Abstract
The thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) are the main virulence factors of Vibrio parahaemolyticus. We isolated V. parahaemolyticus from seawater, fish, and oysters obtained from the Pueblo Viejo Lagoon in Veracruz, determined the serogroups, phenotypically and genotypically characterized TDH and TRH, and investigated the presence of the toxR gene. A total of 46 V. parahaemolyticus strains were isolated, and all of them amplified the 368-bp toxR gene fragment. The trh gene was not identified in any of the strains; 4 of the 46 strains were Kanagawa phenomenon (KP) positive and amplified the 251-bp tdh gene fragment. The most frequent serogroup was serogroup O3. This is the first report of the presence of KP-positive tdh-positive environmental V. parahaemolyticus strains in Mexico.
Vibrio parahaemolyticus is a halophilic gram-negative bacterium that is widely distributed in coastal waters worldwide and is associated with gastroenteritis, wound infections, and septicemia (5). V. parahaemolyticus infections are frequently reported in coastal areas, apparently because of the high consumption of sea products and direct contact with estuarine waters (19).
Epidemiological studies have revealed an association between the Kanagawa phenomenon (KP) and gastroenteritis (23, 25). KP is a type of beta-hemolysis induced by the thermostable direct hemolysin (TDH) in Wagatsuma agar. Most (90%) of the strains isolated from clinical cases show this type of hemolysis, while only 1 to 2% of the strains of environmental origin are KP positive (20).
Several cases of gastroenteritis caused by hemolytic but KP-negative TDH-negative V. parahaemolyticus strains were reported in the 1980s (11), which led to identification of a new hemolysin known as TDH-related hemolysin (TRH) (12, 13, 30). TDH and TRH are encoded by the tdh and trh genes, respectively; these two genes both have 567-bp open reading frames, and they show 68.6% sequence similarity. These hemolysins are considered the main virulence factors of this microorganism (20).
In the present study we determined the prevalence of V. parahaemolyticus in seawater, oyster, and fish samples collected from the Pueblo Viejo Lagoon in Veracruz, an important estuary on the coastline of the Gulf of Mexico. The strains isolated were serotyped and screened for hemolytic activity and for the presence of the toxR, tdh, and trh genes.
MATERIALS AND METHODS
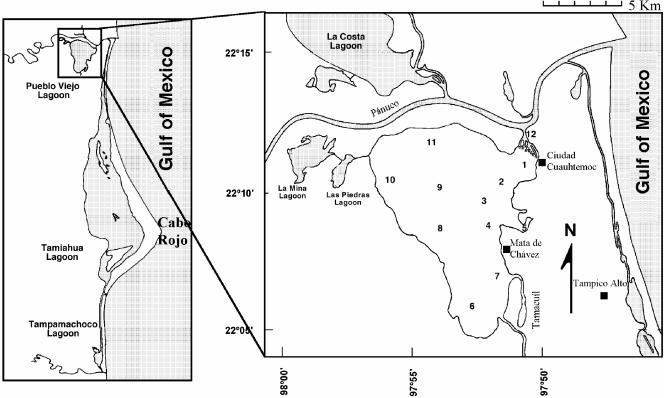
A total of 266 seawater, oyster, and fish samples were collected from 12 different sites in the Pueblo Viejo Lagoon (Fig. 1) in June, August, September, and November 2001, as well as in February; March, April, May, and June 2002.
FIG. 1.
Distribution of the 12 sampling sites in the Pueblo Viejo Lagoon, Veracruz. Site 1, El Bajo; site 2, Punto de Buda; site 3, Santa Clara; site 4, Ensenada; site 5, El Ciruelo; site 6, Cruz de Piedra; site 7, Isleta de Coralillo; site 8, Isleta Tomate; site 9, Medianía; site 10, Tamales; site 11, Punta de Mala Gana; site 12, Puente Hondo. Modified from reference 3.
The fish and oysters were transported in individually labeled and sealed plastic bags to avoid contamination. Seawater samples were collected in labeled plastic jars. The samples were placed in sealed containers with dry ice and transported frozen to the laboratory for analysis. The time between sample collection and analysis was approximately 24 h.
