Abstract
The need for protozoa for the proliferation of Legionella pneumophila in aquatic habitats is still not fully understood and is even questioned by some investigators. This study shows the in vivo growth of L. pneumophila in protozoa in aquatic biofilms developing at high concentrations on plasticized polyvinyl chloride in a batch system with autoclaved tap water. The inoculum, a mixed microbial community including indigenous L. pneumophila originating from a tap water system, was added in an unfiltered as well as filtered (cellulose nitrate, 3.0-μm pore size) state. Both the attached and suspended biomasses were examined for their total amounts of ATP, for culturable L. pneumophila, and for their concentrations of protozoa. L. pneumophila grew to high numbers (6.3 log CFU/cm2) only in flasks with an unfiltered inoculum. Filtration obviously removed the growth-supporting factor, but it did not affect biofilm formation, as determined by measuring ATP. Cultivation, direct counting, and 18S ribosomal DNA-targeted PCR with subsequent sequencing revealed the presence of Hartmannella vermiformis in all flasks in which L. pneumophila multiplied and also when cycloheximide had been added. Fluorescent in situ hybridization clearly demonstrated the intracellular growth of L. pneumophila in trophozoites of H. vermiformis, with 25.9% ± 10.5% of the trophozoites containing L. pneumophila on day 10 and >90% containing L. pneumophila on day 14. Calculations confirmed that intracellular growth was most likely the only way for L. pneumophila to proliferate within the biofilm. Higher biofilm concentrations, measured as amounts of ATP, gave higher L. pneumophila concentrations, and therefore the growth of L. pneumophila within engineered water systems can be limited by controlling biofilm formation.
Legionella pneumophila is widespread in natural freshwater environments, despite its fastidious nature (16). The bacterium has also frequently been observed in engineered water systems such as warm water distributing systems, cooling towers, humidifiers, and fountains (25, 43, 50). The multiplication of the organism in water systems poses a potential human health risk when aerosolization can occur (12).
Legionellae grow in vitro only in complex media with supplements of cysteine and iron salts (11). For their multiplication in vivo, other microorganisms are required, but different species of heterotrophic bacteria (Pseudomonas aeruginosa, Klebsiella pneumoniae, and Flavobacterium sp.) alone do not support the growth of L. pneumophila (31, 45). In vitro studies using cocultures have repeatedly demonstrated the intracellular multiplication of L. pneumophila in amoebae (Acanthamoeba, Echinamoeba, Hartmannella, Naegleria, Vahlkampfia, and Dictyostelium) and in a ciliated protozoon (Tetrahymena pyriformis) (12, 15, 21, 39). Amoebae have been observed in water systems associated with Legionnaires' disease (5, 7), and L. pneumophila can recolonize water distributing systems within a few weeks after disinfection (26, 27). Batch experiments with tap water showed that L. pneumophila did not multiply in the absence of protozoa, and growth in the presence of a protozoon (Hartmannella) was confirmed by the use of cocultures (14, 45).
Rogers and coworkers (35, 36) found that the growth of L. pneumophila on a variety of materials in contact with tap water is coupled with biofilm development. However, they concluded that (i) the numbers of L. pneumophila cells in the biofilms were unrelated to the total numbers of microorganisms present and (ii) the growth of L. pneumophila coincided with undetectable numbers of potential hosts on some materials. Furthermore, it had been concluded from several studies that the intracellular growth of L. pneumophila in protozoa is not always essential for the multiplication of L. pneumophila within a mixed bacterial consortium in water (37, 42).
This paper describes the use of a batch test system for the development of different concentrations of aquatic biofilms by using polyvinyl chloride, in both unplasticized (PVCu) and plasticized (PVCp) versions, as a support and as a source of energy and carbon. In vivo observations revealed that the intracellular multiplication of L. pneumophila in Hartmannella vermiformis was the mechanism of proliferation for L. pneumophila in biofilms on PVCp, which contained a high concentration of heterotrophic bacteria.
MATERIALS AND METHODS
Batch test system.
Biofilms were developed on pieces of PVCu (about 9.5 cm2 of surface area for each) and pieces of PVCp tubes (diameter, 0.80 cm; about 6.2 cm2 of surface area for each) in a static test using heat-cleaned (550°C) Erlenmeyer flasks with a volume of 1 liter. All material pieces were pasteurized at 70°C for 30 min and then incubated in 600 ml of sterilized tap water (pH 7.8) at surface-to-volume ratios of 0.24 and 0.16 cm−1 for PVCu and PVCp, respectively. These surface-to-volume ratios were kept constant during the test; after the collection of a piece of PVCu or PVCp, a proportional volume of water was also removed from the flask. Nitrate and phosphate from autoclaved stock solutions were added to the flasks at final concentrations of 72.5 and 13.5 μM, respectively, to prevent growth limitation by these nutrients. A mixed microbial community including indigenous L. pneumophila serotype 1, originating from a plumbing system in The Netherlands, was used as the inoculum. Before its use as an inoculum, this mixed microbial community had been maintained in tap water containing pieces of silicone tubing at 37°C, followed by storage at −80°C. The inoculum was added in an unfiltered as well as a filtered state (3.0-μm-pore-size cellulose nitrate filter; Sartorius, Goettingen, Germany). Filtration reduced the indigenous L. pneumophila concentration in the inoculum from 4.1 to 2.4 log CFU/ml and the concentration of the heterotrophic bacteria from 5.1 to 4.2 log CFU/ml. Compared to that of the unfiltered inoculum, a 10-fold larger volume of the filtered inoculum was added to the test flasks to partly correct for these differences. The protozoon concentration in the unfiltered inoculum, as determined by a cultivation method, was 1.4 log protozoa/ml; no protozoa were detected in the filtered inoculum. A pure culture of the same indigenous L. pneumophila serotype 1 strain was also used as an inoculum after two passages on buffered charcoal yeast extract agar (11) and subsequent suspension in sterile tap water. Filter-sterilized (pore size, 0.2 μm; Schleicher & Schuell, Dassel, Germany) cycloheximide was added at a final concentration of 100 μM to two flasks with the unfiltered inoculum to prevent the growth of protozoa. All tests were conducted in duplicate, and incubation took place at 37°C without shaking for several weeks.
