Abstract
Based on the hypothesis that intestinal clostridia play a role in late-onset autism, we have been characterizing clostridia from stools of autistic and control children. We applied the TaqMan real-time PCR procedure to detect and quantitate three Clostridium clusters and one Clostridium species, C. bolteae, in stool specimens. Group- and species-specific primers targeting the 16S rRNA genes were designed, and specificity of the primers was confirmed with DNA from related bacterial strains. In this procedure, a linear relationship exists between the threshold cycle (CT) fluorescence value and the number of bacterial cells (CFU). The assay showed high sensitivity: as few as 2 cells of members of cluster I, 6 cells of cluster XI, 4 cells of cluster XIVab, and 0.6 cell of C. bolteae could be detected per PCR. Analysis of the real-time PCR data indicated that the cell count differences between autistic and control children for C. bolteae and the following Clostridium groups were statistically significant: mean counts of C. bolteae and clusters I and XI in autistic children were 46-fold (P = 0.01), 9.0-fold (P = 0.014), and 3.5-fold (P = 0.004) greater than those in control children, respectively, but not for cluster XIVab (2.6 × 108 CFU/g in autistic children and 4.8 × 108 CFU/g in controls; respectively). More subjects need to be studied. The assay is a rapid and reliable method, and it should have great potential for quantitation of other bacteria in the intestinal tract.
Autism is a complex disease with unclear causes. Many autistic subjects exhibit a range of gut disorders, which include constipation, diarrhea, retention of gas, and abdominal pain and discomfort. Abnormal gut microflora may play a role in these problems. Research into the characteristics of the gut flora in autism has been limited. In our initial studies that characterized the fecal bacterial composition by culturing, we noted abnormalities in the fecal bacterial composition of children with autism compared to age- and sex-matched controls. We found higher counts of clostridia overall and more species of clostridia in stools of autistic children than in healthy children (11). In particular, Clostridium bolteae, a novel species that we described previously (29; called Clostridium clostridioforme in reference 11), caught our attention because it was cultured from 5 of 15 autistic children, but none of 8 controls. However, it is well known that traditional culture-based methods, while very important, result in a significant underestimation of bacteria present in fecal samples (14, 19, 30).
Molecular techniques introduced in microbial ecology have made it possible to study the composition of intestinal flora in a culture-independent way based on the detection of rRNA genes. Although these methods, such as fluorescent in situ hybridization (12, 13, 19), denaturing gradient gel electrophoresis, temperature gradient gel electrophoresis (8, 10, 28), and the 16S rRNA gene clone library method (15, 27, 30), have been applied successfully for studying the ecology of intestinal flora, PCR analysis using specific primers achieves the most sensitive results as well as providing ease of use and speed (23, 24, 32). Most recently, real-time quantitative PCR has been used for the specific detection and quantitation of selected bacteria from fecal DNA (1, 2, 4, 9, 16, 18, 21, 22, 25, 33).
Few studies have reported on using real-time PCR for quantitation of clostridia in different environments. Belanger et al. (2) and Kimura et al. (17) reported on the successful quantitation of Clostridium difficile in feces and Clostridium botulinum type E in fish samples using specific primers and probes targeted to toxin genes, respectively. A very recent study investigated the feasibility of using 16S rRNA gene-targeted specific primers and probes for quantitation of major intestinal bacteria, including certain Clostridum species by real-time PCR (26). In this study, we evaluated the suitability of a real-time PCR (5′ nuclease PCR assay) to detect and quantitate C. bolteae and some Clostridium groups (clusters) in fecal specimens of autistic and control children.
MATERIALS AND METHODS
Bacterial strains and fecal specimens.
All reference strains used in this study are listed in Table 1. The strains were obtained from different sources, as indicated in Table 1: ATCC is the American Type Culture Collection, CCUG is the Culture Collection, University of Göteborg, DSM is the Deutsche Sammlung von Mikroorganismen und Zellkulturen, and WAL is the Wadsworth Anaerobe Laboratory. All the strains were cultivated on brucella agar (Anaerobe Systems, Morgan Hill, Calif.) supplemented with 5% sheep blood and incubated anaerobically at 37°C under N2 (86%), H2 (7%), and CO2 (7%) gas phase in an anaerobic incubator. The WAL strains are human fecal isolates; they were identified by phenotypic testing and 16S rRNA sequencing (11).
TABLE 1.
