Abstract
The heterologous expression of DNA extracted directly from environmental samples (environmental DNA [eDNA]) in easily cultured hosts provides access to natural products produced by previously inaccessible microorganisms. When eDNA cosmid libraries were screened in Escherichia coli for antibacterially active clones, long-chain N-acyltyrosine-producing clones were found in every eDNA library. These apparently common natural products have not been previously described from screening extracts of cultured bacteria for biologically active natural products. Of the 11 long-chain N-acyl amino acid synthases (NASs) that were characterized, 10 are unique sequences. A predicted protein of previously unknown function from Nitrosomonas europaea, a gram-negative nitrifying beta-proteobacterium, is 14 to 37% identical to eDNA NASs. When cloned into E. coli, this open reading frame confers the production of long-chain N-acyltyrosines to the host and is therefore the first NAS from a cultured bacterium to be functionally characterized. Understanding the role that long-chain N-acyl amino acids play in soil microbial communities should now be feasible with the identification of a cultured organism that has the genetic capacity to produce these compounds.
Many lines of evidence now suggest that only a tiny and unrepresentative minority of soil microbes are cultured by using conventional approaches (2, 10, 11, 14, 17, 18, 20). Soil microbes that have not yet been cultured outnumber their cultured counterparts by 2 to 3 orders of magnitude, and this uncultured majority no doubt produces secondary metabolites that could serve as molecular probes of biological processes and therapeutic agents. Unfortunately, no generally useful methods of culturing these uncultured and hence unstudied microorganisms have been identified. We and others have worked on a general approach for accessing the natural products of uncultured microorganisms that omits culturing the producing organism (3-5, 7, 19). Heterologous expression of large fragments of microbial DNA extracted directly from environmental samples (environmental DNA [eDNA]) in easily cultured hosts should provide access to at least some currently inaccessible natural products (9).
In our approach, which is designed to find small-molecule antibiotics, eDNA is used to prepare cosmid libraries in Escherichia coli, and antibacterially active eDNA clones are found by using a double-antibiotic selection screen that was developed to identify and recover antibacterially active clones directly from the original library selection plates. Two active clones, CSL12 and CSLC2, were initially characterized, and both were found to produce long-chain N-acyltyrosine antibiotics (3, 5). The long-chain N-acyl amino acid synthase (NAS) from CSLC2 is part of a biosynthetic gene cluster of 13 open reading frames (ORFs) that codes for the production of long-chain N-acyltyrosines as well as two additional families of long-chain acyl phenols that are related to N-acyltyrosines but were not found to be antibacterially active (3, 6).
The presence of multiple different N-acyltyrosine-producing clones in the initial two eDNA libraries that were screened suggested that NAS genes might occur at a high frequency in eDNA. However, the rapid identification of additional NAS sequences from eDNA by hybridization or PCR-based strategies was not feasible because the eDNA NASs showed very little overall sequence similarity. The use of an expression-based screening strategy was the only feasible approach to identify additional NASs in eDNA samples. Clones from seven independent soil samples were screened for the production of antibacterial activities, and cultures of these clones were assayed for the presence of long-chain N-acyltyrosines. Environmental DNA clones that produce long-chain N-acyltyrosine antibiotics were found in all seven eDNA libraries that were screened. The routine identification of long-chain N-acyltyrosine-producing clones in eDNA libraries constructed from multiple geographically distinct environmental samples suggests that these compounds may play an important, if undefined, role for soil microbes. To explore the genetic diversity underlying the production of these apparently common natural products, nine additional long-chain N-acyltyrosine-producing clones were characterized.
MATERIALS AND METHODS
Sample library construction protocol.