Biomass measurements in biofilm and planktonic phase.
The attached biomass (biofilm) and the suspended biomass (planktonic phase) were analyzed for their total ATP concentrations and for the numbers of Legionella, heterotrophic bacteria, and protozoa at different time points during the experiments. The microorganisms in the biofilm were removed from the material pieces by six 2-min sonication steps in 10 ml of sterilized tap water (U950D; Ultrawave Limited, Cardiff, United Kingdom) at a frequency of 30 kHz and an average power input of 0.11 W/ml. The total ATP concentrations, representing the active biomasses, in the biofilm and planktonic phase were determined by ATP analysis with a commercially available ATP kit (Celsis International B.V., Landgraaf, The Netherlands). Legionella concentrations were determined by plating on buffered charcoal yeast extract agar and incubation at 37°C for 7 days (11). The heterotrophic bacterial concentration was determined by the heterotrophic plate count (HPC) method with R2A agar (34). Protozoon concentrations were determined by cultivation, direct counting with the fluorochrome primulin, and/or fluorescent in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Cultivation was done according to the method described by Darbyshire and coworkers, adjusted for water samples by the addition of 50 μl of the biofilm or planktonic phase to the first wells (10). Pseudomonas fluorescens (about 7 log CFU/ml) was used as a food source for the protozoa. The protozoon concentration was calculated with GenStat, 6th ed. (VSN International Ltd., Oxford, United Kingdom). The cultivated protozoa were identified by use of An Illustrated Key to Freshwater and Soil Amoebae (32). The fluorochrome primulin (Aldrich Chemical Company, Inc., Milwaukee, Wis.) was applied to stain the protozoa, followed by direct counting as described elsewhere (6). Direct counting with FISH is described below.
FISH.
At each time point, 30 ml of the planktonic phase and 50 ml of the biofilm phase were filtered over a 0.4-μm-pore-size HTTP Isopore membrane (Millipore, Bedford, Mass.) in a vacuum not exceeding 3 kPa. The samples were fixed on the filter with a 4% paraformaldehyde-phosphate-buffered saline solution at room temperature for 30 min, washed with distilled water, and dehydrated at 46°C for 5 min. The filters were placed on glass slides for hybridization by the application of 40 μl of hybridization buffer (20% [vol/vol] formamide, 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 15 mM Tris-HCl [pH 7.4]), 200 ng of the fluorescein-labeled probe EUK516 (Table 1) (1), 225 ng of the Cy3-labeled probe LEGPNE1 (Table 1) (19), 38 ng of poly(U), and 76 ng of bovine serum albumin and then were covered with a coverslip. Incubation was performed at 46°C for 3 h in an isotonically humid chamber. The labeled oligonucleotides were removed by incubating the filters upside down in preheated washing buffer (40 mM NaCl, 15 mM Tris-HCl [pH 7.4], 0.01% sodium dodecyl sulfate). After incubation for 30 min at 46°C, the filters were rinsed with distilled water (room temperature) and dried at 46°C for 5 min. The filters were mounted in Vectorshield (Vector Laboratories, Inc., Burlingame, Calif.) on a glass slide. Fluorescence was detected by use of a Leica DMRXA fluorescence microscope supplied with a COHU high-performance charge-coupled device digital camera. Leica Q Fluoro, v. V1.0b, software was used to record the pictures.
TABLE 1.
Primer sequences used for this study
| Primer | Sequence | Reference |
|---|---|---|
| 5′→3′ directed primers | ||
| Euk 516 | ACCAGACTTGCCCTCC | 1 |
| LEGPNE1 | ATCTGACCGTCCCAGGTT | 19 |
| EUKf | ACCTGGTTGATCCTGCCAG | 30 |
| 373C | GATTCCGGAGAGGGAGCCTGA | 49 |
| 892Ca | GTCAGAGGTGAAATTCTAGG | 40 |
| 1262C | GTGGTGCATGGCCGTTCTTA | 49 |
| 3′→5′ directed primers | ||
| EUKr | TGATCCTTCYGCAGG TTCAC | 30 |
| 373 | TCAGGCTCCCTCTCCGGAATC | 49 |
| 892a | CCTAGAATTTCACCTCTGAC | 40 |
| 1262 | GAACGGCCATGCACCAC | 49 |
The primer sequence was slightly modified, being adjusted to H. vermiformis Nijmegen (49).
PCR for detection of eukaryotes.
All Erlenmeyer flasks with PVCp test pieces were checked for the presence of eukaryotes by use of a PCR assay. At each time point, 30 ml of the planktonic phase and 50 ml of the biofilm phase were filtered over a 0.2-μm-pore-size GTTP Isopore membrane (Millipore). DNA extraction of the filters was done by use of a FastDNA spin kit for soil (BIO 101, Carlsbad, Calif.) according to the instructions supplied by the manufacturer. The 18S rRNA gene was amplified by a PCR with the oligonucleotide primers EUKf and EUKr (Table 1), which were complementary to regions of conserved sequences located proximal to the 5′ and 3′ termini of known 18S rRNA genes (30). The PCR mixtures (50 μl) contained about 100 ng of template DNA, each primer at a concentration of 0.5 μM, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 2.5 U of Taq DNA polymerase (Invitrogen, Life Technologies, Breda, The Netherlands), and the PCR buffer supplied with the enzyme. PCR cycling was performed in an UNO II thermocycler Biometra (Westburg, Leusden, The Netherlands) with the following program: predenaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 61°C for 60 s, and extension at 72°C for 90 s; and a final extension step at 72°C for 10 min.
Cloning of PCR-amplified products.