Strains used in this study
| Strain | Sourcea |
|---|---|
| Anaerococcus spp. | |
| A. lactolyticus | CCUG 31351T |
| A. octavius | CCUG 38493T |
| A. prevotii | CCUG 41932T |
| A. tetradius | CCUG 46590T |
| A. vaginalis | CCUG 31349T |
| Clostridium spp. | |
| C. beijerinckii | ATCC 25752T |
| C. bifermentans | ATCC 638T |
| C. bifermentans | WAL 16469 |
| C. bolteae | ATCC BAA-613T |
| C. bolteae | WAL 14578 |
| C. bolteae | WAL 14510 |
| C. bolteae | WAL 16099 |
| C. bolteae | WAL 12258 |
| C. bolteae | WAL 7642 |
| C. butyricum | ATCC 13732T |
| C. butyricum | WAL 16108 |
| C. cadaveris | ATCC 25783T |
| C. carnis | ATCC 25777T |
| C. celerecrescens | DSMZ 5628T |
| C. fallax | ATCC 19400T |
| C. glycolicum | ATCC 14880T |
| C. glycolicum | WAL 14424 |
| C. indolis | ATCC 25771T |
| C. irregulare | ATCC 25756T |
| C. limosum | ATCC 25620T |
| C. paraputrificum | ATCC 25780T |
| C. paraputrificum | WAL 14502 |
| C. pasteurianum | ATCC 6013T |
| C. perfringens | ATCC 13124T |
| C. perfringens | WAL 14572 |
| C. putrificum | ATCC 25784T |
| C. septicum | ATCC 12464T |
| C. sordellii | ATCC 9714T |
| C. subterminale | ATCC 25774T |
| C. subterminale | WAL 16483 |
| C. symbiosum | ATCC 14940T |
| C. symbiosum | WAL 14673 |
| C. tertium | ATCC 14573T |
| C. tertium | WAL 16200 |
| C. tyrobutyricum | ATCC 25755T |
| C. xylanolyticum | DSMZ 6555T |
| C. acetobutylicum | ATCC 824T |
| C. baratii | ATCC 27638T |
| C. bolteae | WAL 16351 |
| C. celatum | ATCC 27791T |
| C. clostridioforme | ATCC 25537T |
| C. clostridioforme | WAL 12260 |
| C. clostridioforme | WAL 7855 |
| C. coccoides | ATCC 29236T |
| C. cochlearium | ATCC 17787T |
| C. difficile | ATCC 9689T |
| C. difficile | WAL 14186 |
| C. ghonii | ATCC 25757T |
| C. malenominatum | ATCC 25776T |
| C. methoxybenzovorans | DSMZ 12182T |
| C. novyi | ATCC 27606 |
| C. oroticum | ATCC 13619T |
| C. sphenoides | ATCC 19403T |
| C. sporogenes | ATCC 19404 |
| Eubacterium spp. | |
| E. tenue | ATCC 25553T |
| Finegoldia spp. | |
| F. magna | CCUG 17636T |
| Peptoniphilus spp. | |
| P. anaerobius | CCUG 7835T |
| P. asaccharolyticus | CCUG 9988T |
| P. harei | CCUG 38491T |
| P. indolicus | CCUG 17639T |
| P. ivorii | CCUG 38492T |
| P. lacrimalis | CCUG 31350T |
| Peptostreptococcus spp. | |
| P. micros | CCUG 46357T |
| Ruminococcus spp. | |
| R. flavefaciens | WAL 16464 |
| R. gnavus | CCUG 43222T |
| R. gnavus | WAL 16098 |
| R. lactaris | WAL 14694 |
| R. productus | CCUG 10976T |
| R. torques | WAL 14161 |
| Sarcina spp. | |
| S. maxima | CCUG 45682T |
| S. ventriculi | WAL 16376 |
All WAL strains were identified by 16S rDNA sequencing.
Fecal specimens were obtained at Rush Children's Hospital, Chicago, Ill., under the jurisdiction of its Institutional Review Board (IRB) and with written informed consent by a parent or guardian. Our IRB approved receipt of these specimens by our laboratory. Stool specimens were packed in dry ice and shipped to our laboratory by overnight air express. The entire fecal specimen was homogenized by use of a sterile stainless steel Waring blender, and aliquots of each specimen were frozen at −80°C until DNA was extracted.
Extraction of DNA from pure cultures and feces.
Genomic DNA from all bacterial strains was purified from cultures with a QIAamp DNA extraction kit, (QIAGEN, Valencia, Calif.), and the concentration was determined by spectrophotometer (A260). Duplicate samples of genomic DNA (20 ng) from each bacterium were amplified for 40 cycles with each of four real-time PCR primer/probe sets (Table 2) to assess specificity.
TABLE 2.
Sequences of oligonucleotide primers and probe
| Target bacteria and primer or probea | Oligonucleotide sequence (5′→3′) | Tm (°C)b |
|---|---|---|
| Probe-I | GTGCCAGCAGCCGCGGTAATACG | 72.4 |
| Group I (Clostridium cluster I) | ||
| Forward primer, CI-F1 | TACCHRAGGAGGAAGCCAC | 54.6 |
| Reverse primer, CI-R2 | GTTCTTCCTAATCTCTACGCAT | 53.0 |
| Group II (Clostridium cluster XI) | ||
| Forward primer, CXI-F1 | ACGCTACTTGAGGAGGA | 46.5 |
| Reverse primer, CXI-R2 | GAGCCGTAGCCTTTCACT | 52.3 |
| Group III (Clostridium cluster XIVab) | ||
| Forward primer, CXIV-F1 | GAWGAAGTATYTCGGTATGT | 46.2 |
| Reverse primer, CXIV-R2 | CTACGCWCCCTTTACAC | 44.8 |
| Probe-II (C. bolteae) | CAGGTGGTGCATGGTTGTCGTCAG | 69.2 |
| Forward primer | CCTCTTGACCGGCGTGT | 56.7 |
| Reverse primer | CAGGTAGAGCTGGGCACTCTAGG | 62.1 |
Probe-I is an internal universal probe that corresponds to a region of the 16S rRNA gene that is conserved in all eubacteria (5); it was used with three sets of cluster-specific primers. Probe-II was used with C. bolteae-specific primer set. Clostridium clusters are as described in reference 7.
The Tm of DNA was determined with NetPrimer software (PREMIER Biosoft International).
One-milliliter aliquots of stool (previously diluted 1:3 in sterile bi-distilled water and thoroughly homogenized under anaerobic conditions in an anaerobic chamber) were centrifuged at 14,000 × g for 3 min to pellet fecal bacterial cells. The supernatant was carefully removed and discarded. Two hundred milligrams of cell pellet was transferred to a fresh tube and subjected to DNA extraction by using a commercial extraction system (QIAamp DNA stool mini kit; QIAGEN) according to the instructions of the manufacturer. Our pilot studies have shown that this QIAamp product produces high-quality DNA free of PCR-inhibiting substances. DNA extraction was performed in duplicate.
