Abstract
Background:
The aim of this study was to identify the Trichomonas vaginalis strains/haplotypes based on identifying their probable variations in asymptomatic patients referred to Tabriz health centers, northwestern Iran.
Methods:
Sampling was taken from 50-suspected women to T. vaginalis in northwestern Iran. The obtained samples were smeared and cultured. Fifty DNA samples were extracted, amplified and identified by nested polymerase chain reaction and PCR-RFLP of actin gene using two endonuclease enzymes: MseI and RsaI. To reconfirm, the amplicons of actin gene were directly sequenced in order to identify the strains/haplotypes.
Results:
PCR-RFLP patterns, sequencing and phylogenetic analyses revealed definitely the presence of the G (n=22; 73.4%) and E (n=8; 26.6%) strains. Multiple alignments findings of genotype G showed five haplotypes and two amino acid substitutions in codons 192 and 211 although, no remarkable unique haplotype was found in genotype E.
Conclusion:
The accurate identification of T. vaginalis strains based on discrimination of their unknown haplotypes particularly those which are impacted on protein translation should be considered in parasite status, drug resistance, mixed infection with HIV and monitoring of asymptomatic trichomoniasis in the region.
Keywords: Trichomonas vaginalis, Actin gene, Genotypes, Asymptomatic infection
Introduction
Trichomonas vaginalis, the etiologic agent of human trichomoniasis is an anaerobic flagellated protozoan parasite, which is presented a broad spectrum of clinical manifestations ranging from asymptomatic to severe cervicitis, urethritis and vulvovaginitis. Trichomoniasis enhances the risk of human immunodeficiency virus transmission, pelvic inflammatory disorder, infertility and adverse pregnancy outcome (1–5).
The global incidence of the parasite is more than 170 million cases, inclusive 2–8% in Iran, 25% in United States, 11–25% in Africa and 6.8% in India (6, 7).
The clinical presentations in symptomatic patients are principally included itching, leukorrhea and macular colpitis (4, 5). However, the asymptomatic women may also experience abdominal pain, irritation, and discomfort. Definitely, at least part of mentioned pathological signs in asymptomatic patients could be due to the genetic diversity of the parasite strains to express virulence factors, which increase cytoadherence, cytolysis and cell detaching (6). Hence, rapid accurate identification of virulent and non-virulent strains of T. vaginalis along with characterization of their unknown mutants could be genetically distinguished and evaluated by making use of well-known molecular analyses.
Determination of heterogeneity traits (transcription and/or translations levels) of T. vaginalis strains especially in who are carrier individuals and asymptomatic patients could be prognosed a real drawing of virulence/pathogenicity intensity, re/emergent strains and taxonomic status of parasite in the region (8, 9).
The early techniques including morphological and/or biochemical characters chiefly isoenzyme analysis were utilized for Trichomonas isolates (4, 5). However, a rapid well known method for detecting T. vaginalis isolates would be a very useful adjunct technique in indigenous regions where a range of asymptomatic to symptomatic trichomoniasis are circulating unambiguously.
The development of DNA-based techniques have been employed for demonstration of genetic diversity of T. vaginalis strains, include random amplified polymorphic DNA (RAPD), PCR–hybridization, multilocus sequence typing (MLST(, microsatellite (MS) genotyping, restriction fragment length polymorphism method (RFLP) however, each of them have their own specific difficulties in during experiment(10–14).
On the one hand, some studies have introduced the sequencing as a firm method able to detect the heterogeneity/homogeneity traits in both DNA (haplotype) and protein (amino acid) levels (15, 16).
The various genes used in molecular typing of T. vaginalis isolates, for example ITS1, HSP70, 18srRNA, and actin genes (12, 17–19). It is noteworthy that the actin gene as a semi-conserved marker can apply as an appropriate candidate in typing of T. vaginalis isolates based on designed profiles of RFLP method (19).
The aim of this study was to characterize the genotypes of T. vaginalis isolates based on RFLP and phylogenetic analyses of actin gene in asymptomatic women of northwestern Iran along with identification of their probable variations to get a better approach of parasite taxonomy, pathogenicity rate, monitoring and surveillance of asymptomatic trichomoniasis in the region.
Materials and Methods
Ethics Statement
All human participants were enrolled in conformity with informed consent, privacy and confidentiality of patients who were sampled and analyzed anonymously during study (No. 92-95).