V. parahaemolyticus strain isolation and identification.
V. parahaemolyticus was isolated and identified as described in the Bacteriological Analytical Manual of the Food and Drug Administration (8). All commercial reagents used were obtained from Difco unless indicated otherwise. The oysters were shucked in aseptic conditions. The heads and tails of the fish were cut off, and the guts were removed. The oyster and fish samples were then homogenized in blenders, and 50 g of each homogenate was placed in 450 ml of alkaline peptone water (APW) to obtain a 10−1 dilution. Seawater samples (25 ml) were added to 225 ml of APW, and 10−2 and 10−3 APW dilutions were prepared in duplicate and incubated at 37 and 42°C for 6 to 24 h. Enrichment broth was streaked onto thiosulfate-citrate-bile salts-sucrose agar plates and incubated at 37°C for 18 to 24 h. Two or three suspect V. parahaemolyticus colonies (round, 1 to 2 mm in diameter, humid, shiny, sucrose negative) were selected. Halophilism tests were performed on NaCl-tryptone agar (T1N0, T1N3, T1N6, T1N8, and T1N10). Additional characterization tests included Gram staining, cytochrome oxidase activity tests, ornithine indole motility biochemical tests, triple sugar iron tests, lysine iron agar tests, arginine dehydrolase tests, urea tests, tests for glucose oxidation-fermentation in Hugh-Leifson broth, and tests for arabinose, lactose, mannitol, mannose, and sucrose fermentation.
Serotyping.
Serogroups were determined as described by Elliot et al. (8) by using commercial antisera (Denka Seiken, Co., Ltd., Tokyo Japan).
Hemolytic activity.
V. parahaemolyticus strains were seeded in Wagatsuma agar prepared as described by Elliot et al. (9); this agar contained 3 g of yeast extract, 10 g of peptone, 70 g of NaCl, 5 g of K2HPO4, 10 g of mannitol, 0.001 g of violet crystal, 15 g of agar, 1 liter of distilled water, and 50 ml of human anticoagulated blood. The bacteria were incubated at 37°C for 18 h. In order to characterize the TRH phenotype, strains were seeded in heart infusion broth and SPP broth (5 g of NaCl, 5 g of NaHPO4, and 2 g of glucose in 1 liter of distilled water; pH 7.6). Hemolytic activity was determined as described by Honda et al. (13) and Kishishita et al. (18). Positive and negative controls were included in all assays (Table 1).
TABLE 1.
Reference strains
| Strain | Serotype | Genes
|
Reference or source | ||
|---|---|---|---|---|---|
| tdh | trh1 | trh2 | |||
| V. parahaemolyticus VPAQ4037 | O3:K6 | − | + | − | 22 |
| V. parahaemolyticus VPAT4 | O4:K37 | − | − | + | 18 |
| V. parahaemolyticus VPWP1 | O4:K12 | + | − | − | 21 |
| V. parahaemolyticus VP17802 | O1 | − | − | − | ATCCa |
| V. alginolyticus VA-219 | − | − | − | Mitsuaki Nishibuchi, Kyoto, Japan | |
ATCC, American Type Culture Collection.
DNA purification.
V. parahaemolyticus strains were seeded in Luria-Bertani agar supplemented with 1% NaCl and incubated at 37°C for 24 h. Five colonies were selected, resuspended in 100 μl of filtered sterile distilled water, and boiled for 15 to 20 min to liberate the nucleic acid.
PCR.