eDNA used to construct cosmid libraries was isolated directly from samples collected in Ithaca, N.Y., Boston, Mass., and Costa Rica. DNA was extracted by heating environmental samples in lysis buffer (100 mM Tris-HCl, 100 mM sodium EDTA, 100 mM sodium phosphate, 1.5 M NaCl, 1% hexacetyltrimethylammonium bromide, 2% sodium dodecyl sulfate, adjusted to pH 8.0) (23) at 70°C for 2 h. The suspension was then extracted once with an equal volume of chloroform (optional), and eDNA was precipitated from the centrifuge-clarified aqueous phase with the addition of 0.6 volumes of isopropanol. The resulting pellet was resuspended overnight in sterile H2O and then run on a preparative 0.7% agarose gel. After 1 h at 100 V and an additional 14 to 20 h at 20 V, the edges of the gel were removed and stained with ethidium bromide. High-molecular-weight (HMW) eDNA, which separated from the remaining soil debris and smaller sheared eDNA, was electroeluted (100 V, three times for 45 min) from a slice of the remaining unstained preparative gel. The purified HMW eDNA was either partially digested with BamHI and dephosphorylated with calf intestinal alkaline phosphatase (New England Biolabs [NEB]) or blunt ended (end-repair enzyme mix; Epicentre) to yield large fragments of ligation-ready eDNA. Twenty-microliter ligation reactions containing 1 μg of the Supercos I vector treated in a stepwise fashion with XbaI, calf intestinal alkaline phosphatase and BamHI, 0.5 of 2.0 μg of HMW eDNA partially digested by BamHI, and 2,000 U of T4 DNA ligase were incubated overnight at 16°C. Aliquots of the ligation mix were subsequently packaged into Gigapack III Gold packaging extracts (Stratagene) and used to transform E. coli XL1-Blue MR cells. Blunt-ended HMW eDNA was ligated into the pWEB vector prepared to be ready for cloning as described by the vendor (Epicentre). The ligation reactions were then packaged into MaxPlax packaging extracts (Epicentre) and used to transform E. coli EC 100 cells.
Transposon mutagenesis and sequence analysis.
The cosmids isolated from cultures of active clones were randomly transposon mutagenized with donor plasmid pGPS2.1 by using the genome priming system (NEB), or alternatively, an antibacterially active NotI subclone in pBC (Stratagene) was randomly transposon mutagenized with donor plasmid pGPS1.1 (NEB). E. coli transformed with the transposon-mutagenized cosmids was selected on Luria-Bertani (LB) agar plates containing kanamycin (30 μg/ml), ampicillin (50 μg/ml; optional), and chloramphenicol (20 μg/ml). The triply resistant transformants were then patched in duplicate onto LB agar plates containing both kanamycin and chloramphenicol and incubated at 30°C for 2 to 3 days. One set of the duplicate plates was overlayed with top agar containing Bacillus subtilis (BR151/pPL608) (22) to identify the mutagenized cosmid clones that no longer produced antibacterial activity. Cosmids were isolated from inactive colonies that were picked from the undisturbed plate, and the eDNA sequence surrounding the site of the transposon insertion was determined with primers S and N (NEB) that are specific for the genome priming system transposon. The sequence derived from transposon insertions that disrupted the production of antibacterial activity was used to construct the eDNA sequence responsible for the observed antibacterial activity. Primer walking was used to complete each sequence and confirm the sequence obtained from the transposon mutagenesis experiments. Sequencing was performed at the Cornell University BioResource Center. The cladogram of NAS sequences was constructed with MacVector (version 7.1; Oxford Molecular Ltd.) by using ClustalW (16).
GST fusion proteins.
The predicted NASs from Nitrosomonas europaea and Desulfovibrio vulgaris were amplified from genomic DNA (American Type Culture Collection) by using the FailSafe PCR system from Epicentre and primer pairs 5′-GCGCTGATCATGCAAATTGCGAATCATCTTCAATC-3′ and 5′-GCGCGAATTCCGGTACGATGAAATTGGTTCCGA-3′ (BclI and EcoRI) for N. europaea and 5′-GCGAGGGATCCATGCTCGCCATCGCCGACAAGCGCCGT-3′ and 5′-GTCACGAAGCTTCTCGCCCTCACTCGGCAGGCAGCAGGA-3′ (BamHI and HindIII) for D. vulgaris. Each PCR product was gel purified (QIAquick; QIAGEN), digested with the appropriate restriction endonucleases (as indicated), and ligated into gel-purified pGEX-3X (Pharmacia Biotech) or pET28a (Novagen) to give pNeGST and pDvulHIS, respectively. The genes for N-acyltyrosine synthase 2 (NasY2) and NasY9 were amplified from pCSL132 (5′CCGGATCCCCATGATGTCTCTACCTGCTTACCACT-3′ and 5′-GGAATTCTGCATTTCTGGCGTTTGATCTGCTT-3′ [BamHI and EcoRI]) and pCSLG10 (5′-CGCGGGATCcATGAAGCTTATACCAGGTTCT-3′ and 5′-GCGAATTCCGAACAATATACCGGCTGACCG-3′ [BamHI and EcoRI]), respectively. Each PCR product was gel purified (QIAquick; QIAGEN), digested with the appropriate restriction endonucleases (as indicated), and ligated into gel-purified pGEX-3X or pETMAL to give pNasY2GST and pNasY9MAL, respectively.