PCR amplicons were purified by the use of a QIAquick PCR purification kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). PCR products were cloned into Escherichia coli JM109 by use of the Promega pGEM-T easy vector system (Promega, Madison, Wis.). PCRs were performed on cell lysates of ampicillin-resistant transformants, using the pGEM-T-specific primers T7 and Sp6 to confirm the sizes of the inserts. To establish the eukaryotic diversity between the clones obtained from the aquatic biofilm with L. pneumophila, we subjected the amplicons to restriction fragment length polymorphism analysis with the restriction enzymes MspI, AluI, and CfoI. The amplicons derived from plasmids containing unique inserts were purified by use of a QIAquick PCR purification kit and were subjected to DNA sequence analysis.
Sequence analysis.
Sequence analysis was done by MWG-Biotech (Ebersberg, Germany). The primers used to sequence the total 18S rRNA gene are given in Table 1 (40, 49).
Phylogenetic analysis.
The determined 18S rRNA gene sequences were aligned by use of the ARB software package (28). Different phylogenetic trees were constructed by different methods and by using different filters as implemented in the ARB software package.
Nucleotide sequence accession numbers.
The 18S ribosomal DNA (rDNA) sequences from H. vermiformis KWR-1, KWR-2, and KWR-3 have been deposited in GenBank under accession numbers AY502959, AY502960, and AY502961, respectively.
RESULTS
Batch test system.
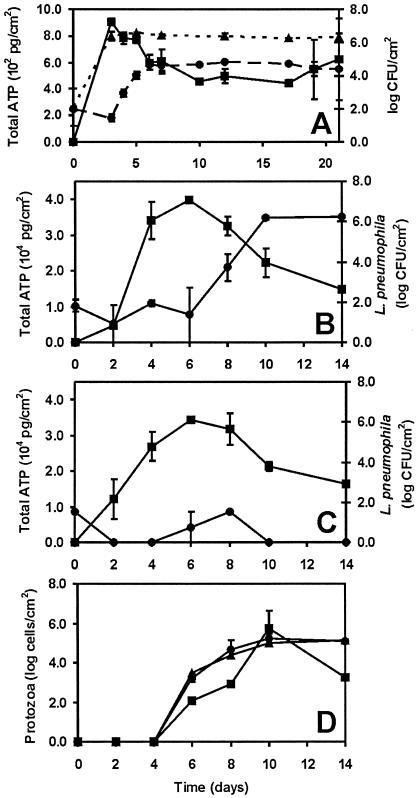
A maximum ATP concentration of 9.1 × 102 ± 2.8 × 101 pg/cm2 was reached within 3 days in the batch test system with PVCu. The HPC values showed that exponential growth occurred up to day 3. With PVCp, a maximum ATP concentration of 3.8 × 104 ± 2.7 × 103 pg/cm2 was reached within 6 days. The total ATP concentrations declined to about 4.6 × 102 ± 0.5 × 101 pg/cm2 on day 10 for PVCu and to about 1.6 × 104 ± 9.8 × 102 pg/cm2 on day 14 for PVCp (Fig. 1A to C) and remained stable when we continued the experiment until day 40 (data not shown). Also, the HPC values observed with PVCu remained at a constant level during prolonged incubations.
FIG. 1.
(A) Concentrations of total ATP (▪), L. pneumophila (•), and heterotrophic bacteria (▴) in flasks with the unfiltered inoculum and PVCu as the biofilm carrier. Each point represents the mean concentration from two experiments, and bars indicate standard errors. (B) Concentrations of total ATP (▪) and L. pneumophila (•) in flasks with the unfiltered inoculum and PVCp as the biofilm carrier. Each point represents the mean concentration from two experiments, and bars indicate standard errors. (C) Concentrations of total ATP (▪) and L. pneumophila (•) in flasks with the filtered inoculum and PVCp as the biofilm carrier. Each point represents the mean concentration from two experiments, and bars indicate standard errors. (D) Protozoon concentrations in flasks with the unfiltered inoculum. Squares (▪) represent the concentrations determined by the cultivation method; circles (•) and triangles (▴) represent the concentrations determined by direct counting methods using primulin and FISH, respectively. Bars indicate standard errors. No protozoa were detected in flasks with the filtered inoculum.
The L. pneumophila concentration increased exponentially in flasks with the unfiltered inoculum when the ATP concentrations and HPC values had reached maximal levels. Prolonged incubations showed that the Legionella concentration remained stable until the end of the experiment (Fig. 1A and B). The maximum L. pneumophila concentration for the experiments with PVCp was 6.3 ± 0.1 log CFU/cm2 and was lower for experiments with PVCu (4.9 ± 0.1 log CFU/cm2).
The percentage of biomass present in the biofilm relative to that in the planktonic phase depended on the material used for biofilm development. For PVCu, 57% ± 6% of the total ATP, 77% ± 6% of L. pneumophila cells, and 38% ± 11% of the HPC value were present in the biofilm. For PVCp, these values were 37% ± 9% for total ATP and 52% ± 15% for L. pneumophila. The concentrations of total ATP and L. pneumophila were highest with PVCp, and this material was used in further experiments for the identification of a growth-supporting factor(s) for L. pneumophila.
Identification of growth-supporting factor(s) for L. pneumophila.
Growth of L. pneumophila was detected in the flasks with the unfiltered inoculum (with or without cycloheximide). No proliferation of L. pneumophila was observed in the flasks with the filtered inoculum, although the total ATP concentrations under both conditions were similar (Fig. 1B and C). Also, no growth of L. pneumophila was observed when a pure culture of L. pneumophila serotype 1 was used as the inoculum. The addition of an untreated filter with its retentate to the flasks gave similar results to those for flasks with the unfiltered inoculum, but the addition of a pasteurized (30 min, 70°C) filter with its retentate to the flasks with the filtered inoculum did not induce the growth of L. pneumophila (data not shown).
Protozoa were detected by cultivation and direct counting in all of the flasks in which the growth of L. pneumophila was observed. No protozoa were detected (n < 1.6 log protozoa/cm2 for cultivation and n < 2.1 log protozoa/cm2 for direct counting) in the flasks in which no proliferation of L. pneumophila occurred. Based on their morphology, all of the protozoa cultivated in the 96-well plates were identified as H. vermiformis. The direct counting methods also revealed that only one type of protozoa with the typical features of H. vermiformis was present in the Erlenmeyer flasks.