The presence of the toxR gene was determined by using the following primers described by Kim et al. (17): 5′-GTCTTCTGACGCAATCGTTG-3′ (forward) and 5′-ATACGAGTGGTTGCTGTCATG-3′ (reverse), which produced a 368-bp amplicon. The PCR conditions were as follows: the reaction mixture (final volume, 25 μl) consisted of 3 μl of the solution containing DNA, 2.5 μl of 10× reaction buffer (Applied Biosystems), 6 μl of 25 mM MgCl2, 1 μl of Taq polymerase (5 U/μl), 4 μl of deoxynucleoside triphosphates (1 μM), 1 μl of each primer (0.0125 ng/μl), and 6.5 μl of distilled water. The reactions were performed with a Gene Amp PCR system 2700 thermocycler (Applied Biosystems) as follows: 1 min of initial denaturation at 94°C, followed by 20 cycles of denaturation at 94°C for 1 min, alignment at 63°C for 1.5 min, and extension at 72°C for 1.5 min and a final extension at 72°C for 7 min.
The tdh and trh genes were amplified with the following primer sets: 5′-GGTACTAAATGGCTGACATC-3′ (forward) and 5′-CCACTACCACTCTCATATGC-3′ (reverse) for tdh; and 5′-GGCTCAAAATGGTTAAGCG-3′ (forward) and 5′-CATTTCCGCTCTCATATGC-3′ (reverse) for trh (26). These primer sets produced 251- and 250-bp amplicons, respectively. The reaction mixtures (final volume, 25 μl) contained 3 μl of the solution containing DNA, 2.5 μl of 10× reaction buffer (Applied Biosystems), 4 μl of 25 mM MgCl2, 1 μl of Taq polymerase (5 U/μl), 4 μl of deoxynucleoside triphosphates (1 μmol), 1 μl of each primer (0.025 ng/μl), and 8.5 μl of distilled water. The reactions were performed with a Gene Amp PCR system 2700 thermocycler (Applied Biosystems) as follows: 1 min of initial denaturation at 94°C, followed by 35 cycles of denaturation at 94°C for 1 min, alignment at 55°C for 1 min, and extension at 72°C for 1 min and a final extension at 72°C for 7 min.
Positive and negative DNA controls were included in all assays (Table 1). Amplified products were separated by electrophoresis in ethidium bromide-stained 2% agarose gels in Tris-borate-EDTA buffer at 120 V for 30 min. The gels were visualized with a UV transilluminator (Eagle Eye; Stratagene).
RESULTS
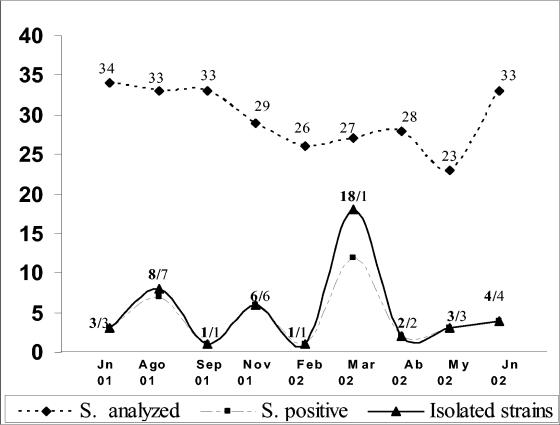
A total of 266 samples were analyzed; these samples included 103 seawater samples, 75 oyster samples, and 88 fish samples. Overall, V. parahaemolyticus strains were isolated from 15% of the samples (Fig. 2 and Table 2). Of the 46 strains isolated, 20 (43%) were obtained from seawater samples, 17 (37%) were obtained from fish, and 9 (20%) were obtained from oysters.
FIG. 2.
Number of samples (S) from the Pueblo Viejo Lagoon positive for V. parahaemolyticus over the study period. Jn, June; Ago, August; Sep, September; Nov, November; Feb, February; Mar, March; Ab, April; My, May.
TABLE 2.