Design of oligonucleotide primers and probe.
Sequences of the 16S rRNA genes of the organisms of interest and of closely related bacteria were aligned with CLUSTAL-W (http://genome.kribb.re.kr) and inspected for regions of conserved and variable sequences. Based on the multialignment analysis data, four pairs of 16S rRNA gene-targeted cluster- and species-specific primers were designed, one each for Clostridium clusters I, XI, and XIVab (7) and C. bolteae, respectively. They were designed from a collection of 128 Clostridium 16S rRNA sequences obtained from the GenBank database. Potential candidates for PCR primers were compared to the aligned SSU_rRNA database of the Ribosomal Database Project (RDP) using the CHECK_PROBE utility (20) and were compared to all available 16S ribosomal DNA (rDNA) sequences by using the BLAST database search program (www.ncbi.nlm.nih.gov/BLAST) (3). The primers were used in combination with internal nonspecific probes (Table 2). The primers' target regions were located as near as possible to the probe in order to enable an efficient hydrolysis of the probe during primer elongation by the polymerase. The primer and probe sequences were analyzed for Tm (melting temperature), secondary structure formation, G+C content, and primer-dimer formation with NetPrimer analysis software (http://www.premierbiosoft.com/netprimer). The probe was 5′ labeled with carboxyfluorescein (FAM) as the reporter dye and Black Hole quencher (BHQ) as the 3′ quencher dye. The BHQ dye was used as the quencher dye because of its broad quenching spectrum and a lower signal/noise ratio than that of other quenching dyes.
SmartCycler real-time PCR assay.
The amplification reactions were carried out in a total volume of 25 μl, which consisted of 2 μl of DNA samples and 23 μl of the master mixture. The latter contained 1× puReTaq Ready-To Go PCR bead (2.5 U of puReTaq DNA polymerase, 10 mM Tris-HCl [pH 9.0 at room temperature], 50 mM KCl, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate [dNTP], and stabilizer bovine serum albumin [Amersham Biosciences, Piscataway, N.J.]), 300 nM each primer, 200 nM fluorescence-labeled probe, and 2.5 mM MgCl2, made up to 23 μl with distilled water. The assay was performed with the Cepheid (Sunnyvale, Calif.) SmartCycler instrument in a protocol comprising 1 cycle of 2 min at 95°C (hot start) followed by 45 cycles of 95°C for 20 s for denaturation. Annealing was performed for 30 s at 58°C for C. bolteae, 63°C for Clostridium cluster I, 58°C for Clostridium cluster XI, and 52°C for Clostridium cluster XIVab. Extension was performed at 72°C for 45 s for all. Data analysis was performed with Cepheid software. The CT values (i.e., the threshold cycles in which exponential amplification of PCR products was first detected) were determined on the basis of the mean baseline signals during the early cycles of amplification.
Standard curve.
The standard curve for C. bolteae was constructed by using DNA recovered from fecal samples spiked with different numbers of C. bolteae type strain ATCC BAA-613 (30 to 3.0 × 107 CFU ml−1). Briefly, C. bolteae ATCC BAA-613 was collected from thioglycolate broth cultures while in logarithmic growth phase. Ten-fold dilutions were made in dilution blanks (Anaerobe Systems), and the number of CFU of bacteria in each dilution was determined by plating on brucella blood agar plates. All determinations were performed in triplicate. The average titer (CFU per milliliter) of three replicates was determined. One milliliter of each dilution was added to tubes containing 200-mg aliquots of a stool sample that was verified to be C. bolteae negative by both bacteriological and PCR techniques and pelleted by centrifugation. The pellets with different numbers of C. bolteae cells were subjected to DNA extraction by using a QIAamp DNA Stool Mini kit. The DNA was used to establish a standard curve.
To generate standard curves for each of the three Clostridium clusters, the CT values were plotted relative to the corresponding serial 10-fold dilutions of template DNA extracted from cultures of representative clostridial stool isolates of different Clostridium clusters: a mixed culture of C. subterminale WAL 16483, C. perfringens WAL 14572, and C. paraputrificum WAL 14502 for cluster I; a mixed culture of C. bifermentans WAL 16496, C. difficile WAL 14186, and C. glycolicum WAL 14424 for cluster XI; and a mixed culture of C. symbiosum WAL 14673, C. bolteae WAL 16351 and Ruminococcus gnavus WAL 16098 for cluster XIVab. Briefly, 1 ml of the overnight thioglycolate broth cultures of these bacterial strains was subjected to DNA extraction by using the QIAamp DNA stool mini kit. Ten-fold serial dilutions of these bacterial DNAs were prepared. The corresponding CFU per PCR were calculated based on plate counts, as described above.
For comparison of PCR amplification efficiencies and detection sensitivities among different experiments, slopes of the standard curves were calculated by performing a linear regression analysis with the SmartCycler software. A mixture of all PCR reagents without any DNA was used as a negative control. Amplification efficiency (E) was estimated by using the slope of the standard curve and the formula E = (10−1slope)−1. A reaction with 100% efficiency will generate a slope of −3.32. The standard curves were used for determining the detection limits of the assays and enumeration of C. bolteae and members of each of the three Clostridium clusters in stool samples.
Enumeration of Clostridium in stool samples by real-time PCR.