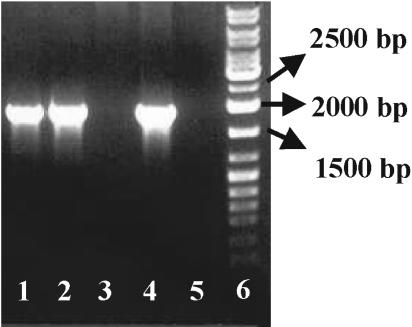
From day 6 on, eukaryotic DNAs were detected by 18S rDNA-targeted PCRs in the biofilm and the planktonic phase in flasks that showed growth of L. pneumophila (with and without cycloheximide). Although protozoa were present in the flasks with the unfiltered inoculum before day 6, their DNAs were not detectable by PCR, indicating that the initial protozoon concentration was below the detection threshold. Cloning and subsequent sequencing confirmed that H. vermiformis was the only protozoon species present. No eukaryotic DNAs were detected in the flasks with the filtered inoculum (Fig. 2).
FIG. 2.
Detection of eukaryotic DNA. Lane 1, DNA from biofilm from flasks with unfiltered inoculum in the presence of 100 μM cycloheximide; lanes 2 and 4, DNAs from biofilms from flasks with unfiltered inoculum; lanes 3 and 5, DNAs from biofilms from flasks with filtered inoculum; lane 6, marker (sm 0333 gene ruler DNA ladder mix; Fermentas, St. Leon-Rot, Germany).
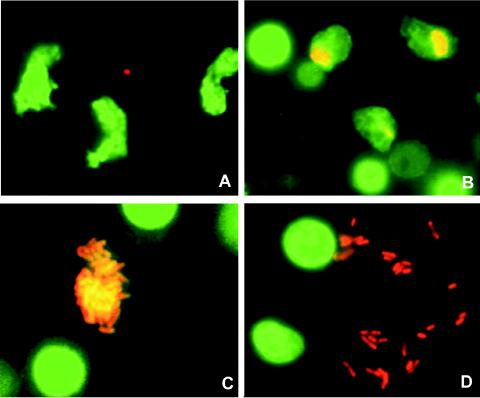
The FISH technique revealed the different stages of the intracellular proliferation of L. pneumophila within protozoa (Fig. 3).
FIG. 3.
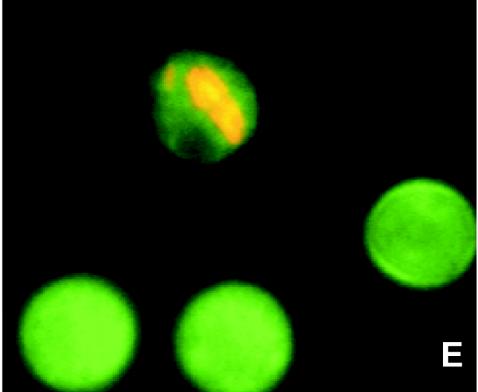
Different stages of intracellular proliferation of L. pneumophila within amoebae. All images were made by using material from the biofilm phase, since amoebae were mainly present in this phase. (A) Trophozoite amoebae on day 6 of the experiment. Magnification, ×1,000. (B) Trophozoite amoebae that were just infected with L. pneumophila (day 10). Magnification, ×1,000. (C) Heavily infected amoeba (day 14). Magnification, ×1,600. (D) Free L. pneumophila bacteria, just escaped from a lysed amoeba (day 14). Magnification, ×1,600. (E) Cysts of amoebae, with one infected by L. pneumophila (day 14). Magnification, ×1,600.
Quantitative relationship between L. pneumophila and H. vermiformis.
Protozoa were first detected on day 6 (Fig. 1D). The cultivation method revealed that the concentration of protozoa increased to 5.8 ± 0.9 log protozoa/cm2 by day 10 and decreased afterwards. The results of the direct counting methods using primulin and FISH were similar to the results of the cultivation method (Fig. 1D), but with these methods no decline in protozoon concentration was detected after day 10. Furthermore, it was found that 93% ± 4% of the protozoa were present in the biofilm rather than in the planktonic phase. During the experiments, the amoebae changed from the trophozoite form on day 6 (Fig. 3A) to the cyst form on day 14 (Fig. 3E). Table 2 gives the different states of the protozoa as percentages of the total concentrations of protozoa observed by direct cell counting using primulin and FISH.
TABLE 2.
Numbers of protozoa, as determined by direct counting using primulin and FISH, in a batch test system with PVCp as the biofilm carrier in duplicate flasks
| Time point (day) | Total concn of protozoa (log cells/cm2)a | % of protozoa in indicated statea
|
Trophozoites (%) containing L. pneumophilaa | |
|---|---|---|---|---|
| Trophozoite | Cyst | |||
| 0 | ND | ND | ND | ND |
| 2 | ND | ND | ND | ND |
| 4 | ND | ND | ND | ND |
| 6 | 3.4 ± 0.2 | 100 ± 0.0 | 0 ± 0.0 | ND |
| 8 | 4.6 ± 0.2 | 93.9 ± 6.1 | 6.1 ± 6.1 | ND |
| 10 | 5.1 ± 0.1 | 5.7 ± 3.0 | 94.3 ± 3.0 | 25.9 ± 10.5 |
| 14 | 5.1 ± 0.1 | 0.2 ± 0.1 | 99.8 ± 0.1 | 92.3b |
ND, not detected: the concentration was below the detection limit (n < 2.1 log protozoa/cm2).
A total of 13 trophozoites were observed in three different tests, and 12 of them contained Legionella.
For a time interval of T (hours), the specific growth rate μ (per hour) of the protozoa was calculated as follows:
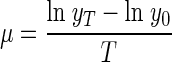 |
(1) |
where y0 and yT are the protozoon numbers at the beginning and end of the interval. During exponential growth, these numbers are related by the equation
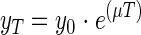 |
(2) |
The maximum growth rate of the H. vermiformis strain, as observed with the cultivation method, was 0.14 h−1. With both direct counting methods, a maximum growth rate of 0.15 h−1 was calculated.