Sampling and culturing
Fifty suspected women samples to trichomoniasis aged over 18 yr were collected from both vaginal infections and urine alluvium referred to Tabriz health centers, northwestern Iran from 2013–2014. Wet mounts examinations were done for all samples and all were cultured in 50 ml of Diamond broth medium at 37 °C under aerobic conditions. The pH of the medium was adjusted according to the requirement of the respective trichomonad.
DNA extraction and PCR-RFLP
The genomic DNA was extracted by the extraction kit (Takapouzist) from medium following the manufacturer’s instructions. The target of nested-PCR was chosen within the actin gene with outer (OPs) and inner (IPs) primers (19). The OPs used were TV8S (5′-TCTGGAATGGCTGAAGAAGACG-3′) and TV9R (5′-CAGGGTACATCGTATTGGTC-3′) and IPs used were TV10S (5′-CAGACACTCGGTTATCG-3′) and Tv11R (5′-CGGTGAACGATGGATG-3′). The size of the target was 1100 bp, which is only 28 bp shorter than the full length of the open reading frame of the actin gene. The volume was adjusted to 20μL with distilled water, and 10μL of DNA extract was added to each reaction mixture for the amplification with the outer primers with inner primers, the volume was adjusted to 20 μL with distilled water too, and 8 μL of amplified products was added to each reaction mixture. PCR amplification was performed in two stages in a thermocycler (19). The first stage consisted of ten cycles. Each cycle consisted of 30 s of denaturation at 94 °C, 30 s of annealing at 55 °C, and a 3 min extension at 72 °C. The first cycle was followed by 5 min of denaturation at 95 °C. The second stage consisted of 25 cycles with the same denaturation and annealing steps. The extension step was extended by 5 s per cycle. The last cycle was followed by a 7 min final extension at 72 °C. PCR products were subjected to electrophoresis in 2% agarose gel and were observed under ultraviolet light after staining for 15min with (0.5 g/mL) ethidium bromide. In order to conduct the PCR-RFLP method, all PCR amplicons were digested for 4 h at 37 °C with restriction enzymes RsaI and MesI using buffer recommended by the manufacturer (Fermentas, Vilnius, Lithuania) (19).
Endonuclease reaction of actin gene was performed in a volume of 30 μl containing 1 μl of RsaI (blind-end digestion) with cut site GT↓AC, 1 μl of MesI (sticky-end digestion) with cut site TT↓AA, 10 μl of PCR products, 3 μl of 10× buffers, and 16 μl of distilled water. Restriction fragments of amplicons were electrophorezed using a 3 % (w/v) agarose gel at 100 V for 60 min.
Sequencing and Phylogenetic analysis
To reconfirm the RFLP results, 30 amplified PCR products were directly sequenced by targeting actin gene in both directions using the inner primers by ABIPRISMTM 3130 Genetic Analyzer automated sequencer (Applied Bio-system, USA). Ambiguous heterozygous) sites were coded using the standard IUPAC codes for combinations of two or more bases. Contigs from all samples were aligned and edited in consensus positions compared to GenBank sequences of all regional species in order to homology of novel haplotypes with the using Sequencher Tmv.4.1.4 Software for PC (Gene Codes Corporation) (http://multalin.toulouse.inra.fr/multalin). DNA sequences after translation to protein were analyzed in order to identify the probable amino acid substitutions. Maximum likelihood (ML) tree was constructed via MEGA v5.05 for showing the phylogenetic position of common and new haplotypes of the actin gene sequences based on the Kimura 2-parameter model of nucleotide substitution search by stepwise addition of 100 random replicates and bootstrap values with 1000 replicates (20).
Results
Actin-PCR-RFLP analysis
Of 50-suspected T. vaginalis isolates, 30 were examined by PCR-RFLP analysis of the nuclear actin region. The fragment size of PCR amplicons (1100 bp) was digested by restriction enzymes RsaI and MseI. The order of frequency, genotypes G (n=22; 73.4%) and E (n=8; 26.6%) were unequivocally discriminated.
The genotype G with fragment sizes of 581 and 519 bp was digested by MseI also, the genotype E with fragment sizes of 103, 87, 236, 568, 106 bp was digested by RsaI in asymptomatic patients (Fig. 1) however, no other expected genotypes were distinguished in the region.