Purified DNA from stool samples was used with optimized PCR conditions and an appropriate standard curve to enumerate the load of C. bolteae and members of different Clostridium clusters in the stool samples. Each stool sample was subjected to four PCR runs (2 PCR repetitions × 2 DNA extractions; n = 4). The amount of DNA measured by real-time PCR was converted to cell numbers to allow comparison with the CFU data. This was accomplished by using the standard curve that was generated by plotting the CT against CFU. This approach was used because, for stool samples, it is easier to understand results in actual CFU numbers than in DNA concentrations or copy numbers.
The CT standard deviations were calculated as shown in Table 3. The CFU of C. bolteae and the members of each cluster were determined from CT values by using the standard curves (Fig. 1 and 2).
TABLE 3.
Clostridium detection in stool samples of autistic and control children by real-time PCRa
| Sampleb |
C. bolteae
|
Clostridium cluster I
|
Clostridium cluster XI
|
Clostridium cluster XIVab
|
||||
|---|---|---|---|---|---|---|---|---|
| Mean CT (SD) | CFU/g of fecal specimen | Mean CT (SD) | CFU/g of fecal specimen | Mean CT (SD) | CFU/g of fecal specimen | Mean CT (SD) | CFU/g of fecal specimen | |
| Control | ||||||||
| 98-1306C | 33.6 (0.22) | (4.0 ± 0.3) × 102 | 28.6 (0.31) | (3.1 ± 0.4) × 105 | 25.4 (0.21) | (3.5 ± 0.3) × 106 | 21.7 (0.33) | (5.9 ± 0.3) × 107 |
| 98-1332C | 35.2 (0.21) | (2.8 ± 0.2) × 102 | 31.6 (0.46) | (4.9 ± 0.4) × 104 | 26.3 (0.27) | (1.7 ± 0.3) × 106 | 18.5 (0.22) | (2.5 ± 0.4) × 108 |
| 98-1333C | 36.9 (0.29) | (9.2 ± 0.4) × 101 | 27.6 (0.45) | (6.6 ± 0.4) × 105 | 24.6 (0.25) | (6.2 ± 0.5) × 106 | 21.0 (0.23) | (7.5 ± 0.2) × 107 |
| 98-1334C | 31.4 (0.18) | (2.7 ± 0.2) × 103 | 28.4 (0.25) | (3.9 ± 0.3) × 105 | 26.4 (0.31) | (1.7 ± 0.2) × 106 | 18.0 (0.35) | (3.5 ± 0.3) × 108 |
| 98-1335C | 31.3 (0.24) | (3.3 ± 0.4) × 103 | 27.6 (0.46) | (6.6 ± 0.3) × 105 | 26.6 (0.21) | (1.5 ± 0.2) × 106 | 20.5 (0.26) | (9.6 ± 0.2) × 107 |
| 98-1342C | 30.8 (0.22) | (5.6 ± 0.4) × 103 | 31.9 (0.39) | (4.3 ± 0.3) × 104 | 25.7 (0.13) | (2.6 ± 0.4) × 106 | 18.1 (0.31) | (3.4 ± 0.3) × 108 |
| 00-1318C | <DL | <DL | 28.5 (0.23) | (3.5 ± 0.4) × 105 | 25.5 (0.12) | (2.9 ± 0.4) × 106 | 22.0 (0.25) | (4.6 ± 0.3) × 107 |
| 00-1319C | 28.6 (0.15) | (1.9 ± 0.1) × 104 | 27.1 (0.21) | (8.5 ± 0.5) × 105 | 23.6 (0.11) | (1.2 ± 0.1) × 107 | 16.5 (0.38) | (8.8 ± 0.2) × 108 |
| Avg | (3.9 ± 0.3) × 103 | (4.1 ± 0.3) × 105 | (4.0 ± 0.4) × 106 | (2.6 ± 0.2) × 108 | ||||
| Autistic | ||||||||
| 97-1301A | 31.5 (0.17) | (2.7 ± 0.2) × 103 | 24.6 (0.22) | (5.5 ± 0.2) × 106 | 21.4 (0.08) | (6.7 ± 0.2) × 107 | 18.5 (0.29) | (2.5 ± 0.3) × 108 |
| 97-1316A | 30.0 (0.16) | (7.6 ± 0.2) × 103 | 24.4 (0.24) | (6.6 ± 0.2) × 106 | 22.9 (0.17) | (2.2 ± 0.3) × 107 | 17.4 (0.41) | (4.9 ± 0.3) × 108 |
| 97-1321A | 24.1 (0.10) | (3.9 ± 0.1) × 105 | 25.1 (0.32) | (3.3 ± 0.3) × 106 | 22.5 (0.13) | (3.0 ± 0.3) × 107 | 18.6 (0.25) | (2.4 ± 0.4) × 108 |
| 98-1301A | 28.4 (0.18) | (2.0 ± 0.3) × 104 | 30.9 (0.44) | (8.2 ± 0.6) × 104 | 25.3 (0.23) | (3.8 ± 0.5) × 106 | 22.4 (0.09) | (3.9 ± 0.1) × 107 |