Isolation of N-acyl amino acids.
The original eDNA cosmid clones were grown in LB agar (30 μg of kanamycin/ml) for 3 days at room temperature or 30°C. E. coli BL21(DE3) transformed with pNeGST, pDvulHIS, pNasY2GST, or pNasY9MAL was grown at 37°C to an optical density at 600 nm of 0.7, at which point the temperature was reduced to 20°C and the cultures were induced with 10 μM isopropyl-β-d-thiogalactopyranoside for an additional 18 h at 20°C. The mature cultures were neutralized with 2N HCl and extracted twice with an equal volume of ethyl acetate. The dried organic extracts were then concentrated in vacuo and analyzed directly by fast atom bombardment mass spectrometry (FABMS), or alternatively, the resulting extracts were either analyzed directly or partitioned by silica gel flash chromatography (step gradient CHCl3-methanol modified with 0.1% HOAc), and the active material that eluted from the column was analyzed by mass spectrometry. The FABMS analysis was performed by the University of Illinois (Urbana) Mass Spectrometry Facility.
Bioautography.
Crude ethyl acetate extracts and column fractions were chromatographed on silica gel thin-layer chromatography (TLC) plates (90:10 CHCl3-methanol), and then the dried TLC plate was overlayed onto an LB agar plate containing B. subtilis. After 1 to 2 h, the TLC plate was removed from the agar surface, and the agar plate containing the transferred extract was incubated at 30°C overnight. The pattern of growth inhibition in the bacterial lawn was then recorded the following day.
Nucleotide sequence accession numbers.
The eDNA NASs have been deposited in GenBank under to the following accession numbers: AF324335 (CSL12, NasY1) (5), AY214921 (CSL132, NasY2), AY214922 (CSL144, NasY3), AY214923 (CSL147, NasY1), AY128669 (CSLC2, FeeM) (3), AY214924 (CSLC3, NasY5), AY214925 (CSLD10, NasY6), AY214926 (CSLF42, NasY7), AY214927 (CSLF43, NasY8), AY653031 (CSLG7, NasY9), and AY653030 (CSLG10, NasY10).
RESULTS AND DISCUSSION
Library construction and screening.
E. coli-based cosmid libraries were constructed from DNA isolated from seven different environmental samples, five collected in Ithaca, N.Y., one collected in Boston, Mass., and one collected in Costa Rica (Table 1). Clones from each of these libraries were then screened for the production of antibacterial activities by using a double-antibiotic selection scheme. Cosmid clones initially selected on LB plates containing kanamycin were allowed to incubate at 30°C for 24 h, and then after an additional 2 to 4 days at room temperature, the mature colonies were overlayed with top agar containing kanamycin-resistant B. subtilis (BR151/pPL608) (22). Colonies that produced zones of growth inhibition in the B. subtilis lawn, indicating the production of antibacterial activity, were picked through the top agar and cleared of the ampicillin-sensitive assay strain by streaking the active colonies onto LB plates containing ampicillin. Individual colonies were picked from the ampicillin plates and reassayed for antibacterial activity with the same B. subtilis top agar overlay assay. A single active clone from each secondary screen was then archived and characterized in further detail. In the original overlay assay, approximately 1 in every 10,000 to 20,000 eDNA clones produced a zone of growth inhibition against the B. subtilis assay strain. Antibacterially active clones found in the initial overlay assay were grown in liquid culture, and the ethyl acetate extracts obtained from these cultures were assayed for antibiosis by using bioautography.
TABLE 1.