The L. pneumophila concentrations observed in amoebae ranged from 1 cell per amoeba when they were just infected (Fig. 3B) to a maximum of about 100 cells per amoeba just before lysis (Fig. 3C). All but one infected amoebae were present as trophozoites (Fig. 3B and C), and only one infected cyst was detected (0.02%) (Fig. 3E). Table 2 gives the numbers of protozoa that were infected with L. pneumophila as percentages of all protozoa and as percentages of the trophozoites.
With the different methods for determining the protozoon concentration (cultivation and direct counting using primulin and FISH), a maximal protozoon concentration of 5.3 ± 0.4 log protozoa/cm2 was detected on day 10 (Fig. 1D). On that day, 6.2 ± 0.1 log CFU of L. pneumophila/cm2 were present (Fig. 1B). Lysis of the protozoa occurred after about 100 L. pneumophila cells per lysed amoeba were observed (Fig. 3C). Hence, 4.2 ± 0.1 log protozoa/cm2 should account for the total L. pneumophila concentration. These lysed trophozoites were not included in the number of protozoa detected on day 10. From this estimated number of lysed protozoa, we calculated that an average of 10.5% ± 6.1% (n = 5) of all protozoa had been infected between days 6 and 10. With FISH analysis, the intracellular proliferation of L. pneumophila within trophozoites was observed in 25.9% ± 10.5% (n = 2) of the trophozoites that were present on day 10 (Table 2). This percentage was significantly different from the estimated percentage for days 6 to 10 (P = 0.06 by a t test). On day 14, the percentage of infected trophozoites had increased further (>90%), and only 0.2% of the protozoa were present as trophozoites (Table 2).
Eukaryotic identification.
The 18S rDNA-targeted PCR amplicons were cloned into E. coli JM109 and subsequently subjected to restriction fragment length polymorphism analysis. With this analysis, 19 different restriction patterns were obtained for 50 clones. After partly sequencing their 18S rRNA genes, we identified all 19 sequences as being from H. vermiformis, with ≥98% similarities. These 19 clones could be divided into three groups (KWR-1, KWR-2, and KWR-3). The complete (1,838 bp) 18S rRNA genes for these three different operating taxonomic units were determined and showed G+C contents of 49.62, 49.51, and 49.67% for KWR-1, KWR-2, and KWR-3, respectively.
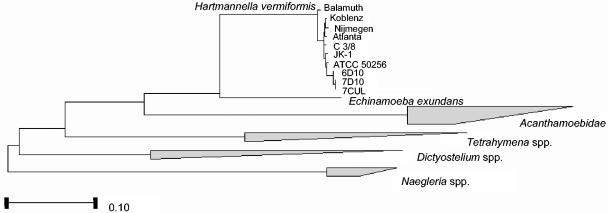
The sequences (KWR-1, KWR-2, and KWR-3) were compared by phylogenetic analysis with those of other protozoa in which the intracellular growth of L. pneumophila had been shown (Fig. 4). The obtained sequences fell into the group of H. vermiformis, consistently forming a subgroup. A total of two nucleotide variations were present in all sequence types and were different from seven other H. vermiformis sequences at positions 1550 and 1628 (nucleotide numbering is according to the H. vermiformis strain Nijmegen sequence [49]). A single base pair insertion (G) was identified between positions 1818 and 1819. In addition to the previously mentioned two mutations, only a few additional single nucleotide polymorphisms were observed among the different H. vermiformis strains. These observations indicate that there is a high degree of sequence similarity between H. vermiformis strains originating from different habitats and different parts of the world.
FIG. 4.
Phylogenetic tree from 18S rRNA gene sequences showing the relationships of the examined sequences to those of other protozoa in which intracellular growth of L. pneumophila was shown (12, 15, 21, 39). Different trees were calculated, and all trees gave the same consensus, as shown in the figure. Bar, 10% sequence divergence. Strains Koblenz (X75514), Nijmegen (X75515), and Atlanta (X75513) were previously sequenced by Weekers et al. (49), strain Balamuth (ATCC 30966, M95168) was sequenced by Gunderson et al. (20), strain C3/8 (AF426157) was sequenced by Walochnik et al. (48), and strains JK-1 and ATCC 50256 were sequenced by Kuchta et al. (24). Clones: 6D10, KWR1; 7D10, KWR2; 7CUL, KWR3.
DISCUSSION
Experimental design.
For the present study, we used a batch test system for the simulation of biofilm development in tap water. This batch test system has previously been compared with a dynamic model of biofilm development on different materials that are in contact with tap water (44). That previous study showed that the batch test system is well suited for the study of biofilm formation on material surfaces that are in contact with tap water. With this model, it is easy to obtain different biofilm concentrations by using different materials. ATP was used to assess the concentrations of active biomasses. The similarity between the total biomass concentrations in tests with a filtered inoculum and with an unfiltered inoculum indicates that ATP represented the bacterial biomasses. The concentrations of total ATP and L. pneumophila were clearly higher in the presence of PVCp than in the presence of PVCu. Obviously, the concentration of growth-supporting factors for L. pneumophila was higher with PVCp, thus facilitating their identification. The high biomass concentrations observed with PVCp were caused by the leaching of biodegradable plastizicers from this material (9). From the obtained percentages of attached and suspended biomasses and the applied surface-to-volume ratios (0.24 to 0.16 cm−1), and assuming a biofilm thickness of 100 μm or less, it can be estimated that the biofilm concentrations were about 100 times higher than the concentrations of suspended biomass. The high proportion (>90%) of protozoa observed in the biofilm on PVCp further indicates that the biofilm was the place where the multiplication of protozoa and L. pneumophila occurred.
