystis aeruginosa encodes an aspartate racemase. Biochem. J. 373:909-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sivonen, K. 1996. Cyanobacterial toxins and toxin production. Phycologia 35:12-24. [Google Scholar]
- 46.Slesarev, A. I., K. V. Mezhevaya, K. S. Makarova, N. N. Polushin, O. V. Shcherbinina, V. V. Shakhova, G. I. Belova, L. Aravind, D. A. Natale, I. B. Rogozin, R. L. Tatusov, Y. I. Wolf, K. O. Stetter, A. G. Malykh, E. V. Koonin, and S. A. Kozyavkin. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA. 99:4644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tillett, D., E. Dittmann, M. Erhard, H. von Dohren, T. Borner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 51.Tillett, D., D. L. Parker, and B. A. Neilan. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ. Microbiol. 67:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuge, K., T. Akiyama, and M. Shoda. 2001. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]