Plasmids and eDNA clones
| Cosmid or protein | eDNA collection site or protein description | NAS | Reference or source |
|---|---|---|---|
| eDNA cosmids | |||
| pCSL12 | Park Park (no. 1), Ithaca, N.Y. | NasY1 | 5 |
| pCSL132 | Sapsucker Woods, Ithaca, N.Y. | NasY2 | This paper |
| pCSL144 | Park Park (no. 2), Ithaca, N.Y. | NasY3 | This paper |
| pCSL147 | Park Park (no. 2), Ithaca, N.Y. | NasY1 | This paper |
| pCSLC2 | Beebe Lake, Ithaca, N.Y. | FeeM | 3 |
| pCSLC3 | Beebe Lake, Ithaca, N.Y. | NasY5 | This paper |
| pCSLD10 | Cornell University, Ithaca, N.Y. | NasY6 | This paper |
| pCSLF42 | Bromeliad tank, “INBio finca,” Costa Rica | NasY7 | This paper |
| pCSLF43 | Bromeliad tank, “INBio finca,” Costa Rica | NasY8 | This paper |
| pCSLG7 | Riverway, Boston, Mass. | NasY9 | This paper |
| pCSLG10 | Riverway, Boston, Mass. | NasY10 | This paper |
| Fusion protein constructs | |||
| pNasY9MAL | pETMAL with NasY9 | This paper | |
| pNasY2GST | pGEX-3X with NasY2 | This paper | |
| pFeeMGST | pGEX-3X with FeeM | This paper | |
| pDvuIHIS | pET28a with NAS-like ORF from D. vulgaris | This paper | |
| pNeGST | pGEX-3X with NAS from N. europaea | This paper |
Two antibacterially active clones that produced antibacterially active ethyl acetate extracts, CSL12 and CSLC2, were initially characterized from the pool of antibacterially active clones found with the B. subtilis overlay assay. Bioassay-guided fractionation of the ethyl acetate extracts from cultures of CSL12 and CSLC2 led to the characterization of long-chain N-acyltyrosine antibiotics that contain both fully saturated and monounsaturated fatty acids ranging from 8 to 18 carbons in length (Fig. 1) (3, 5). Cosmids pCSL12 and pCSLC2 were found to contain single ORFs that are necessary and sufficient to confer the production of these compounds to the E. coli host. To further explore this apparently common family of metabolites, additional clones from a total of seven eDNA libraries were screened for the production of long-chain N-acyltyrosines.
FIG. 1.
Identity matrix (a) and cladogram (b) derived from ClustalW sequence alignments of the conceptual translation for each predicted NAS characterized from eDNA. The eDNA NASs cluster into at least two groups (group I and group II) and possibly a third (NasY3).
Long-chain N-acyltyrosines.
Clones that produced extracts with bioautography patterns similar to those produced by extracts from cultures of CSL12 and CSLC2 were found in all seven of the eDNA libraries that were screened. A random subset of this family of antibacterially active clones (CSL12, CSL132, CSL144, CSL147, CSLC2, CSLC3, CSLD10, CSLF42, CSLF43, CSLG7, and CSLG10) was selected for extensive characterization (Table 1). Extracts from cultures of each clone were analyzed by FABMS and shown to contain m/z peaks corresponding to long-chain N-acyltyrosine antibiotics similar to those found in extracts from cultures of CSL12 and CSLC2 (Table 2). The most frequently encountered derivative of tyrosine is modified with a C14 fatty acid, although derivatives ranging from 8 to 18 carbons in length were also identified in these extracts.
TABLE 2.
Major molecular ions (m/z) observed in the ethyl acetate extract from E. coli cultures transformed with eDNA NAS and the predicted long-chain N-acyltyrosines that correspond to each of the observed masses
Masses were obtained for all of the pure compounds that were isolated by high-performance liquid chromatography (see references 3 [CSLC2] and 5 [CSL12]). In all other examples, only the major components seen by FABMS analysis of crude extracts are reported.
MAL, maltose-binding fusion protein; Ne, NAS from N. europaea.
Long-chain N-acyltyrosine synthase genes.
The nasY gene present in each cosmid was located by transposon mutagenesis and sequenced (Table 1 and Fig. 1). Of the 11 nasY genes that were sequenced, 10 are unique and none had been reported from a cultured organism. The NASs found in CSL12 and CSL147 (NasY1) are 100% identical. They were found in two different libraries and represent the only redundant sequences that were identified in these screens. Environmental DNA libraries provide an opportunity to rapidly screen a much larger pool of genetic diversity than could traditionally be achieved working with individual strains of bacteria in pure culture. Because the vast majority of DNA present in an eDNA library should be derived from previously uncultured bacteria, probing these libraries with robust yet sensitive bioassays should consistently yield high numbers of previously unreported sequences.