The following relationship between L. pneumophila and total ATP concentrations has been reported for the batch test system with incubation at 25°C: log(Legionella growth potential) = 0.81 log(biomass production potential) + 1.1 (44). The concentrations of L. pneumophila and total ATP observed in the present study after 14 days for PVCp and after 21 days for PVCu did not fit this equation. The log(Legionella growth potential) values in the present study were 1.4 and 1.3 times higher for PVCp and PVCu, respectively, than that calculated with the given equation. This difference was attributed to the different temperatures used for incubation of the test flasks because a temperature of 37°C is more favorable for the growth of L. pneumophila than is 25°C (44, 47).
Growth of Legionella in aquatic biofilm only in the presence of H. vermiformis.
Inoculum filtration (3.0-μm pore size) did not influence the total biomass yield (total ATP) but removed a factor that enabled L. pneumophila to grow, despite the presence of this bacterium in the inoculum (Fig. 1C). Also, no growth of L. pneumophila was observed when it was added to the flasks as a pure culture. The inactivation of the growth-supporting factor by pasteurization indicated that a live organism served as the growth-supporting factor. These findings are in complete agreement with the observations of Wadowsky and coworkers (45), who tested the effect of inoculum filtration (pore size, 1.0 μm) on the growth of L. pneumophila in tap water and observed inactivation of the growth-supporting factor at 60°C.
The cultivation method for the detection of protozoa showed only H. vermiformis, and this organism was only present in the flasks with the unfiltered inoculum. FISH analysis revealed the in vivo intracellular multiplication of L. pneumophila in amoebae, which were identified as H. vermiformis by PCR, cloning, and sequence analysis. H. vermiformis has also repeatedly been identified as a growth-supporting factor for L. pneumophila by researchers in the United States (13, 14, 24, 31, 45, 46). Several authors have questioned the fact that protozoa are required as a host for L. pneumophila, and the extracellular growth of L. pneumophila has been reported in two studies. In one study, cycloheximide inhibited the growth of protozoa, but L. pneumophila proliferated (42). The other study suggested that the bacterial consortium within a biofilm supplies sufficient nutrients to enable legionellae to grow extracellularly (37). In the present study, however, growth of L. pneumophila was observed at a similar concentration of cycloheximide, but H. vermiformis was still detected by 18S rDNA-targeted PCR (Fig. 2). Figure 3C shows a microcolony of L. pneumophila inside an amoeba just before lysis. The amoeba itself is hardly visible. After the lysis of amoebae, the bacteria are free in the environment, where they may occasionally cluster together (Fig. 3D).
Intracellular growth of L. pneumophila has been observed in vitro in T. pyriformis, Acanthamoeba castellanii, and H. vermiformis by the use of FISH (18, 19, 29). The protozoon cultures in these studies were inoculated with a high concentration of L. pneumophila (6 to 8 log cells/ml). In the present study, the development of biofilms in tap water was simulated by the use of relatively small inoculum sizes for both L. pneumophila and H. vermiformis (4.1 and 1.4 log cells/ml, respectively), thus more closely resembling the actual situation in water systems.
The present study also demonstrated the presence of L. pneumophila inside a cyst of H. vermiformis (Fig. 3E) in vivo, but the percentage of infected cysts was very low (0.02%). Kuchta et al. (23, 24) did not detect L. pneumophila within cysts of H. vermiformis, and Greub and Raoult (17) observed the bacterium within the cyst wall, but not within vacuoles or the cytoplasm, of a mature cyst of H. vermiformis. L. pneumophila-infected cysts of Acanthamoeba palestinensis have been observed in another study (41), and 2 to 5% of Acanthamoeba polyphaga cysts were found to be infected with L. pneumophila (38). Our observations support earlier suggestions that trophozoites and cysts may both provide a protective environment for L. pneumophila in engineered water systems (22, 41).
Quantitative relationship between total ATP, L. pneumophila, and H. vermiformis.
Heterotrophic bacteria that contributed to the peak of the total ATP concentration (Fig. 1A) most likely served as a food source for H. vermiformis, since protozoa were first detected on day 6, when the maximum total ATP concentration was observed (Fig. 1D). The concentration of L. pneumophila increased exponentially after the first H. vermiformis protozoa were detected. The growth rates of H. vermiformis obtained in this study were 2.9-fold higher than the maximum growth rates of naked amoebae in their natural environment (3) and were similar to the highest growth rates of naked amoebae obtained under laboratory conditions (4, 8). Hence, the aquatic biofilm developing on the pieces of PVCp provided a favorable environment for the growth of H. vermiformis.
Our observations and calculations further showed that under the test conditions used, a significant percentage of the H. vermiformis population was infected with L. pneumophila. No infected protozoa were detected on day 8 of the experiment, when 93.9% ± 6.1% of the protozoa were present as trophozoites. On day 10, only 5.7% ± 3.0% of the protozoa were present as trophozoites, 25.9% ± 10.5% of which contained L. pneumophila. These percentages were even more extreme on day 14, when 0.2% ± 0.1% of the protozoa were present as trophozoites and >90% of the trophozoites were infected (Table 2). After day 10, no further growth of either L. pneumophila or H. vermiformis was observed (Fig. 1B and D). This observation supports the conclusion that the growth of L. pneumophila depended on intracellular proliferation. The high percentage of infected trophozoites on day 14 suggests that Legionella can inhibit the growth of H. vermiformis under batch test conditions. On the other hand, the growth of protozoa had already ceased on day 10, possibly by the depletion of preferred prey bacteria. A clarification of these processes requires further research.
Phylogenetic analysis of H. vermiformis.
Among the protozoa in which intracellular growth of L. pneumophila occurs, the cluster of H. vermiformis showed the highest similarity with Echinamoeba exundans. This was also found by Amaral Zettler et al. (2). In fact, E. exundans was first described as a species of Hartmannella (33). The clones KWR-1, KWR-2, and KWR-3 clearly formed a subgroup within the H. vermiformis group, although there are very few sequence variations within the H. vermiformis cluster (49). This subgroup showed the highest similarity with H. vermiformis strain ATCC 50256, which was originally isolated from a hot water tank in relation with L. pneumophila (24). The obtained phylogenetic tree and the results of Walochnik et al. (48) do not support the previously suggested separation between European and North American strains of H. vermiformis (24, 49).
