Abstract
This study was conducted to understand the seasonal distribution of Vibrio vulnificus in oysters from two estuaries and the effect of environmental factors on the abundance of V. vulnificus in tropical waters. V. vulnificus was detected in 56.6% of the samples tested by colony hybridization with an alkaline phosphatase-labeled oligonucleotide probe (VV-AP), and the counts ranged from <10/g during the summer months to 103/g in the monsoon season at both sites. The density of V. vulnificus appeared to be controlled more by salinity than by temperature. A nested PCR used in this study detected V. vulnificus in 85% of the samples following 18 h of enrichment in alkaline peptone water.
Vibrio vulnificus is widely distributed in coastal and estuarine waters throughout the world (3, 18). This opportunistic pathogen has been identified as being capable of causing life-threatening septicemia and wound infections in individuals with underlying debilitations such as chronic liver disease and in immunocompromised individuals, being responsible for more than 95% of seafood-associated deaths. Infection with V. vulnificus occurs by the ingestion of raw or undercooked shellfish, particularly oysters, or by direct entry through wounds (13, 19).
The ecology of V. vulnificus in temperate waters has been well studied. The salinity and temperature of water strongly influence the density of V. vulnificus (12, 17, 24). Low salinities (5 to 25 ppt) and warm temperatures (20 to 35°C) have been reported to be favorable for this organism. The optimum salinity range is 5 to 25 ppt (17), although it has been isolated from waters with salinities ranging from 1 to 34 ppt. Several studies have linked the abundance of V. vulnificus in oysters to warmer temperatures (20 to 35°C) and low-to-moderate salinity (7 to 16 ppt) (11, 12, 20, 24).
Very few studies on the prevalence of V. vulnificus in tropical waters such as those of India have been carried out (8, 9, 25), and these were based on conventional isolation and identification methods. There is very little information on the abundance and ecology of this organism in tropical waters. Direct enumeration of V. vulnificus in seafoods is greatly facilitated by the use of oligonucleotide probes. Wright et al. (26, 27) described a colony hybridization assay for the enumeration of V. vulnificus with an alkaline phosphatase-labeled oligonucleotide probe (VV-AP). The objectives of the present study were to determine the densities of V. vulnificus in freshly harvested oysters on the southwest coast of India by colony hybridization with the VV-AP probe and to detect its presence by conventional isolation and direct PCR on enrichment broths. The Food and Drug Administration (5) recommends enrichment in alkaline peptone water (APW), followed by isolation on selective agars, thiosulfate citrate bile salt sucrose (TCBS) agar, and modified cellobiose-polymyxin-colistin (mCPC) agar for detection of V. vulnificus. However, Hoi et al. (7) reported high recovery of V. vulnificus on cellobiose-colistin (CC) agar following enrichment in APW with polymyxin B (APWP). To understand the efficacy of these methods in the detection of tropical strains of V. vulnificus, in this study we evaluated the efficiency of isolation of V. vulnificus on TCBS, CPC (cellobiose-polymyxin B-colistin), and mCPC agars after enrichment in APW and on CC agar after enrichment in APW broth containing polymyxin B (APWP).
V. vulnificus abundance determined by colony hybridization.
Oyster samples were collected weekly (from March 2002 to June 2002) and biweekly (from June 2002 to March 2003) at two sites (site 1, Sasthan, Udupi District; site 2, Mulki, Mangalore District) that are about 100 km apart by road. The samples were chilled in ice immediately after collection and held at <10°C during transport, and the analysis was done within 3 h of collection. Water temperature was recorded with a calibrated thermometer, and chlorinity was determined by a titrimetric method (1). Salinity was calculated from chlorinity with the following formula: salinity = 1.80655 × chlorinity.