Sequence analysis.
No absolutely conserved residues were detected in a ClustalW (16) alignment using all of the eDNA-derived NAS sequences. The ClustalW-derived phylogenetic tree produced from this alignment suggests that the conceptually translated proteins from 9 out of the 10 unique eDNA NASs fall into two distinct groups, and the remaining NAS, NasY3, is not significantly related by sequence identity to either group (Fig. 1). Group I contains the original two NASs known to produce long-chain N-acyltyrosines, NasY1 from CSL12 and FeeM from CSLC2, as well as three new NASs, NasY2, NasY5, and NasY7. There are three highly conserved regions in this family of NASs: (G/A)(T/S)(I/L/V)(S/T)(I/L/V), (V/I)NP(R/K)H, and NAPA(V/I)(A/L) (Fig. 2). Although FeeM and NasY7 show only limited overall sequence identity to the other NASs in this large family, they contain the conserved residues seen in the other sequences (Fig. 1 and 2). The low overall sequence similarity observed between FeeM and the other sequences in this group is at least in part due to FeeM being between 58 and 78 residues shorter than each of the other sequences in the group (Fig. 2).
FIG. 2.
ClustalW-derived multiple-sequence alignment of the group I long-chain NASs. Identical residues appear in black boxes, and similar residues appear in open boxes. The three conserved regions in the group I NASs are underlined.
The four sequences that make up group II show high sequence identity (≥40%) and have a large number of absolutely conserved residues. The inclusion of NasY3 in the multiple-sequence alignment of group II results in only 11 absolutely conserved residues, and if NasY3 is included in the group I alignment, only five of the absolutely conserved residues shown in Fig. 2 are still conserved in the new alignment. NasY3 could be distantly related to either group I or group II and would therefore represent a link between groups I and II. It is also possible that NasY3 represents a third group of NASs that is distinct from both groups. Additional NasY sequences are needed to determine the precise relationships within this group of enzymes.
Homserine lactones and AISs.
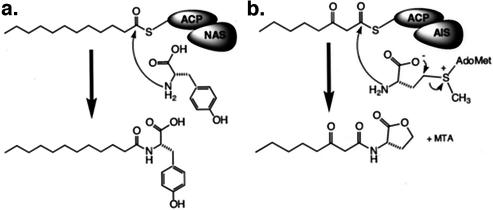
NasY6 and NasY10 from group II show low-level sequence identity (15% identity) to a putative autoinducer synthase (AIS) (GenBank accession number ZP 00004916) from the cultured bacterium Rhodobacter sphaeroides (8, 12). AISs condense S-adenosylmethionine with a variety of acyl carrier protein (ACP)-linked fatty acids to produce medium- and long-chain N-acyl homoserine lactones that are used by gram-negative bacteria as quorum sensors (Fig. 3b) (13). Multiple-sequence alignments with AISs contain only a few highly conserved residues, and no obvious catalytic residues are conserved within this large family of sequences (12, 21). Many of the conserved residues are charged and thought to hold the S-adenosylmethionine in the proper orientation for a reaction with the phosphopantetheinyl-linked fatty acid of the ACP. The low overall sequence conservation within AISs suggests that properly orienting the S-adenosylmethionine with respect to the highly activated incoming ACP-linked fatty acid may be the sole requirement for catalysis by this family of enzymes. Although AISs show only limited sequence identity to N-acyl amino acid biosynthesis genes, both families of enzymes produce long-chain N-acyl amino acid derivatives, and this limited sequence similarity may indicate a shared biosynthetic strategy. The long-chain NASs may therefore catalyze the transfer of a fatty acid directly from the endogenous E. coli ACP-linked fatty acid onto an amino acid (Fig. 3a).
FIG. 3.
Hypothetical scheme for the biosynthesis of long-chain N-acyltyrosines by NASs expressed in E. coli. The proposed use of an ACP-linked fatty acid in the biosynthesis of long-chain N-acyltyrosines (a) is similar to the biosynthesis of acylhomoserine lactones by AISs for cultured bacteria (b).