| 98-1343A | 27.7 (0.22) | (3.2 ± 0.4) × 104 | 25.7 (0.34) | (2.6 ± 0.3) × 106 | 26.6 (0.22) | (1.6 ± 0.3) × 106 | 18.9 (0.17) | (1.5 ± 0.2) × 108 |
| 98-1344A | 29.3 (0.17) | (1.2 ± 0.2) × 104 | 28.2 (0.21) | (4.7 ± 0.4) × 105 | 24.3 (0.20) | (8.6 ± 0.4) × 106 | 15.5 (0.30) | (1.6 ± 0.3) × 109 |
| 00-1311A | 22.9 (0.10) | (7.3 ± 0.1) × 105 | 27.8 (0.44) | (5.9 ± 0.3) × 105 | 21.4 (0.14) | (1.6 ± 0.3) × 106 | 18.9 (0.17) | (1.4 ± 0.2) × 108 |
| 00-1312A | 33.0 (0.13) | (1.2 ± 0.1) × 103 | 26.1 (0.38) | (2.4 ± 0.3) × 106 | 25.5 (0.15) | (3.2 ± 0.6) × 106 | 18.7 (0.24) | (2.2 ± 0.4) × 108 |
| 00-1313A | 26.0 (0.08) | (9.5 ± 0.1) × 104 | 26.6 (0.37) | (1.4 ± 0.3) × 106 | 24.6 (0.22) | (6.1 ± 0.6) × 106 | 17.7 (0.52) | (4.2 ± 0.5) × 108 |
| 00-1314A | 26.3 (0.21) | (5.0 ± 0.4) × 104 | 23.3 (0.24) | (1.3 ± 0.2) × 107 | 22.5 (0.18) | (2.8 ± 0.2) × 107 | 18.5 (0.22) | (2.7 ± 0.2) × 108 |
| 00-1315A | 28.0 (0.15) | (2.7 ± 0.2) × 104 | 29.1 (0.29) | (2.2 ± 0.5) × 105 | 23.8 (0.15) | (1.1 ± 0.1) × 107 | 20.6 (0.25) | (8.4 ± 0.1) × 107 |
| 00-1316A | 31.4 (0.17) | (3.3 ± 0.2) × 103 | 24.8 (0.30) | (4.8 ± 0.3) × 106 | 23.3 (0.18) | (1.5 ± 0.1) × 107 | 15.1 (0.34) | (2.4 ± 0.4) × 109 |
| 00-1317A | 30.4 (0.19) | (7.1 ± 0.3) × 103 | 23.6 (0.24) | (1.1 ± 0.2) × 107 | 23.8 (0.13) | (1.1 ± 0.1) × 107 | 22.1 (0.27) | (4.4 ± 0.3) × 107 |
| 02-1409A | 22.0 (0.10) | (1.3 ± 0.1) × 106 | 29.9 (0.31) | (1.7 ± 0.3) × 105 | 24.6 (0.25) | (6.8 ± 0.3) × 106 | 17.3 (0.42) | (5.1 ± 0.1) × 108 |
| 03-1404A | 26.6 (0.13) | (5.0 ± 0.2) × 104 | 25.4 (0.33) | (2.9 ± 0.2) × 106 | 22.4 (0.07) | (3.0 ± 0.2) × 107 | 18.5 (0.17) | (2.7 ± 0.2) × 108 |
| Avg | (1.8 ± 0.1) × 105 | (3.7 ± 0.4) × 106 | (1.4 ± 0.1) × 107 | (4.8 ± 0.6) × 108 | ||||
Mean CT represents two PCR replicates of two replicate DNA extractions (n = 4). CFU were calculated by using the standard curve. <DL, below the detection limit of 0.6 CFU/reaction; SD, standard deviation.
C, control children; A, autistic children.
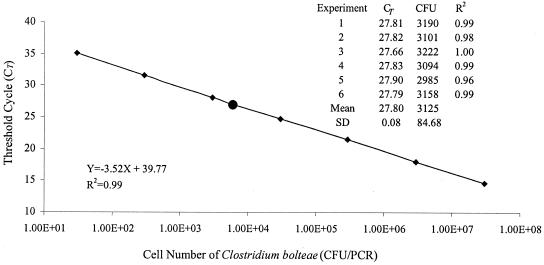
FIG. 1.
Standard curve generated by analysis of a dilution series of C. bolteae cells spiked in stools by real-time PCR. Quantitation was performed by determining the CT. The same experiment, which included a known C. bolteae-positive stool with an unknown count (•), was repeated six times. The CT values, the CFU/PCR determined, and R2 values of each experiment are listed in the inset. Means and the standard deviation were calculated from these six experiments. ⧫, C. bolteae standards.
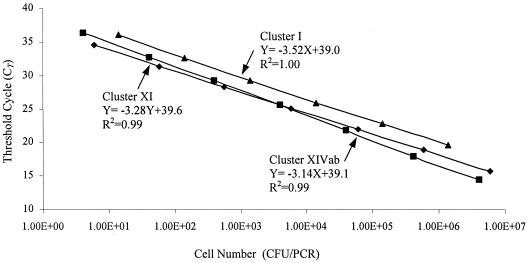
FIG. 2.
Standard curves generated by analysis of a dilution series of DNA extracted from mixed cultures of representative strains for each Clostridium cluster by real-time PCR. The CT values are plotted against the corresponding cell numbers in the PCR. ▴, Clostridium cluster I; ⧫, Clostridium cluster XI; ▪, Clostridium cluster XIVab.
Statistical analysis.
Parametric t tests and multivariate logistic regression and/or classification tree (CART) method (6) was used to compare mean CFU values between autistic and control samples.
RESULTS
Specificity of oligonucleotide primers.