An alkaline phosphatase-labeled oligonucleotide probe (VV-AP; DNA Technologies, Aarhus, Denmark) binding to the V. vulnificus specific cytolysin-hemolysin gene (vvhA) was used for colony hybridization as described previously (4, 26, 27). Oysters were washed and shucked, and the meat from 20 to 25 oysters was pooled and homogenized. The homogenate was then diluted 1:1 and 1:10 in APW by briefly blending 50 g of homogenate in 50 ml of APW and 20 g in 180 ml of APW, respectively. Homogenates (0.2 g from the 1:1 dilution and 0.1 ml from the 1:10 dilution of oyster meat in APW) were spread directly on T1N3 plates (1% tryptone, 3% sodium chloride, 2% agar) in duplicate. The plates were incubated for 18 to 24 h at 37°C. Colony lifts with no. 541 Whatman filters, alkaline lysis, and proteinase K (Bangalore Genei, Bangalore, India) treatment (for 1 h at 42°C) were performed as previously described (26). Hybridization was done at 56°C for 1 h, followed by washing twice in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.5% (wt/vol) sodium dodecyl sulfate for 10 min at 56°C. The washing conditions were modified to overcome nonspecific hybridization signals that were observed when the protocol originally described by Wright et al. (26, 27) was used. The modified conditions were tested with laboratory isolates of V. vulnificus, V. parahaemolyticus, V. harveyi, and V. cholerae before they were adopted for the study. Hybridization-positive colonies were visualized by incubating the filters in BCIP (5-bromo-4-chloro-3-indolylphosphate)-Nitro Blue Tetrazolium (Bangalore Genei) solution. Control strips spotted with control strains of V. vulnificus (ATCC 27562), V. parahaemolyticus (AQ 4037), and V. harveyi (laboratory isolate) were used in each hybridization reaction.
The V. vulnificus counts obtained were subjected to multiple regression analysis against water temperature and salinity with the SYSTAT statistical package (SYSTAT, Richmond, Calif.) to determine the relationship between the counts and the environmental factors. Nondetectable V. vulnificus counts of <10/g were recorded as 5/g for statistical analysis. For simple comparison of counts between the sites, simple arithmetic means of overall counts were calculated.
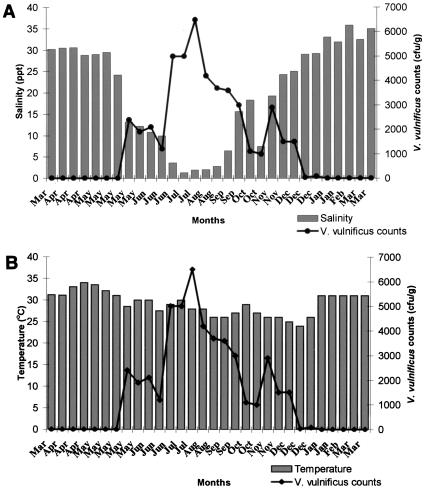
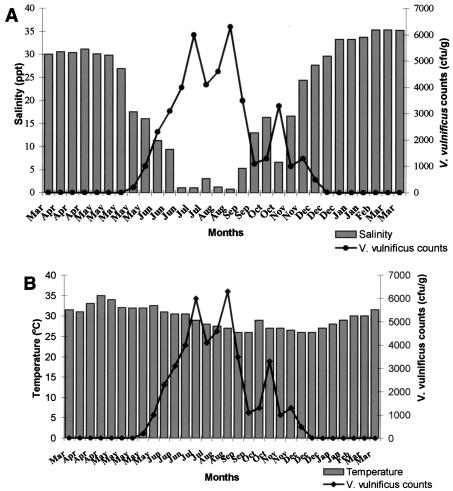
Thirty samples collected from each site over a period of 1 year were analyzed to determine levels of V. vulnificus and the effects of temperature and salinity on its abundance. The V. vulnificus counts at the two sites followed similar patterns (Fig. 1A and B and 2A and B). During the 1-year study period, the organism was detected in 18 samples (60%) from site 1. The counts ranged from <10/g to 6.5 × 103/g, with an average of 1.5 × 103/g. At site 2, 16 samples (53.3%) were positive and the V. vulnificus counts ranged from <10/g to 6.3 × 103/g, with an average of 1.4 × 103/g. The counts demonstrated a seasonal trend, and two definite periods with significant changes in V. vulnificus counts were identified, the monsoon season (June to September), when the counts were the highest, and high summer (March to May), when this organism could not be detected by the colony hybridization method. During the monsoon season, the average count at site 1 was 3.8 × 103/g and the average temperature and salinity were 27.9°C and 6.09 ppt, respectively. At site 2, the average V. vulnificus count was 3.9 × 103/g during the monsoon season and the average temperature and salinity were 28.38°C and 5.14 ppt, respectively. The highest count (6.5 × 103/g) was recorded at site 1 when the salinity and temperature were 1.94 ppt and 28.0°C, respectively. From the last week of December to the third week of May, the count in all of the samples from both sites was <10/g. The decline in counts started in December, when the salinity crossed 25 ppt. No V. vulnificus was detected when the salinity was >30 ppt at both sites.