As with AIS sequence alignments, only a limited number of conserved residues are seen in the multiple-sequence alignment of group I sequences. The highly conserved residues seen in AIS alignments are not readily identifiable in alignments of eDNA-derived NASs. In the proposed biosynthesis of N-acyltyrosines, which is analogous to that of homoserine lactones by AISs, fatty acids are presented to an NAS as preactivated phosphopantetheinyl-derivatized ACP linked substrates. The use of an ACP-activated fatty acid by NASs and AISs could explain the absence of any highly conserved catalytic residues. In addition, the limited fatty acid selectivity by the two systems could help explain the absence of additional highly conserved residues within these families of enzymes. The limited number of absolutely conserved residues seen in the NAS group I alignment may therefore be involved in specifically binding the amino acid. These conserved residues likely control the selective binding of tyrosine as well as hold it in the proper orientation to react with incoming ACP-linked fatty acids. At least one three-dimensional structure analysis, in conjunction with site-directed mutagenesis, is needed to confirm the role these residues play in the biosynthesis of long-chain N-acyltyrosines.
NASs are sufficient to biosynthesize long-chain N-acyl amino acids in E. coli.
At least one representative enzyme from each of the two major groups of NASs (NasY2 and FeeM from group I and NasY9 from group II) was subcloned as a glutathione S-transferase (GST) fusion protein and assayed for the ability to produce N-acyl amino acids in E. coli. FABMS analysis of ethyl acetate extracts from cultures of E. coli transformed with each GST fusion construct confirmed the production of long-chain N-acyltyrosines by the NAS fusion proteins (Table 2). Although variations in the array of fatty acids conjugated onto the amino acid head group were observed in extracts from different fermentation conditions and from cultures containing the GST fusion proteins, long-chain N-acyltyrosines were consistently produced by cultures transformed with eDNA NASs.
N. europaea.
Although long-chain N-acyltyrosines have never been reported from the extensive screening of extracts from cultured organisms for antibacterial activity, three potential NASs were identified in BLAST searches using group I eDNA NAS sequences. NasY3 and group II NASs show no significant sequence identity (<20%) to sequences derived from cultured bacteria. The group I NASs are similar to a predicted protein of unknown function from N. europaea (GenBank accession number ZP 00003821) and to the N-terminal ∼250 amino acids of two predicted proteins of unknown function from Desulfovibrio vulgaris (989 amino acids) and Desulfovibrio desulfuricans (1,006 amino acids) (accession number ZP 00130011) that are 51% identical. The central region of the two large Desulfovibrio proteins is similar to the ThiF/MoeB (MeoW)/HesA (1, 15) family of nucleotide-binding proteins that are thought to activate carboxylic acids, while the C-terminal ends of these predicted proteins show no significant similarity to any deposited sequences of known function.
To assess the ability of these ORFs to confer the production of long-chain N-acyltyrosines to E. coli, the proposed NAS from D. vulgaris, a representative of the two Desulfovibrio ORFs, and the proposed NAS from N. europaea were subcloned into E. coli as fusion proteins. No clone-specific compounds were seen in ethyl acetate extracts from E. coli cultures containing a His6 fusion of the proposed NAS from D. vulgaris (pDvulHIS). Although the overexpression of this large ORF as a His6 fusion protein did not lead to the production of detectable levels of N-acyl amino acids under the conditions tested, this does not rule out the possibility that it could produce N-acyl amino acids in E. coli or D. vulgaris under alternative expression conditions. The proposed NAS domain in conjunction with the two additional domains in the large Desulfovibrio ORFs could also be used in the biosynthesis of an as-yet-undefined N-acyl amino acid-based natural product. Ethyl acetate extracts from cultures of E. coli containing the N. europaea GST (pNeGST) fusion protein contained long-chain N-acyltyrosines (m/z of 390, 392) as seen with the eDNA NASs. This N. europaea ORF is the first NAS from a cultured bacterium that has been functionally characterized. Understanding the role that long-chain N-acyltyrosines play in soil microbial communities should now be feasible with the identification of a cultured organism that has the genetic capacity to produce these compounds.
Conclusions.
Environmental DNA clones that produce long-chain N-acyltyrosine antibiotics were found in all seven eDNA libraries that were screened, five constructed from soil collected in New York, N.Y., one from soil collected in Boston, Mass., and one from sediment collected in Costa Rica. Of the 11 eDNA NASs that have been characterized, 10 are unique sequences and none have been previously described from cultured organisms. NASs derived from eDNA show low-level (<20%) sequence identity to AISs, the biosynthetic enzymes for homoserine lactones. Long-chain N-acyltyrosines may therefore be biosynthesized from the condensation of an ACP-linked fatty acid with the primary amine of an amino acid in a mechanism similar to that used in the biosynthesis of homoserine lactones. The identification of NAS-like sequences in three recently determined bacterial genomes led to the functional annotation of an ORF of previously unknown function from N. europaea as an NAS. Understanding the role that long-chain N-acyl amino acids play in soil microbial communities should now be feasible with the identification of a cultured organism that has the genetic capacity to produce these compounds.