The results of comprehensive cross-specificity checks on the RDP database enabled the final selection of four 16S rDNA-targeted PCR primer pairs theoretically specific for C. bolteae or each of three Clostridium clusters (Table 2). Degenerate PCR primers were used in order to broaden the specificity within each cluster. The primer pair CI-F1 and CI-R2 is specific for cluster I, which includes most species of the Clostridium cluster I and three species of cluster II. Primer pair CXI-F1 and CXI-R2 is specific for cluster XI, which includes part of Clostridium cluster XI. Finally, primer pair CXIV-F1 and CXIV-R2 is specific for cluster XIVab, which includes most species belonging to Clostridium cluster XIVab. The specificity of species- and cluster-specific primers was further confirmed by amplifying genomic DNA from target and phylogenetically related nontarget bacterial strains (Table 1) with each of the four sets of primers by conventional PCR and real-time PCR. In conventional PCR, the primer pairs were specific for their target species or cluster at the appropriate annealing temperatures and yielded PCR products of the expected size only from their targets. The optimized conventional PCR conditions were applied to real-time PCR. In real-time PCR, no fluorescent signal was detected from non-target bacterial DNA (data not shown). Although our TaqMan probe and reverse primer for C. bolteae also showed 100% homology with 16S rDNA of its most closely related species, C. clostridioforme, the forward primer did not show any major sequence homology (70.5% only); there was no signal detected from DNA from C. clostridioforme strains.
Standard curve and detection limits.
In real-time PCR assay, the CT at which the fluorescent signal is statistically significant above background is measured. The CT value is proportional to the amount of target DNA and hence the number of bacteria in the samples. The detection sensitivity for C. bolteae was determined by two sets of genomic DNA prepared from 10-fold serial dilutions (30 to 3.0 × 10 7 CFU/ml) of C. bolteae type strain ATCC BAA-613 spiked into a C. bolteae-negative stool sample. The detection sensitivities for three Clostridium clusters were determined by two sets of 10-fold serial dilutions of genomic DNA prepared from representatives of each cluster. Representative data are shown in Fig. 1 and 2. The results are reported as the CT numbers versus CFU numbers.
As shown in Fig. 1 and 2, plotting the obtained CT values relative to the CFU of bacteria resulted in a linear correlation with square regression coefficient of ≥0.99, demonstrating that quantification of the target DNA was possible. To evaluate the robustness of the assays, the same experiment, which included an unknown stool sample, was repeated six times on different days with two different sets of DNA. The result for C. bolteae is given in the inset in Fig. 1. The standard deviation of CT values for the unknown sample based on the six PCR runs (3 PCR repetitions × 2 DNA extractions; n = 6) were 0.08, 0.37, 0.22, and 0.52 (Table 3); determined by the C. bolteae and cluster I, XI, and XIVab standard curves, respectively. This showed that both DNA extraction by the QIAamp stool DNA extraction kit and the real-time PCR assay were highly reproducible.
The function describing the relationship between CT values and CFU for the assay has been calculated based on the mean values obtained from the six PCR runs (Fig. 1 and 2). Average slopes of −3.52, −3.52, −3.28, and −3.14 were generated for C. bolteae and clusters I, XI, and XIVab, respectively. According to the formula log E = slope−1, the standard curves generated from the PCR data resulted in a reaction efficiency of above 96.5% for all four PCR assays.
The estimated detection limits for the primer and probe sets specific for C. bolteae and clusters I, XI, and XIVab were 0.6, 2, 6, and 4 bacteria per PCR, respectively. All nontemplate controls and all dilutions with DNA corresponding to less than these numbers of cells per reaction gave negative results.
Enumeration of Clostridium in stool samples by real-time PCR.
The bacterial load of C. bolteae and each Clostridium cluster in stools was estimated by real-time PCR with DNA extracted from the 23 samples (Table 3). The reproducibility of the assay was high, with the standard deviation of CT values for stool samples ranging from 0.08 to 0.29 for C. bolteae, 0.21 to 0.46 for cluster I, 0.07 to 0.31 for cluster XI, and 0.09 to 0.52 for cluster XIVab. The mean counts (CFU per gram [wet weight of centrifuged specimens] of feces) of C. bolteae and members of clusters I, XI, and XIVab were 1.8 × 105 ± 0.1 × 105, 3.7 × 106 ± 0.4 × 106, 1.4 × 107 ± 0.1 × 107, and 4.8 × 108 ± 0.6 × 108 in autistic children, respectively, and were 3.9 × 103 ± 0.3 × 103, 4.1 × 105 ± 0.3 × 105, 4.0 × 106 ± 0.4 × 106, and 2.6 × 108 ± 0.2 × 108 in controls, respectively. Analysis of the real-time PCR data indicated that the cell count differences between autistic and control children in C. bolteae and the following Clostridium groups were statistically significant: mean counts of C. bolteae and clusters I and XI in autistic children were 46-fold (P = 0.01), 9.0-fold (P = 0.014), and 3.5-fold (P = 0.004) greater than those in control children, respectively, but the differences in cluster XIVab between autistic and control children were not statistically significant (P = 0.3). More subjects need to be studied for all groups.
DISCUSSION
Real-time PCR has several advantages over conventional PCR. It focuses on the logarithmic phase of product accumulation rather than on the end product abundance. Therefore, it is more accurate since it is less affected by amplification efficiency or depletion of a reagent. In addition, it has an increased dynamic range for quantification of target sequence (at least 5 orders of magnitude). Furthermore, without any post-PCR manipulation of the samples, cross-contamination between samples is greatly reduced. Finally, PCR results can be obtained within 1 h. In the present study, we developed quantitative real-time TaqMan PCR assays targeting the 16S rRNA for detection and quantitation of C. bolteae and members of three Clostridium clusters (I, XI, and XIVab) in stool samples. The methods were successful in detecting and quantitating Clostridium in pure culture and in stool samples from autistic children and control children.