FIG. 1.
(A) Effect of salinity on V. vulnificus counts of samples from site 1. (B) Effect of temperature on V. vulnificus counts of samples from site 1.
FIG. 2.
(A) Effect of salinity on V. vulnificus counts of samples from site 2. (B) Effect of temperature on V. vulnificus counts of samples from site 2.
Understanding the seasonal abundance of V. vulnificus would be important in predicting the risk of illness to oyster consumers. In temperate waters, V. vulnificus has been recovered from oysters at levels of 103 to 104/g in summer months but not during winter months (27). In tropical waters, the seasonal variation in temperature is not so marked; therefore, it is interesting to study the ecology of this organism in such environments. The present study clearly demonstrates that V. vulnificus is present in oysters in India and that the levels are high when the salinity of the water is low. Counts could be recorded from the third week of May to the end of December at both sites. In Mangalore, the southwest monsoon starts during the third week of May. The highest counts recorded were during and after the southwest monsoon. Rivers on the southwest coast of India are seasonal, and the inflow from these rivers into the estuaries diminishes from January until the onset of the southwest monsoon in the last week of May, resulting in a gradual rise in salinity. From January until the second week of May, the salinity values were above 30 ppt. During this period, the samples from both sites did not harbor V. vulnificus. V. vulnificus counts were maximum (6.5 × 103/g) at 1.94 ppt. These observations strongly suggest the influence of environmental factors such as salinity and monsoon on the abundance of V. vulnificus on the Mangalore coast. Several investigators have previously reported the effect of salinity on the presence of V. vulnificus (17). Although the bacterium has been isolated from salinities ranging from 1 to 34 ppt, the preferred range is 5 to 25 ppt (17). Salinities greater than 25 ppt have an adverse effect on the survival of the organism (10, 16). In our study, counts of 103/g were recorded even when the salinity was below 5 ppt.
The V. vulnificus counts were found to be dependent less on temperature than on salinity. During the 1-year study period, the mean temperature at the two sites ranged between 24 and 34°C. The highest concentration of V. vulnificus has been reported during warmer months (3, 22). V. vulnificus has been recovered, although sporadically and usually in small numbers, from oysters harvested from cooler environments such as the New England coast (21) and the Pacific coast (11). The ecology in Indian waters is different from that of temperate and subtropical waters. The water temperature on our coast rarely goes below 25°C, and therefore the effect of temperature is considerably less than that in temperate countries, where most of the studies on the abundance of V. vulnificus have been conducted. In tropical waters the temperature fluctuates less and is therefore favorable for V. vulnificus year round but during the summer season salinity is typically above the optimum for this organism. Previous reports from India (8, 9) have described V. vulnificus counts of 102 to 104/g in fish and shellfish samples collected from open markets and an incidence rate of 25% in fresh and processed samples. However, these studies were based on random samples of different varieties of fish and shellfish analyzed by conventional culture-based techniques and there are no comprehensive reports on the seasonality of V. vulnificus abundance in any particular species from tropical waters. To our knowledge, this is the first report on measurements of the density and seasonal variations of V. vulnificus in oysters from tropical waters by molecular methods.
Comparison of selective agars for isolation of V. vulnificus.
Twenty-five grams of oyster meat was inoculated into 225 ml of APW, and another 25 g was inoculated into APWP (2.0 × 104 U/liter) and incubated for 18 h at 37°C. Following 16 to 18 h of incubation in APW, a loopful was streaked onto TCBS, CPC, and mCPC agars. CC agar was streaked from an APWP enrichment. All of the plates except the TCBS plate were incubated at 40°C, and the TCBS plate was incubated at 37°C. A minimum of five typical colonies were selected from each enrichment plate and subjected to biochemical identification (5). Strains identified as V. vulnificus were further confirmed by hybridization with the VV-AP probe and a PCR assay with vvhA-specific primers.