Origins and serotypes of the V. parahaemolyticus strains isolated at different times
| Month | No. of samples analyzed | No. of samples positive | No. of colonies assessed | No. of colonies confirmed | Sampling site | Sample type | No. of strains | Serogroup |
|---|---|---|---|---|---|---|---|---|
| June 2001 | 34 | 3 | 99 | 3 | Santa Clara | Fish | 1 | O3 |
| El Ciruelo | Fish | 1 | O3 | |||||
| El Bajo | Seawater | 1 | O3 | |||||
| August 2001 | 33 | 7 | 102 | 8 | Isleta de Coralillo | Fish | 2 | O4; O4 |
| El Ciruelo | Fish | 1 | O3 | |||||
| Mediania | Fish | 1 | O1 | |||||
| Seawater | 1 | O10 | ||||||
| Cruz de Piedra | Seawater | 1 | O10 | |||||
| Punta de Buda | Seawater | 1 | O3 | |||||
| Santa Clara | Seawater | 1 | O10 | |||||
| September 2001 | 33 | 1 | 69 | 1 | Ensenada | Seawater | 1 | O3 |
| November 2001 | 29 | 6 | 81 | 6 | Isleta Tomates | Oyster | 1 | O1 |
| Seawater | 1 | O3 | ||||||
| Ensenada | Fish | 1 | O1 | |||||
| El Bajo | Fish | 1 | O1 | |||||
| El Ciruelo | Fish | 1 | O4 | |||||
| Cruz de Piedra | Seawater | 1 | O10 | |||||
| February 2002 | 26 | 1 | 104 | 1 | Mediania | Seawater | 1 | O4 |
| March 2002 | 27 | 12 | 122 | 18 | Punta de Malagana | Seawater | 1 | O2 |
| Puente Hondo | Seawater | 1 | O5 | |||||
| El Bajo | Seawater | 3 | O5; O3; O3 | |||||
| Fish | 1 | O1 | ||||||
| El Ciruelo | Seawater | 1 | O1 | |||||
| Fish | 1 | O3 | ||||||
| Cruz de Piedra | Oyster | 1 | O3 | |||||
| Punta de Buda | Fish | 3 | O2; O3; O5 | |||||
| Oyster | 1 | O1 | ||||||
| Mediania | Fish | 1 | O3 | |||||
| Santa Clara | Oyster | 3 | O1; O1; O10 | |||||
| Isleta Tomates | Seawater | 1 | O2 | |||||
| April 2002 | 28 | 2 | 96 | 2 | Punta de Buda | Oyster | 1 | O1 |
| Santa Clara | Oyster | 1 | O1 | |||||
| May 2002 | 23 | 3 | 108 | 3 | Mediania | Seawater | 1 | O10 |
| Punta de Malagana | Seawater | 1 | O2 | |||||
| Santa Clara | Oyster | 1 | O5 | |||||
| June 2002 | 33 | 4 | 101 | 4 | Santa Clara | Fish | 1 | O5 |
| Punta de Buda | Seawater | 1 | O3 | |||||
| El Bajo | Fish | 1 | O5 | |||||
| Ensenada | Seawater | 1 | O5 | |||||
| Total | 266 | 39 | 882 | 46 | 46 |
V. parahaemolyticus serotyping was performed for epidemiological purposes. The 46 strains were identified as members of 6 of the 11 acknowledged serogroups; serotype O3 was the most frequent serotype (30%) (Table 3).
TABLE 3.
Serotyping of the 46 strains isolated
| Serogroup | No. of strains |
|---|---|
| O1 | 11 |
| O2 | 4 |
| O3 | 14 |
| O4 | 4 |
| O5 | 7 |
| O10 | 6 |
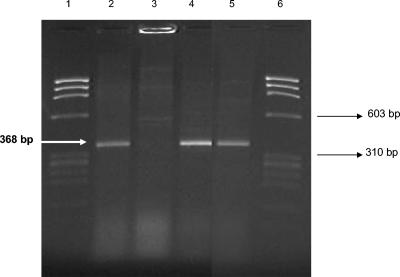
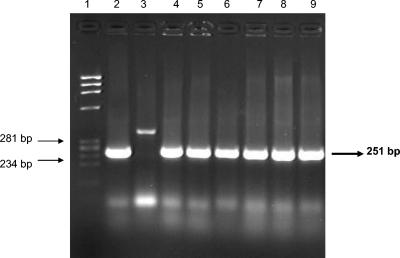
Only four strains showed hemolytic activity in Wagatsuma agar (KP+), the TRH phenotype was not identified in any of the 46 strains, and all strains were positive for the toxR gene amplifying the 368-bp fragment (Fig. 3). The presence of the tdh and trh genes is indicative of the pathogenic potential of V. parahaemolyticus. Only 4 of the 46 strains (9%) amplified the 251-bp tdh gene fragment, which correlated with positive hemolytic activity (KP+ strains) (Fig. 4). These four strains were serotype O3. The trh gene fragment was not amplified in any of the strains analyzed.