Good detection methods share the following features: specificity, sensitivity, and reproducibility. Real-time PCR appears to possess all these features. In the present study, specificity, a key factor for accurate quantitation of the bacteria of interest, was determined both by homology search in nucleotide databases and by testing bacterial strains with different degrees of relatedness to the target species based on 16S rRNA comparative analysis. Although theoretical cross-specificity analysis of the primers indicated that the cluster XIVab-specific primer pair may cross-react with some nontarget bacteria (only one side primer contained 2-bp mismatches), such as Anaeroplasma bactoclasticum, Anaeroplasma varium, and Mycoplasma feliminutum, these bacteria have never been reported as being isolated from human gut. Since the primers were designed on the basis of currently available 16S rRNA sequences, the specificity is not guaranteed for unknown gut bacteria. It is an advantage, then, that these probes are designed on a phylogenetic basis and that related species are more likely to have the same target sequences.
The reason we used 16S rDNA as a target to design the probe and primers is that the large 16S rDNA database available allows the identification of sequences exclusive to our target species. As we know, the complex microflora of the human gut is difficult to study with probes or primers on a species level due to the diversity of the ecosystem. Therefore, it is more convenient to have probes or primers specific for genera or groups present in the gut. For intertwined genera such as Clostridium, genus-specific probes or primers cannot be designed and primers and/or probes for phylogenetic clusters have to be considered. Analysis of Clostridium 16S rRNA sequences has enabled the phylogeny of the genus Clostridium to be described and major Clostridium clusters to be identified (7). This in turn has enabled the design and development of cluster-specific 16S rDNA-targeted oligonucleotide probes and PCR primers (12, 13, 23, 31). The major disadvantage of this approach to design species-specific primers is that the variable regions within the 16S rDNA can be almost identical for closely related bacteria. To overcome this problem, stringent conditions such as the hot-start PCR technique and high annealing temperature, were used to exclude amplification of organisms closely related to C. bolteae. Thus, even though the TaqMan probe and the reverse primer show 100% homology with the corresponding regions of 16S rRNA of C. clostridioforme, no signal was generated when the real-time PCR was performed with this set of oligonucleotides. It is also unlikely that other non-target closely related DNA in stool specimens would be amplified.
The TaqMan assays we developed proved very sensitive and reproducible. The standard curves showed that the estimated detection limit for primer and probe sets specific for C. bolteae and Clostridium clusters I, XI, and XIVab were 0.6, 2, 6, and 4 CFU per PCR, respectively. We found that the reproducibility of the quantitative experiments was over 99%, based on multiple replicate PCR runs. When quantifying targets using real-time PCR, both the variability attributable to DNA yields between extractions and the PCR assay itself need to be considered. Commercially available kits for the isolation of genomic DNA from stool samples are desirable due to the high throughput and elimination of phenol, which requires special disposal protocols and can affect PCR results. In this study, the QIAamp stool DNA extraction kit was used for stool DNA extraction. It proved to be good in providing DNA suitable for the assay. The small standard deviation of mean CT values (Table 3) confirmed the reproducibility of the DNA extraction method and was associated with only a small amount of error during the PCR analysis.
A challenge in using the real-time PCR method is to convert measurements of fluorescent signal into target cell densities. The approach used in this study is to directly relate the signal to cell numbers by a plate counting method. It is therefore necessary to exercise some caution in selecting which Clostridium strains are used to establish standard curves. It is known that the final determination of bacterial load by real-time PCR in a multispecies population will be influenced by the variation in the number of rRNA operons in a given species. Unfortunately, information on the number of 16S rRNA genes per genome in Clostridium species is very limited (rRNA Operon Copy Number Database; http://rrndb.cme.msu.edu/rrndb/servlet/controller/controller?page = home), so only presumptions can be made about the size of the different clostridial cluster populations. However, in a complex ecosystem like a stool sample, other methodologies are likely to be far less sensitive or precise. In this study, several Clostridium stool isolates in each cluster (C. butyricum, C. paraputrificum, C. perfringens, C. subterminale, and Clostridium tertium in cluster I; C. bifermentans, C. difficile, and C. glycolicum in cluster XI; and C. symbiosum, C. bolteae, R. gnavus, R. torques, and R. lactaris in cluster XIVab) were tested for constructing a standard curve. The standard curves obtained from the different strains within the same cluster were in good agreement (data not shown). This suggests that the sensitivity of measurement by these assays may not vary greatly depending on the species or strain. We used three Clostridium strains, which were isolated from stool specimens of autistic or control children in another study (11), for each cluster to construct the standard curves.
Since stool samples contain diverse bacterial communities, the amplification of C. bolteae DNA to construct a standard curve was performed with the background of stool community DNA. We spiked the dilution series of C. bolteae cells into one C. bolteae-negative stool sample to determine the PCR efficiency and reliability under the usual stool community DNA conditions. However, there was no stool sample that was Clostridium negative; therefore, we used DNA extracted from pure cultures of representative strains from each cluster to construct their standard curves.
Our previous study compared culture methodologies with PCR for the detection of C. bolteae from stool samples, with the conclusion that C. bolteae detection by PCR is more sensitive and more reliable than culture (29). The findings in this study by real-time PCR are in agreement with the previous observations and provide reliable quantitative information. Analysis of the real-time PCR data indicated that the cell count differences between autistic and control children in C. bolteae and the Clostridium clusters I and XI were statistically significant; however, we feel that to be confident about it, we need to study large numbers of samples.
In summary, we developed a new method based on real-time PCR that allowed for fast, reliable quantitation of C. bolteae and three Clostridium clusters in stool samples. C. bolteae is also a pathogen encountered in clinically significant infections in humans. Therefore, this procedure may also be useful in recovering C. bolteae from clinical specimens with complicated mixed floras. To our knowledge, this study demonstrates for the first time the potential of the real-time PCR technique for quantifying Clostridium clusters in human stool. Investigation of the intestinal microflora is crucial for obtaining an understanding of the role of the gut microflora in health and also an understanding of the role of the microflora in disease (e.g., inflammatory bowel disease—Crohn's disease and ulcerative colitis). The assay is rapid and reliable, and we assume that it has great potential for quantitation of other bacteria in the intestinal tract. A major advantage of this method is that it lends itself to high throughput.