Fig. 1:
A: PCR-RFLP observation in isolated samples based on actin gene. Lane 1; M: 50bp size marker, lane 2: genotype G digested by Mse 1, lane 3: genotype E digested by Rsa 1, lane 4: -Ve :negative control. B: Amplified actin gene with size 1100 bp. M: 100bp size marker
Analysis of the actin gene
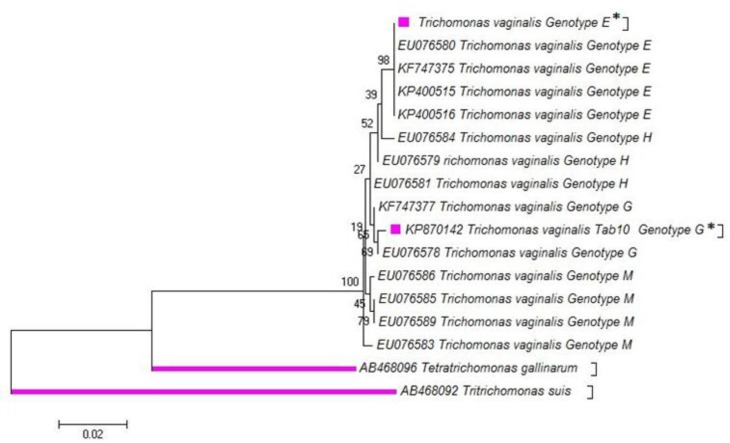
Partial nucleotide sequences obtained for the actin gene of thirty T. vaginalis isolates were edited and aligned with published sequences for the G and E genotypes. Twenty-two isolates produced identical sequence to the genotype G while eight isolates had identical sequence to the genotype E. In total, five different single nucleotide changes in the open reading frame (ORF) of the actin gene were detected. The multiple alignment of two (Tab 10) of 22 genotype G showed the amino acid substitutions in codons of 192 [Aspartic acid; (D) replaced with Glycine; (G)] and 211 [Arginine; (R) replaced with Leucine; (L)] (Fig. 2), although no other genotypes G and E showed any significant nucleotide substitution and amino acid replacement. Phylogenetic analysis was conducted using our new (Tab 10: GenBank accession no. KP870142) and common haplotypes of actin genes with those, which were submitted in GenBank using MEGA v5.05 (Fig. 3). Tetratrichomonas gallinarum and Tritrichomonas suis were considered as out group branches (GenBank Accession nos. AB468096 and AB4698092).
Fig. 2:
Amino acid sequence alignments of actin gene of genotype G based on new detected amino acids and reference sequences in the study
Fig. 3:
The Phylogeny of Trichomonas genotypes according to the maximum-likelihood (ML), tree was conducted based on the multiple sequence alignment (haplotypes) of actin gene by MEGA5.05. Only bootstrap values of higher than 70% are indicated on each branch. Distance represents the number of base substitutions per site. Tetratrichomonas gallinarum and Tritrichomonas suis were considered as out group branches (GenBank Accession nos. AB468096 and AB4698092). *= Identified genotypes in this study
Discussion
Trichomoniasis is noticed as an emerging sexual disease, which its association with HIV infection has led to a renewed attention in asymptomatic women (21). It is still not obvious why a number of individuals infected with T. vaginalis become symptomatic whilst the rest remain asymptomatic.
The identifying molecular variation by strain typing techniques and function of host factors play a crucial role in leading to asymptomatic infection which can be used to study modes of transmission, epidemiology, virulence factors of parasite (cytoadherence, cytolysis and cell detaching) and drug resistance (5, 6).
In this study, the genotypes G and E of T. vaginalis isolates were unequivocally identified by standardized profile of RFLP method containing amino acid substitution of genotype G in asymptomatic trichomoniasis of Iranian isolates where there was no comprehensive molecular typing study on genotypic traits of parasite. The PCR-RFLP method described here allows T. vaginalis isolates to be easily and rapidly distinguished using size and sequence of the nuclear genomic actin gene (12, 19).