FIG. 3.
Amplification of the toxR gene by PCR. The primers amplified a 368-bp fragment. Lanes 1 and 6, molecular weight marker φX147/HaeIII; lane 2, control strain V. parahaemolyticus ATCC 17802; lane 3, negative control strain Va-219; lanes 4 and 5, experimental strains.
FIG. 4.
Amplification of the tdh gene by PCR. The primers amplified a 251-bp fragment. Lane 1, molecular weight marker φX174/HaeIII; lane 2, control strain V. parahaemolyticus WP1; lane 3, negative control strain Va-219; lanes 4 to 9, experimental strains.
DISCUSSION
V. parahaemolyticus is distributed in water environments and is associated with gastroenteritis, wound infections, and septicemia. Diseases caused by this pathogen are frequently reported in coastal states and are caused by seafood consumption and direct contact with estuarine waters (19).
In the present study we demonstrated the presence of V. parahaemolyticus in 15% of the seawater, fish, and oyster samples analyzed. This study was the first research study to investigate isolation and the distribution of this pathogen in a Mexican coastal lagoon. There have been reports of V. parahaemolyticus isolation in Canada (16), France (10), Asia (2, 28), the United States (6), and Mexico (27). The factors affecting the incidence and distribution of V. parahaemolyticus in the environment include water temperature, salt and oxygen concentrations, interactions with the plankton, the presence of sediment, the organic matter in suspension, fish, and seafood, as well as the incorporation and tidal action of estuarine waters. The presence of V. parahaemolyticus seems to be constant in the Gulf of Mexico because the temperature does not drop below 11.6°C, unlike what occurs in Japan, Europe, Australia, and the United States, where isolation of this pathogen decreases during the winter months (14).
We found V. parahaemolyticus in 11 of the 12 sampling sites; the only site at which we did not find this pathogen was the Tamales site, which was located in the northeast part of the lagoon. Strains were serotyped for epidemiological purposes, and serogroup O3 was the most frequent serogroup (30%) (Table 2). Several researchers have pointed out that certain serogroups are constant in some geographical areas (1, 24). Serotypes O3:K6, O4:K68, and O1:KUT (K untypeable) have been related to the majority of the infections caused by this pathogen in Asia and the United States during the last 5 years (4, 5).
We observed positive tdh gene amplification in 4 of the 46 strains analyzed, which correlated with positive hemolytic activity in Wagatsuma agar (KP+). Two of these strains were isolated from fish, and two were isolated from water samples. Most environmental strains are known to be KP−, and only 1 to 2% are KP+ (14, 20). This implies that there is a source of human fecal contamination in the estuarine waters of Pueblo Viejo Lagoon. The lagoon is surrounded by small rural populations, some of which use latrines that discharge feces into the lagoon. Furthermore, the water is very shallow (only 1.5 m deep during the rainy season), so fisherman, dogs, and other domestic animals can easily walk through it, which is an additional source of contamination.
Although it is known that 0 to 5% of the environmental strains produce TRH (10, 29), the trh gene and the TRH phenotype were not identified in any of the strains isolated here. The possibility of false-negative results is unlikely, as the positive control strain (ATCC 17802) (Table 1) amplified the 368-bp trh fragment in all assays performed. There have been few reports of the presence of the tdh and trh genes in V. parahaemolyticus strains of environmental origin; only 0 to 6% of the samples analyzed from the coasts of the United States (6, 7, 15), Europe (10), and Asia (2, 28) contained tdh-positive V. parahaemolyticus strains. The toxR gene fragment was amplified in all the strains isolated here, confirming that they were in fact V. parahaemolyticus, as this gene can be used to identify the species (17).