The highest rate of V. vulnificus isolation from the site 1 samples was on CC agar at 56.6% (after enrichment in APWP), followed by mCPC (50%), CPC (33.3%), and TCBS (0%). The highest rate of V. vulnificus isolation from the site 2 samples was on CC agar at 46.6%, followed by mCPC (43.3%), CPC (30.0%), and TCBS (1.3%). Overall, V. vulnificus could be isolated from 60% of the samples from site 1 and 53.33% of the samples from site 2 after 18 h of enrichment.
The plating efficiency of CC agar with APWP enrichment was found to be better than that of the other three. Hoi et al. (7) reported better performance of CC agar than that of mCPC, CPC, and TCBS agars. The enrichment in APWP prior to streaking onto CC may have provided an edge over APW enrichment. All of the strains isolated in our study were found to belong to biotype 1 on the basis of biochemical reactions such as indole production, growth at 42°C, ornithine decarboxylation, and acid production from mannitol.
Detection of V. vulnificus by PCR on enrichment broths.
A nested PCR amplifying the cytolysin-hemolysin (vvhA) gene (15) was used for direct detection of V. vulnificus in enrichment broth lysates. Crude lysates from enrichment broths were prepared at 0, 6, and 18 h of incubation in APW. A 1.5-ml portion of the culture supernatant was centrifuged at 800 × g for 5 min (Biofuge; Heraeus, Hanau, Germany) to remove larger meat particles, and the supernatant was centrifuged at 10,000 × g for 10 min. The bacterial pellet was washed once in distilled water, centrifuged, resuspended in 100 μl of sterile distilled water, boiled for 10 min, and snap-cooled on ice. The lysates were centrifuged at 10,000 × g for 5 min, and 5 μl of the supernatant was used in a 50-μl reaction mixture. PCR conditions were as described previously by Lee et al. (14, 15) with the first-step primers JY-1 (5′-GACTATCGCATCAACAACCG-3′) and JY-2 (5′-AGGTAGCGAGTATTACTGCC-3′) and the second-step (nested) primers JY-3 (5′-GCTATTTCACCGCCGCTCAC-3′) and JY-4 (5′-CCGCAGAGCCGTAAACCGAA-3′). For the second-step reactions, 5 μl of the first-step PCR product from PCR-negative samples was used as template DNA. The PCR products were separated on 2% gel, stained with ethidium bromide (0.5 μg/ml), and photographed with a gel documentation system (Hero Lab, Wiesloch, Germany).
The results of direct PCR on enrichment broth lysates are shown in Table 1. Only 1 sample (3.3%) from site 1 was positive for V. vulnificus at 0 h, 8 samples (26.66%) were positive at 6 h, and 14 samples (46.66%) were positive at 18 h of enrichment in APW in the first step (Table 1). However, nested PCR could detect V. vulnificus in 13.3% of the samples tested at 0 h while 6 and 18 h of enrichment raised this percentage to 70 and 86.6%, respectively. V. vulnificus was detectable by nested PCR in six samples from which no counts were obtained by colony hybridization during the period of high salinity during the summer months (March to May). No sample from site 2 was positive in the first step at 0 h of enrichment, but 6 samples (20%) were positive at 6 h and 12 samples (40%) were positive at 18 h in the first step. In the second step, only one sample was positive at 0 h but 70% of the samples were positive after 6 h of enrichment and 83.3% were positive after 18 h of enrichment.
TABLE 1.
Summary of results of colony hybridization, conventional isolation, and PCR for V. vulnificus
| Site | No. (%) of samples positive by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Conventional isolation | Colony hybridization | 1-Step PCRa
|
2-Step PCRa
|
|||||
| 0 h | 6 h | 18 h | 0 h | 6 h | 18 h | |||
| 1 | 18 (60) | 18 (60) | 1 (3.3) | 8 (26.6) | 14 (46.6) | 4 (13.3) | 21 (70.0) | 26 (86.6) |
| 2 | 16 (53.3) | 16 (53.3) | 0 (0) | 6 (20.0) | 12 (40) | 1 (3.3) | 21 (70.0) | 25 (83.3) |
Lysates for PCR were prepared from APW enrichment broths.