Until now, the hopeful molecular typing strategies have been described for discriminating T. vaginalis isolates based on RAPD technique used five random primers (OPD1–OPD5) and two primers complementary, RFLP with a heat shock protein 70 (HSP70) and actin genes (12, 19 , 22). RAPD pattern can detect the trichomoniasis symptomatic and asymptomatic in women as well as the metronidazole resistance and genetic variability of T. vaginalis isolates in clinical presentation. The findings of the current investigation is similarity supported by a study which had been comprehensively accomplished the identifying the different genotypes of T. vaginalis in symptomatic patients based on RFLP method of actin gene (19).
In this investigation, no more unique haplotypes were found among sequences of genotypes G and E. This can be described by high copy number of actin gene and its diploid trait (18). However, the identified unique haplotypes and amino acid substitutions in genotype G can be potentially alerted to presence of emergent species/strains of T. vaginalis, pathogenicity rate and drug resistance against metronidazole, which should be considered in follow-up, and surveillance of patients in the region.
“The PCR-RFLP employed here examines directly the parasite genome; thus, the potential difficulties of environmentally and host-induced phenotypic variations are eliminated during our each experiment, furthermore, unlike methods that depend on chance detection of target DNA by random primers” (23), the applied region of this study is part of the much studied conserved region. “This approach is appropriate for discrimination of other closely related parasite groups, particularly in regions where strains occur sympatrically” (19).
The actin gene as a ubiquitous protein played a key role in morphological changes, formation of filaments, cell motility, virulence, and the importance of cytoskeletal integrity for T. vaginalis cytopathogenicity (24–26). In addition, due to existence of semi-conserved region of actin gene it is noticed as a well-known marker in distinguishing of closely related Trichomonas genotypes while the internal transcribed spacer region of the rDNA was not broadly considered the appropriate target for the PCR-RFLP. This can justified by lack of showing polymorphism range and to be its conserved (18).
Conclusion
The present study provides invaluable method regarding the identifying of T. vaginalis genotypes from asymptomatic infections based on RFLP method and phylogenetic analyses. Findings reveal that the accurate identification of heterogeneity traits of T. vaginalis isolates particularly those which are affected on protein level should be considered in drug resistance, mixed infection with HIV, monitoring and surveillance of asymptomatic trichomoniasis in the region. Since Trichomonas is the most prevalent non-viral STD, following strains within potential patents is an ideal epidemiologic tool to study sexual networks and the transmission dynamics of STDs.
Acknowledgements
This study was financially supported by Immunology Research Center, Tabriz University of Medical Sciences, Iran. This article is derived from the master’s thesis of the second author (Thesis No. 92-95). The authors declare that there is no conflict of interest.
References
- 1. World Health Organization Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates. Geneva, Switzerland: WHO, 2001. [Google Scholar]
- 2. Fouts A, Kraus SJ. Trichomonas vaginalis: re evaluation of its clinical presentation and laboratory diagnosis. J Infect Dis. 1980; 141: 137– 143. [DOI] [PubMed] [Google Scholar]
- 3. Cates W, Joesoef RJ, Goldman M. Atypical pelvic inflammatory disease: can we identify clinical predictors? Am J Obstet Gynecol. 1993; 169: 341– 346. [DOI] [PubMed] [Google Scholar]
- 4. Cotch MF, Pastorek JG, Nugent RP, et al. Trichomonas vaginalis associated with low birth weight and pre-term delivery. Sex Transm Dis 1997; 24: 353– 360. [DOI] [PubMed] [Google Scholar]
- 5. Sutton MY, Sternberg M, Nsuami M, Behets F, Nelson AM, St Louis M. Trichomoniasis in pregnant human immunodeficiency virus-infected and human immunodeficiency virus-uninfected Congolese women: prevalence, risk factors, and association with low birth weight. Am J Obstet Gynecol. 1999; 181: 656– 662. [DOI] [PubMed] [Google Scholar]