The data here presented are part of the first studies on the isolation, distribution, and detection of virulence factors of V. parahaemolyticus in Mexico. The presence of potentially pathogenic V. parahaemolyticus strains in the Pueblo Viejo Lagoon is matter of concern for sanitary authorities, because this organism has not been considered a health problem in spite of the information on infection outbreaks along the coastline of the Gulf of Mexico. This information may be important for preventing sanitary problems that might affect the health of the population.
Acknowledgments
We thank Francisco Flores Pedroche for his assistance in modifying the Pueblo Viejo Lagoon area map.
REFERENCES
- 1.Abbot, S. L., C. Powers, C. A. Kaysner, Y. Takeda, M. Ishibashi, S. W. Joseph, and J. M. Janda. 1989. Emergence of a restricted bioserovar of Vibrio parahaemolyticus as the predominant cause of Vibrio-associated gastroenteritis on the west coast of the United States and Mexico. J. Clin. Microbiol. 27:2891-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, M. J., K. Tomochika, S. Miyoshi, and S. Shinoda. 2002. Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208:83-87. [DOI] [PubMed] [Google Scholar]
- 3.Castillo-Rivera, M., R. Zárate, and L. Sanvicente-Añorve. 2003. Patrones de la diversidad de peces en la laguna de Pueblo Viejo, Veracruz, México. Hidrobiologica 13:289-298. [Google Scholar]
- 4.Chiou, C., S. Hsu, S. Chiu, T. Wang, and C. Chao. 2000. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J. Clin. Microbiol. 38:4621-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniels, N. A., L. MacKinnon, R. Bishop, S. Altekruse, B. Ray, R. M. Hammond, S. Thompson, S. Wilson, N. H. Bean, P. M. Griffin, and L. Slutsker. 2000. Vibrio parahaemolyticus infection in the United States, 1973-1998. J. Infect. Dis. 181:1661-1666. [DOI] [PubMed] [Google Scholar]
- 6.DePaola, A., J. L. Nordstrom, J. C. Bowers, J. G. Wells, and D. W. Cook. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69:1521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DePaola, A., C. A. Kaysner, J. Bowers, and D. W. Cook. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66:4649-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliot, E. L., C. A. Kaysner, L. Jackson, and M. L. Tamplin. 1995. V. cholerae, V. parahaemolyticus, V. vulnificus and other Vibrio spp., p. 9.01-9.27. In USFDA bacteriological analytical manual, 8th ed. AOAC International, Arlington, Va.
- 9.Elliot, E. L., C. A. Kaysner, and M. L. Tamplin. 1992. Appendix 3. Media and reagents, p. 508. In USFDA bacteriological analytical manual, 7th ed. AOAC International, Arlington, Va.
- 10.Hervio-Heath, D., R. R. Colwell, A. Derrien, A. Robert-Pillot, J. M. Fournier, and M. Pommepuy. 2002. Occurrence of pathogenic vibrios in coastal areas of France. J. Appl. Microbiol. 92:1123-1135. [DOI] [PubMed] [Google Scholar]
- 11.Honda, S., I. Goto, I. Minematsu, N. Ikeda, N. Asano, M. Ishibashi, Y. Kinoshita, M. Nishibuchi, T. Honda, and T. Miwatani. 1987. Gastroenteritis due to Kanagawa negative Vibrio parahaemolyticus. Lancet i:331-332. [DOI] [PubMed] [Google Scholar]
- 12.Honda, T., Y. Ni, and T. Miwatani. 1989. Purification of a TDH-related hemolysin produced by a Kanagawa phenomenon-negative clinical isolate of Vibrio parahaemolyticus O6:K46. FEMS Microbiol. Lett. 57:241-246. [DOI] [PubMed] [Google Scholar]
- 13.Honda, T., Y. Ni, and T. Miwatani. 1988. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56:961-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joseph, S. W., R. R. Colwell, and J. B. Kaper. 1982. Vibrio parahaemolyticus and related halophilic vibrios. Crit. Rev. Microbiol. 10:77-124. [DOI] [PubMed] [Google Scholar]