We compared the effect of length of enrichment on the detection of V. vulnificus by a nested PCR from enrichment broths. Results in Table 1 show that positivity increased with an increase in enrichment time. Without enrichment (0 h), only 3.3% of the samples were positive but 86.6 and 83.3% of those from sites 1 and 2, respectively, were positive after 18 h. For detection by the colony hybridization method, the organism should be found at a minimum level of 10 organisms per g. During the summer months, no V. vulnificus could be detected by colony hybridization and therefore we wanted to use a more sensitive technique for the detection of this organism. A few samples that had hybridization counts of >103/g were positive at 0 and 6 h by PCR but negative after 18 h of enrichment in APW. This was due to the presence of a high target DNA concentration inhibiting the PCR, as confirmed by PCR positivity in a 100-fold dilution of these lysates. According to Lee et al. (14), the nested primers described by them and used in this study can detect as little as 1 CFU of V. vulnificus per g when the guanidinium isothiocyanate method is used for DNA extraction. In our study we used crude lysates and yet obtained a high percentage of positivity in the nested reaction. Samples that had counts of >103/g were positive in the first-step PCR, indicating that a higher number of organisms is required for PCR detection in the absence of a reliable DNA extraction procedure when a single-stage PCR is used. Out of a total of 60 samples (from both sites), the nested PCR could detect V. vulnificus in 12 samples (6 from each site) even when no isolations were done on any of the four selective agars used. The possible explanation could be that the organism was present at too low a number and might have been overgrown by other organisms in the selective agar or it was unable to grow on the medium (nonculturable). However, a nonculturable state in V. vulnificus is not known to be influenced by higher salinity and temperature. More studies are needed to understand the ecological factors influencing the survival of V. vulnificus apart from temperature and salinity in tropical waters.
To conclude, this is the first comprehensive report on the abundance of V. vulnificus in Indian estuaries with molecular techniques. Our study demonstrates the influence of environmental factors such as temperature and salinity on the presence and densities of this organism in oysters. The clinical significance of the presence of V. vulnificus in Indian waters is not known, as no epidemiological studies have been conducted in India, except for a single report on the involvement of V. vulnificus in wound infection (23). Further genetic characterization of V. vulnificus strains isolated from oysters in this study is in progress.
Acknowledgments
We thank the Food & Nutrition Division, Food and Agriculture Organization, for providing financial support for the purchase of the oligonucleotide probes and reagents used for colony hybridization in this study. Financial support by the Department of Biotechnology, Government of India, and the donation of the PCR machine by the Alexander von Humboldt Foundation, Bonn, Germany, are gratefully acknowledged.
REFERENCES
- 1.American Public Health Association. 1985. APHA standard methods, 16th edition, section 210C. American Public Health Association, Washington, D.C.
- 2.Cook, D. W. 1994. Effect of time and temperature on multiplication of Vibrio vulnificus in postharvest Gulf Coast shellstock oysters. Appl. Environ. Microbiol. 60:3483-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DePaola, A., G. M. Capers, and D. Alexander. 1994. Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast. Appl. Environ. Microbiol. 60:984-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DePaola, A., M. L. Motes, D. W. Cook, J. Veazey, W. E. Garthwright, and R. Blodgett. 1997. Evaluation of alkaline phosphatase-labeled DNA probe for the enumeration of V. vulnificus in Gulf Coast oysters. J. Microbiol. Methods 29:115-120. [Google Scholar]
- 5.Food and Drug Administration. 2000. Bacteriological analytical manual, Food and Drug Administration, 7th edition, p. 383-419. AOAC International, Arlington, Va.