- 6. Petrin D, Delgaty K, Gaiber Bhatt R, Garber G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin Microbiol Rev. 1998; 11: 300– 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization An overview of selected curable sexually transmitted diseases. In: World Health Organization editor. Global programme on AIDS. Geneva, Switzerland: World Health Organization, 1995. [Google Scholar]
- 8. Alderete JF, Kasmala L, Metcalfe E, Garza GE. Phenotypic variation and diversity among Trichomonas vaginalis isolates and correlation of phenotype with trichomonal virulence determinants. Infect Immun. 1986; 53: 285– 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nash TE, McCutchan T, Keister D, Dame JB, Conrad JD, Gilin FD. Restriction endonuclease analysis of DNA from 15 Giardia isolates obtained from humans and animals. J Infect Dis. 1985; 152: 64– 73. [DOI] [PubMed] [Google Scholar]
- 10. Singh B. Molecular methods for diagnosis and epidemiological studies of parasitic infections. Int J Parasitol. 1997; 27: 1135– 1145. [DOI] [PubMed] [Google Scholar]
- 11. Vanacova S, Tachezy J, Kulda J, Flegr J. Characterization of trichomonad species and strains by PCR fingerprinting. J Euk Microbiol. 1997; 44: 545– 552. [DOI] [PubMed] [Google Scholar]
- 12. Stiles JK, Shar PH, Xue L, Meade JC, Lushbaugh WB, Cleary JD, Finley RW. Molecular typing of Trichomonas vaginalis isolates by HSP70 restriction fragment length polymorphism. Am J Trop Med Hyg. 2000; 62: 441– 445. [DOI] [PubMed] [Google Scholar]
- 13. Hampl V, Vanacova S, Kulda J, Flegr J. Concordance between genetic relatedness and phenotypic similarities of Trichomonas vaginalis strains. BMC Evol Biol. 2001; 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fraga J, Rojas L, Sariego I, Sarría CA. Optimization of random amplified polymorphic DNA techniques for use in genetic studies of Trichomonas vaginalis isolates. Infect Genet Evol. 2002; 2: 73– 75. [DOI] [PubMed] [Google Scholar]
- 15. Paces J, Urbankova V, Urbanek P. Cloning and characterization of a repetitive DNA sequence specific for Trichomonas vaginalis. Mol Biochem Parasitol. 1992; 54: 247– 55. [DOI] [PubMed] [Google Scholar]
- 16. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. 2nd ed. Cold Spring Harbor, New York: Cold spring Harbor Laboratory Press, 1989. [Google Scholar]
- 17. Snipes LJ, Gamard PM, Narcisi EM, Beard CB, Lehmann T, Secor WE. Molecular epidemiology of metronidazole resistance in a population of Trichomonas vaginalis clinical isolates. J Clin Microbiol. 2000; 38: 3004– 3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simões-Barbosa A, Lobo TT, Xavier J, Carvalho SE, Leornadecz E. Trichomonas vaginalis: intrastrain polymorphisms within the ribosomal intergenic spacer do not correlate with clinical presentation. Exp Parasitol. 2005; 110: 108– 113. [DOI] [PubMed] [Google Scholar]
- 19. Crucitti T, Abdellati S, Van Dyck E, Buvé A. Molecular typing of the actin gene of Trichomonas vaginalis isolates by PCR–restriction fragment length polymorphism. Clin Microbiol Infec. 2008; 14 (9): 844– 852. [DOI] [PubMed] [Google Scholar]
- 20. Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28: 2731– 2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laga M, Nzilla N, Goeman J. The interrelationship of sexually transmitted diseases and HIV infection implications for control of both epidemics in Africa. AIDS. 1991; 5 (Suppl. 1): S55– S63. [PubMed] [Google Scholar]
- 22. Kaul P, Gupta I, Sehgal R, Malla N. Trichomonas vaginalis: random amplified polymorphic DNA analysis of isolates from symptomatic and asymptomatic women in India. Parasitol Int. 2004; 53: 255– 262. [DOI] [PubMed] [Google Scholar]
- 23. Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990; 18: 6531– 6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arroyo R, Gonzalez-Robles A, Martinez-Palomo A, Alderette JF. Signaling of Trichomonas vaginalis for amoeboid transformation and adhesion synthesis follows cytoadherence. Mol Microbiol. 1993; 7: 299– 309. [DOI] [PubMed] [Google Scholar]
- 25. Benchimol M. Trichomonads under microscopy. Microsc Microanal. 2004; 10: 528– 550. [DOI] [PubMed] [Google Scholar]
- 26. Juliano C, Monaco G, Badeira P, Tedde G, Cappuccinelli P. Action of anticytoskeletal compounds on in vitro cytopathic effect phagocytosis, and adhesiveness of Trichomonas vaginalis. Genitourin Med. 1987; 63: 256– 263. [DOI] [PMC free article] [PubMed] [Google Scholar]