- 15.Kaysner, C. A., C. Abeyta, Jr., R. F. Stott, J. L. Lilja, and M. M. Wekell. 1990. Incidence of urea-hydrolyzing Vibrio parahaemolyticus in Willapa Bay, Washington. Appl. Environ. Microbiol. 56:904-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly, M. T., and E. M. D. Stroh. 1988. Temporal relationship of Vibrio parahaemolyticus in patients and the environment. J. Clin. Microbiol. 26:1754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, Y. B., J. Okuda, C. Matsumoto, N. Takahashi, S. Hashimoto, and M. Nishibuchi. 1999. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37:1173-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishishita. M., N. Matsuoka, K. Kumagai, S. Yamasaki, Y. Takeda, and M. Nishibuchi. 1992. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 58:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marano, N. N., N. A. Daniels, A. N. Easton, A. McShan, B. Ray, J. G. Wells, P. M. Griffin, and F. J. Angulo. 2000. A survey of stool culturing practices for Vibrio species at clinical laboratories in Gulf Coast states. J. Clin. Microbiol. 38:2267-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishibuchi, M., and J. B. Kaper. 1995. Thermostable Direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63:2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishibuchi, M., and J. B. Kaper. 1990. Duplication and variation of the thermostable direct hemolysin (tdh) gene in Vibrio parahaemolyticus. Mol. Microbiol. 4:87-99. [DOI] [PubMed] [Google Scholar]
- 22.Nishibuchi, M., T. Taniguchi, T. Misawa, V. Khaeomanee-Iam, T. Honda, and T. Miwatani. 1989. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect. Immun. 57:2691-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuda, J., M. Ishibashi, S. L. Abbott, J. M. Janda, and M. Nishibuchi. 1997. Analysis of the thermostable direct hemolysin (TDH) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J. Clin. Microbiol. 35:1965-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osawa, R., T. Okitsu, H. Morozumi, and S. Yamai. 1996. Occurrence of urease-positive Vibrio parahaemolyticus in Kanagawa, Japan, with specific reference to presence of thermostable direct hemolysin (TDH) and the TDH-related-hemolysin genes. Appl. Environ. Microbiol. 62:725-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirai, H., H. Ito, T. Hirayama, Y. Nakamoto, N. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tada, J., T. Ohashi, N. Nishimura, Y. Shirasaki, H. Ozaki, S. Fukushima, J. Takano, M. Nishibuchi, and Y. Takeda. 1992. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol. Cell. Probes 6:477-487. [DOI] [PubMed] [Google Scholar]
- 27.Torres-Vitela, M. R., and E. Fernández-Escartín. 1993. Incidencia de Vibrio parahaemolyticus en Pescado, Ostión, y Camarón crudos. Rev. Lat. Am. Microbiol. 35:267-272. [PubMed] [Google Scholar]
- 28.Vuddhakul, V., A. Chowdhury, V. Laohaprertthisan, P. Pungrasamme, N. Patararungrong, P. Thianmontri, M. Ishibashi, C. Matsumoto, and M. Nishibuchi. 2000. Isolation of a pandemic O3:K6 clone of Vibrio parahaemolyticus strains from environmental and clinical sources in Thailand. Appl. Environ. Microbiol. 66:2685-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong, H. C., S. H. Liu, L. W. Ku, I. Y. Lee, T. K. Wang, Y. S. Lee, C. L. Lee, L. P. Kuo, and D. Y. Shih. 2000. Characterization of Vibrio parahaemolyticus isolates obtained from foodborne illness outbreaks during 1992 through 1995 in Taiwan. J. Food Prot. 63:900-906. [DOI] [PubMed] [Google Scholar]
- 30.Yoh, M., T. Miwatani, and T. Honda. 1992. Comparison of Vibrio parahaemolyticus hemolysin (Vp-TRH) produced by environmental and clinical isolates. FEMS Microbiol. Lett. 71:157-161. [DOI] [PubMed] [Google Scholar]