In conclusion, this study demonstrated that L. pneumophila multiplies in trophozoites of H. vermiformis that are present in aquatic biofilms developing on plastic materials in contact with tap water in a batch test system. Different materials (PVCu and PVCp) gave different biofilm concentrations (total ATP concentrations), and higher total ATP concentrations gave higher concentrations of L. pneumophila. The proliferation of L. pneumophila was due to its intracellular multiplication in H. vermiformis in the presence of a high concentration of heterotrophic bacteria in the biofilm. Prevention of the growth of L. pneumophila within engineered water systems thus requires the prevention of growth of protozoa in biofilms. This objective can be achieved by the removal of biodegradable compounds from the water, the selection of materials that do not support biofilm formation, and/or the maintenance of a disinfectant residual.
Acknowledgments
This study was financed by the Water Supply Companies in The Netherlands in the framework of the Joint Research Program.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, D. Richard, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral Zettler, L. A., T. A. Nerad, C. J. O'Kelly, M. T. Peglar, P. M. Gillevet, J. D. Silberman, and M. L. Sogin. 2000. A molecular reassessment of the leptomyxid amoebae. Protist 151:275-282. [DOI] [PubMed] [Google Scholar]
- 3.Arndt, H. 1993. A critical review of the importance of rhizopods (naked and testate amoebae) and actinopods (heliozoa) in lake plankton. Mar. Microb. Food Webs 7:3-29. [Google Scholar]
- 4.Baldock, B. M., J. H. Baker, and M. A. Sleigh. 1980. Laboratory growth rates of six species of freshwater gymnamoebia. Oecologia 47:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Barbaree, J. M., B. S. Fields, J. C. Feeley, G. W. Gorman, and W. T. Martin. 1985. Isolation of protozoa from water associated with a legionellosis outbreak and demonstration of intracellular multiplication of Legionella pneumophila. Appl. Environ. Microbiol. 51:422-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloem, J., M.-J. B. Bär-Gilissen, and T. E. Cappenberg. 1986. Fixation, counting, and manipulation of heterotrophic nanoflagellates. Appl. Environ. Microbiol. 52:1266-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breiman, R. F., B. S. Fields, G. N. Sanden, L. Volmer, A. Meier, and J. S. Spika. 1990. Association of shower use with Legionnaires' disease. Possible role of amoebae. JAMA 263:2924-2926. [PubMed] [Google Scholar]
- 8.Butler, H., and A. Rogerson. 1996. Growth potential, production efficiency and annual production of marine benthic naked amoebae (gymnamoebae) inhabiting sediments of the Clyde Sea area, Scotland. Aquat. Microb. Ecol. 10:123-129. [Google Scholar]
- 9.Colbourne, J. S. 1985. Materials usage and their effects on the microbiological quality of water supplies. Soc. Appl. Bacteriol. Symp. Ser. 14:47S-59S. [DOI] [PubMed] [Google Scholar]
- 10.Darbyshire, J. F., R. E. Wheatley, M. P. Greaves, and R. H. E. Inkson. 1974. A rapid method for estimating bacterial and protozoan concentrations in soil. Rev. Ecol. Biol. Sol 11:465-475. [Google Scholar]
- 11.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 13.Fields, B. S., T. A. Nerad, T. K. Sawyer, H. King, J. M. Barbaree, W. T. Martin, W. E. Morrill, and G. N. Sanden. 1990. Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial legionellosis. J. Protozool. 37:581-583. [DOI] [PubMed] [Google Scholar]
- 14.Fields, B. S., G. N. Sanden, J. M. Barbaree, W. E. Morrill, R. M. Wadowsky, E. H. White, and J. C. Feeley. 1989. Intracellular multiplication of Legionella pneumophila in amoebae isolated from hospital hot water tanks. Curr. Microbiol. 18:131-137. [Google Scholar]
- 15.Fields, B. S., E. B. J. Shotts, J. C. Feeley, G. W. Gorman, and W. T. Martin. 1984. Proliferation of Legionella pneumophila as an intracellular parasite of the ciliated protozoan Tetrahymena pyriformis. Appl. Environ. Microbiol. 47:467-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greub, G., and D. Raoult. 2003. Morphology of Legionella pneumophila according to their location within Hartmanella vermiformis. Res. Microbiol. 154:619-621. [DOI] [PubMed] [Google Scholar]
- 18.Grimm, D., W. Ludwig, B. C. Brandt, R. Michel, K.-H. Schleifer, J. Hacker, and M. Steinert. 2001. Development of 18S rRNA-targeted oligonucleotide probes for specific detection of Hartmannella and Naegleria in Legionella-positive environmental samples. Syst. Appl. Microbiol. 24:76-82. [DOI] [PubMed] [Google Scholar]
- 19.Grimm, D., H. Merkert, W. Ludwig, K.-H. Schleifer, J. Hacker, and B. C. Brand. 1998. Specific detection of Legionella pneumophila: construction of a new 16S rRNA-targeted oligonucleotide probe. Appl. Environ. Microbiol. 64:2686-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunderson, J. H., S. J. Goss, and M. L. Sogin. 1994. The sequence of the Hartmannella vermiformis small subunit rRNA coding region. J. Eukaryot. Microbiol. 41:481-482. [DOI] [PubMed] [Google Scholar]
- 21.Hagele, S., R. Kohler, H. Merkert, M. Schleicher, J. Hacker, and M. Steinert. 2000. Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell Microbiol. 2:165-171. [DOI] [PubMed] [Google Scholar]
- 22.Harf, C., and H. Monteil. 1988. Interactions between free-living amoebae and Legionella in the environment. Water Sci. Technol. 11/12:235-239. [Google Scholar]
- 23.Kuchta, J. M., J. S. Navratil, M. E. Shepherd, R. M. Wadowsky, J. N. Dowling, S. J. States, and R. B. Yee. 1993. Impact of chlorine and heat on the survival of Hartmannella vermiformis and subsequent growth of Legionella pneumophila. Appl. Environ. Microbiol. 59:4096-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchta, J. M., S. J. States, R. M. Wadowsky, and T. J. Byers. 1998. Interactions of Legionella pneumophila with Hartmannella vermiformis including the efficacy of chlorine or copper and silver ions to disrupt the intra-amoebic multiplication of L. pneumophila. Recent Res. Dev. Microbiol. 2:405-425. [Google Scholar]