- 6.Hlady, W. G., and K. C. Klontz. 1996. The epidemiology of Vibrio infections in Florida, 1981-1993. J. Infect. Dis. 173:1176. [DOI] [PubMed] [Google Scholar]
- 7.Hoi, L., I. Dalsgaard, and A. Dalsgaard. 1998. Improved isolation of Vibrio vulnificus from seawater and sediment with cellobiose-colistin agar. Appl. Environ. Microbiol. 64:1721-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karunasagar, I., M. Susheela, and I. Karunasagar. 1987. Vibrio vulnificus in fish and clams in Mangalore water, west coast of India. Mar. Sci. 16:136-137. [Google Scholar]
- 9.Karunasagar, I., M. Susheela, G. R. Malathi, and I. Karunasagar. 1990. Incidence of human pathogenic vibrios in seafoods harvested along the coast of Karnataka (India). FAO Fish. Rep. 401(Suppl.):53-56. [Google Scholar]
- 10.Kaspar, C. W., and M. L. Tamplin. 1993. Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl. Environ. Microbiol. 59:2425-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaysner, C. A., C. Abeyta, Jr., M. M. Wekell, A. DePaola, Jr., R. F. Stott, and J. M. Leitch. 1987. Virulent strains of Vibrio vulnificus isolated from estuaries of the United States West Coast. Appl. Environ. Microbiol. 53:1349-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly, M. T. 1982. Effect of temperature and salinity on Vibrio (Beneckea) vulnificus occurrence in a Gulf Coast environment. Appl. Environ. Microbiol. 44:820-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klontz, K. C., L. Spencer, M. Schreiber, H. T. Janowski, L. M. Blady, and R. A. Gunn. 1988. Syndromes of Vibrio vulnificus infections: clinical and epidemiologic features in Florida cases, 1981-1987. Ann. Intern. Med. 109:318-323. [DOI] [PubMed] [Google Scholar]
- 14.Lee, J. Y., J. B. Eun, and H. S. Choi. 1997. Improving detection of Vibrio vulnificus in Octopus variabilis by PCR. J. Food Sci. 62:179-182. [Google Scholar]
- 15.Lee, J. Y., Y. B. Bang, J. H. Rhee, and S. H. Choi. 1999. Two stage nested PCR effectiveness for direct detection of Vibrio vulnificus in natural samples. J. Food Sci. 64:157-162. [Google Scholar]
- 16.Motes M. L., and A. DePaola. 1996. Offshore suspension relaying to reduce levels of Vibrio vulnificus in oysters (Crassostrea virginica). Appl. Environ. Microbiol. 62:3875-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motes, M. L., A. DePaola, D. W. Cook, J. E. Veazey, J. C. Hunsucker, W. E. Garthwright, R. J. Blodgett, and S. J. Chirtel. 1998. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver, J. D., R. A. Warner, and D. R. Cleland. 1983. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl. Environ. Microbiol. 45:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliver, J. D. 1989. Vibrio vulnificus, p. 569-599. In M. Doyle (ed.), Food borne pathogens. Marcel Dekker, Inc., New York, N.Y.
- 20.O'Neill, K. R., S. H. Jones, and D. J. Grimes. 1990. Incidence of Vibrio vulnificus in northern New England water and shellfish. FEMS Microbiol. Lett. 72:163-168. [DOI] [PubMed] [Google Scholar]
- 21.O'Neill, K. R., S. H. Jones, and D. J. Grimes. 1992. Seasonal incidence of Vibrio vulnificus in the Great Bay estuary of New Hampshire and Maine. Appl. Environ. Microbiol. 58:3257-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruple, A. D., and D. W. Cook. 1992. Vibrio vulnificus and indicator bacteria in shellstock and commercially processed oysters from the Gulf Coast. J. Food Prot. 55:667-671. [DOI] [PubMed] [Google Scholar]
- 23.Saraswathi, K., C. M. Barve, and L. P. Deodhar. 1989. Septicemia due to V. vulnificus. Trans. R. Soc. Med. Hyg. 83:714. [DOI] [PubMed] [Google Scholar]
- 24.Tamplin, M., G. E. Rodrick, N. J. Blake, and T. Cuba. 1982. Isolation and characterization of Vibrio vulnificus from two Florida estuaries. Appl. Environ. Microbiol. 44:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thampuran, N., and P. K. Surendran. 1998. Occurrence and distribution of Vibrio vulnificus in tropical fish and shellfish from Cochin (India). Lett. Appl. Microbiol. 26:110-112. [DOI] [PubMed] [Google Scholar]
- 26.Wright, A. C., G. A. Miceli, W. L. Landry, J. B. Christy, W. D. Watkins, and J. G. Morris, Jr. 1993. Rapid identification of Vibrio vulnificus on nonselective media with an alkaline phosphatase-labeled oligonucleotide probe. Appl. Environ. Microbiol. 59:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright, A. C., R. T. Hill, J. A. Johnson, M. C. Roghman, R. R. Colwell, and J. G. Morris, Jr. 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]