Acknowledgments
This work was funded, in part, by VA Merit Review funds.
Statistical analysis was performed by Jeffrey A. Gornbein of the Biomath Consulting Clinic, UCLA School of Medicine.
REFERENCES
- 1.Bélanger, S. D., M. Boissinot, N. Clairoux, F. J. Picard, and M. G. Bergeron. 2003. Rapid detection of Clostridium difficile in feces by real-time PCR. J. Clin. Microbiol. 41:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bélanger, S. D., M. Boissinot, C. Ménard, F. J. Picard, and M. G. Bergeron. 2002. Rapid detection of Shiga toxin-producing bacteria in feces by multiplex PCR with molecular beacons on the Smart Cycler. J. Clin. Microbiol. 40:1436-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2004. GenBank: update. Nucleic Acids Res. 32:23-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blessmann, J., H. Buss, P. A. T. Nu, B. T. Dinh, Q. T. Viet Ngo, A. L. Van, M. D. A. Alla, T. F. H. G. Jackson, J. I. Ravdin, and E. Tannich. 2002. Real-time PCR for detection and differentiation of Entamoeba histolytica and Entamoeba dispar in fecal samples. J. Clin. Microbiol. 40:4413-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunde, J., and M. Fitzpatrick. 1993. Estimating the number of species: a review. J. Am. Statistical Assoc. 88:364-373. [Google Scholar]
- 7.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. E. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 8.Donskey, C. J., A. M. Hujer, S. M. Das, N. J. Pultz, R. A. Bonomo, and L. B. Rice. 2003. Use of denaturing gradient gel electrophoresis for analysis of the stool microbiota of hospitalized patients. J. Microbiol. Methods 54:249-256. [DOI] [PubMed] [Google Scholar]
- 9.Fang, Y., W.-H. Wu, J. L. Pepper, J. L. Larsen, S. A. E. Marras, E. A. Nelson, W. B. Epperson, and J. Christopher-Hennings. 2002. Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for detection of Mycobacterium avium subsp. paratuberculosis in bovine fecal samples. J. Clin. Microbiol. 40:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felske, A., A. D. L. Akkermans, and W. M. De Vos. 1998. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finegold, S. M., D. Molitoris, Y. Song, C. Liu, M. L. Vaisanen, E. Bolte, M. McTeague, R. Sandler, H. Wexler, E. M. Marlowe, M. D. Collins, P. A. Lawson, P. Summanen, M. Baysallar, T. J. Tomzynski, E. Read, E. Johnson, R. Rolfe, P. Nasir, H. Shah, D. A. Haake, P. Manning, and A. Kaul. 2002. Gastrointestinal microflora studies in late-onset autism. Clin. Infect. Dis. 35(Suppl. 1):S6-S16. [DOI] [PubMed] [Google Scholar]
- 12.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harmsen, H. J., G. R. Gibson, P. Elfferich, G. C. Raangs, A. C. Wildeboer-Veloo, A. Argaiz, M. B. Roberfroid, and G. W. Welling. 2000. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol. Lett. 183:125-129. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535-548. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Fecal microbial diversity in a strict vegetarian as determined by molecular analysis and cultivation. Microbiol. Immunol. 46:819-831. [DOI] [PubMed] [Google Scholar]
- 16.Huijsdens, X. W., R. K. Linskens, M. Mak, S. G. M. Meuwissen, C. M. J. E. Vandenbroucke-Grauls, and P. H. M. Savelkoul. 2002. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40:4423-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, B., S. Kawasaki, H. Nakano, and T. Fujii. 2001. Rapid, quantitative PCR monitoring of growth of Clostridium botulinum type E in modified-atmosphere-packaged fish. Appl. Environ. Microbiol. 67:206-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurowski, P. B., J. L. Traub-Dargatz, P. S. Morley, and C. R. Gentry-Weeks. 2002. Detection of Salmonella spp in fecal specimens by use of real-time polymerase chain reaction assay. Am. J. Vet. Res. 63:1265-1268. [DOI] [PubMed] [Google Scholar]
- 19.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malinen, E., A. Kassinen, T. Rinttila, and A. Palva. 2003. Comparison of real-time PCR with SYBR Green I or 5′-nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected faecal bacteria. Microbiology 149:269-277. [DOI] [PubMed] [Google Scholar]
- 22.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 26.Ott, S. J., M. Musfeldt, U. Ullmann, J. Hampe, and S. Schreiber. 2004. Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric flora. J. Clin. Microbiol. 42:2566-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 28.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song, Y., C. Liu, D. R. Molitoris, T. J. Tomzynski, P. A. Lawson, M. D. Collins, and S. M. Finegold. 2003. Clostridium bolteae sp. nov., isolated from human sources. Syst. Appl. Microbiol. 26:84-89. [DOI] [PubMed] [Google Scholar]
- 30.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Dyke, M. I., and A. J. McCarthy. 2002. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Appl. Environ. Microbiol. 68:2049-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, R.-F., W.-W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolk, D. M., S. K. Schneider, N. L. Wengenack, L. M. Sloan, and J. E. Rosenblatt. 2002. Real-time PCR method for detection of Encephalitozoon intestinalis from stool specimens. J. Clin. Microbiol. 40:3922-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]