- 25.Leoni, E., P. P. Legnani, M. A. Bucci Sabattini, and F. Righi. 2001. Prevalence of Legionella spp. in swimming pool environment. Water Res. 35:3749-3753. [DOI] [PubMed] [Google Scholar]
- 26.Levin, A. S. S., S. Gobara, C. M. Scarpitta, C. L. Warschauer, S. I. Sinto, E. Rodrigues, C. M. F. Mendes, E. Sabbaga, and M. Boulos. 1995. Electric showers as a control measure for Legionella spp. in a renal transplant unit in Sao Paulo, Brazil. J. Hosp. Infect. 30:133-137. [DOI] [PubMed] [Google Scholar]
- 27.Lin, Y. E., R. D. Vidic, J. E. Stout, and V. L. Yu. 1998. Legionella in water distribution systems. JAWWA 90:112-121. [PubMed] [Google Scholar]
- 28.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Y. Kumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. P. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, B. Arndt, and K.-H. Schleifer. 2003. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manz, W., R. Amann, R. Szewzyk, U. Szewzyk, T.-A. Stenstrom, P. Hutzler, and K.-H. Schleifer. 1995. In situ identification of Legionellaceae using 16S rRNA-targeted oligonucleotide probes and confocal laser scanning microscopy. Microbiology 141:29-39. [DOI] [PubMed] [Google Scholar]
- 30.Moon-Van Der Staay, S.-Y., G. W. M. Van der Staay, L. Guillou, and D. Vaulot. 2000. Abundance and diversity of prymnesiophytes in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol. Oceanogr. 45:98-109. [Google Scholar]
- 31.Murga, R., T. S. Forster, E. Brown, J. M. Pruckler, B. S. Fields, and R. M. Donlan. 2001. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147:3121-3126. [DOI] [PubMed] [Google Scholar]
- 32.Page, F. C. 1976. An illustrated key to freshwater and soil amoebae. Scientific publication no. 34. Freshwater Biological Association, Cumbria, United Kingdom.
- 33.Page, F. C. 1967. Taxonomic criteria for limax amoebae, with descriptions of 3 new species of Hartmannella and 3 of Vahlkampfia. J. Protozool. 14:499-521. [DOI] [PubMed] [Google Scholar]
- 34.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers, J., A. Dowsett, P. Dennis, J. Lee, and C. Keevil. 1994. Influence of plumbing materials on biofilm formation and growth of Legionella pneumophila in potable water systems. Appl. Environ. Microbiol. 60:1842-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers, J., A. Dowsett, P. Dennis, J. Lee, and C. Keevil. 1994. Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers, J., and C. Keevil. 1992. Immunogold and fluorescein immunolabeling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58:2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 39.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroeder, J. M., G. C. Booton, J. Hay, I. A. Niszl, D. V. Seal, M. B. Markus, P. A. Fuerst, and T. J. Byers. 2001. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol. 39:1903-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skinner, A. R., C. M. Anand, A. Malic, and J. B. Kurtz. 1983. Acanthamoebae and environmental spread of Legionella pneumophila. Lancet i:289-290. [DOI] [PubMed] [Google Scholar]
- 42.Surman, S., G. Morton, B. Keevil, and R. B. Fitzgeorge. 2002. Legionella pneumophila proliferation is not dependent on intracellular replication, p. 86-89. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Luck (ed.), Legionella. ASM Press, Washington, D.C.
- 43.Tobin, J. O. H., M. S. Dunnill, M. French, P. J. Morris, J. Beare, S. P. Fischer-Hoch, R. G. Mitchell, and M. F. Muers. 1980. Legionnaires' disease in a transplant unit: isolation of the causative agent from shower baths. Lancet ii:118-121. [DOI] [PubMed] [Google Scholar]
- 44.Van der Kooij, D., H. R. Veenendaal, N. P. G. Slaats, and D. Vonk. 2002. Biofilm formation and multiplication of Legionella on synthetic pipe materials in contact with treated water under static and dynamic conditions, p. 176-180. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Luck (ed.), Legionella. ASM Press, Washington, D.C.
- 45.Wadowsky, R. M., L. J. Butler, M. K. Cook, S. M. Verma, M. A. Paul, B. S. Fields, G. Keleti, J. L. Sykora, and R. B. Yee. 1988. Growth-supporting activity for Legionella pneumophila in tap water cultures and implication of hartmannellid amoebae as growth factors. Appl. Environ. Microbiol. 54:2677-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wadowsky, R. M., T. M. Wilson, N. J. Kapp, A. J. West, J. M. Kuchta, S. J. States, J. N. Dowling, and R. B. Yee. 1991. Multiplication of Legionella spp. in tap water containing Hartmannella vermiformis. Appl. Environ. Microbiol. 57:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wadowsky, R. M., R. Wolford, A. M. McNamara, and R. B. Yee. 1985. Effect of temperature, pH, and oxygen level on the multiplication of naturally occurring Legionella pneumophila in potable water. Appl. Environ. Microbiol. 49:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walochnik, J., R. Michel, and H. Aspöck. 2002. Discrepancy between morphological and molecular biological characters in a strain of Hartmannella vermiformis Page 1967 (Lobosea, Gymnamoebia). Protistology 2:185-188. [Google Scholar]
- 49.Weekers, P. H. H., R. J. Gast, P. A. Fuerst, and T. J. Byers. 1994. Sequence variations in small-subunit ribosomal RNAs of Hartmannella vermiformis and their phylogenetic implications. Mol. Biol. Evol. 11:684-690. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto, H., M. Sugiura, S. Kusunoki, T. Ezaki, M. Ikedo, and E. Yabuuchi. 1992. Factors stimulating propagation of legionellae in cooling tower water. Appl. Environ. Microbiol. 58:1394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]