In the search for bioactive natural products from easily cultured bacteria, even common microbial metabolites may have been overlooked because they are not produced by the small fraction of bacteria that grow easily in the laboratory. The heterologous expression of eDNA in easily cultured hosts should provide access to many of these previously inaccessible natural products.
Acknowledgments
This work was supported by the Ellison Medical Foundation and NIH grant CA59021.
REFERENCES
- 1.Appleyard, M. V. C. L., J. Sloan, G. J. M. Kana'n, I. S. Heck, J. R. Kinghorn, and S. E. Unkles. 1998. The Aspergillus nidulans cnxF gene and its involvement in molybdopterin biosynthesis. J. Biol. Chem. 273:14869-14876. [DOI] [PubMed] [Google Scholar]
- 2.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea in soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady, S. F., C. J. Chao, and J. Clardy. 2002. New natural product families from an environmental DNA (eDNA) gene cluster. J. Am. Chem. Soc. 124:9968-9969. [DOI] [PubMed] [Google Scholar]
- 4.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 5.Brady, S. F., and J. Clardy. 2000. Long-chain N-acyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J. Am. Chem. Soc. 122:12903-12904. [Google Scholar]
- 6.Brady, S. F., and J. Clardy. 2003. Synthesis of long-chain fatty acid enol esters isolated from an environmental DNA clone. Org. Lett. 5:121-124. [DOI] [PubMed] [Google Scholar]
- 7.Gillespie, D. E., S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R. Rondon, J. Clardy, R. M. Goodman, and J. Handelsman. 2002. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray, K. M., and E. P. Greenberg. 1992. Sequencing and analysis of luxR and luxI, the luminescence regulatory genes from the squid light organ symbionts Vibrio fischeri ES114. Mar. Mol. Biol. Biotechnol. 6:413-419. [Google Scholar]
- 9.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biology provides access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 10.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milton, D. L., A. Hardman, M. Camara, M., S. R. Chhabra, B. W. Bycroft, G. S. Stewart, and J. Williams. 1997. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J. Bacteriol. 179:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.More, M. I., L. D. Finger, J. L. Stryker, C. Fuqua, A. Eberhard, and S. C. Winans. 1996. Enzymatic synthesis of a quorum-sensing autoinducer using defined substrates. Science 272:1655-1658. [DOI] [PubMed] [Google Scholar]
- 14.Stackebrandt, E., W. Liesack, and B. M. Goebel. 1993. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 7:232-236. [DOI] [PubMed] [Google Scholar]
- 15.Taylor, S. V., N. L. Kelleher, C. K. Kinsland, H.-J. Chiu, C. A. Costello, A. D. Backstrom, F. W. McLafferty, and T. P. Begley. 1998. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 273:16555-16560. [DOI] [PubMed] [Google Scholar]
- 16.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torsvik, V., K. Salte, R. Sorheim, and J. Goksoyr. 1990. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl. Environ. Microbiol. 56:776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, G. Y., E. Graziani, B. Waters, W. Pan, X. Li, J. McDermott, G. Meurer, G. Saxena, R. J. Anderson, and J. Davies. 2000. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2:2401-2404. [DOI] [PubMed] [Google Scholar]
- 20.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 21.Watson, W. T., T. D. Minogue, D. L. Val, S. Beck von Bodman, and M. E. A. Churchill. 2002. Structural basis of quorum sensing signal generating in bacterial pathogenesis. Mol. Cell 9:685-694. [DOI] [PubMed] [Google Scholar]
- 22.Williams, D. M., R. G. Schoner, E. J. Duvall, L. H. Preis, and P. S. Lovett. 1981. Expression of Escherichia coli trp genes and the mouse dihydrofolate reductase gene cloned in Bacillus subtilis. Gene 16:199-206. [DOI] [PubMed] [Google Scholar]
- 23.Zhou, J., M. A. Bruns, and J. M. Tiedge. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]