Abstract
A number of actinomycetes isolates were recovered from coastal sediments in Aberystwyth (Wales, United Kingdom) with standard isolation techniques. Most of them were putatively assigned to the genera Streptomyces and Micromonospora on the basis of their morphological characteristics, and there appeared to be no difference whether the isolation media contained distilled water or seawater. A group of 20 Micromonospora isolates was selected to undergo further polyphasic taxonomic investigation. Three approaches were used to analyze the diversity of these isolates, 16S rDNA sequencing, fluorescent amplified fragment length polymorphism (AFLP), and Fourier transform infrared spectroscopy (FT-IR). The 16S rDNA sequence analysis confirmed that all of these isolates should be classified to the genus Micromonospora, and they were analyzed with a group of other Micromonospora 16S rDNA sequences available from the Ribosomal Database Project. The relationships of the 20 isolates were observed after hierarchical clustering, and almost identical clusters were obtained with these three techniques. This has obvious implications for high-throughput screening for novel actinomycetes because FT-IR spectroscopy, which is a rapid and reliable whole-organism fingerprinting method, can be applied as a very useful dereplication tool to indicate which environmental isolates have been cultured previously.
Actinomycetes are ubiquitous in many niches such as soil, activated sludge, and water. Actinomycetes, especially Streptomyces spp., are an important group of bacteria, not only as degraders of organic matter in the natural environment, but because they are also producers of antibiotics and other useful compounds of commercial interest (6, 8, 26). Nearly 8,000 actinomycete-derived antibiotics had been described by 1994, of which 80% were from Streptomyces species and 20% from other actinomycete genera. Besides Streptomyces, which has been extensively exploited, members of the other genera such as Micromonospora are also considered of high interest as potential producers of novel bioactive compounds. Micromonospora is the type genus of the family Micromonosporaceae and contains many interesting strains, such as antibiotic producers and degraders of natural rubber (16, 23, 30).
The ability to measure bacterial diversity is prerequisite for the systematic study of bacterial biogeography and community assembly. It is therefore central to the ecology of surface waters, the oceans and soils, waste treatment, agriculture, and global elemental cycles. However, the experimental definition of bacterial diversity has never been undertaken for any naturally occurring bacterial community, and the extent of prokaryotic diversity is widely held to be beyond practical calculation (7). While the gold standard environmental diversity metric is currently based on 16S rDNA sequences (49), it is widely recognized that polyphasic taxonomic approaches are better, since they take into account all available phenotypic and genotypic data and integrate them in a consensus type of classification (44).
The successful isolation and enumeration of actinomycetes from the environment is usually achieved by dilution plate techniques with a medium containing selective nutrients and certain antibiotics (16, 40). The rapid characterization and identification of microbiological isolates are prerequisites for an effective industrial screening program. Detection of duplicates is necessary to reduce redundancy in screening assays, a process referred to as dereplication (3), and the selection of unusual strains can help in the discovery of novel compounds. Phylogenetic characterization of isolates on the basis of 16S rDNA and rRNA sequences and DNA-DNA homology are considered by many microbial taxonomists to be the most robust approaches to microbial identification at the species level. Based on 16S rRNA sequence analysis, Stackebrandt et al. (42) suggested the reclassification at the family level of actinomycetes and found that there were taxon-specific 16S rRNA and rDNA signature nucleotides.
The application of both traditional techniques, such as macro- and micromorphological, chemosystematic, and numerical taxonomy of phenotypic measurements, and the modern molecular systematic techniques were instrumental in clarifying the relationships within the actinomycetes (2, 42), although this process is still unfinished (11). However, 16S rDNA sequencing is time-consuming, and its capacity to differentiate at the subspecies level is often insufficient. As a result, a variety of alternative approaches have been developed for characterizing actinomycetes. These include fatty acid methyl esters (19), fatty acid composition by gas chromatography (9), pyrolysis mass spectrometry of cells (4, 41), random amplified polymorphic DNA method (1), and rep-PCR (51).
Amplified fragment length polymorphism (AFLP) is a relatively new genomic fingerprinting technique that is based on the selective amplification of a subset of restriction fragments (22, 46). There is plenty of evidence to show that AFLP can be applied in bacterial taxonomy for the differentiation of closely related taxa and for diagnostic, epidemiological, and source-tracking applications. Indeed, its particular power lies in the fact that it can discriminate bacteria down to the strain level (5, 13, 21). In addition, fluorescent AFLP analysis can be easily automated, digitized, and standardized for long-term database establishment and even exchange of data between laboratories.
Fourier transform infrared spectroscopy (FT-IR) is a rapid, nondestructive spectroscopic approach for whole-organism fingerprinting (10, 18, 33, 38, 39). FT-IR measures vibrations of functional groups and highly polar bonds (14). Thus, these (bio)chemical fingerprints are made up of the vibrational features of all the sample components, and for microbial samples these will include DNA and RNA, proteins, and membrane and cell wall components. FT-IR spectrometers record the interaction of IR radiation with samples, measuring the frequencies at which the sample absorbs the radiation and the intensities of these absorptions. The resolving power of FT-IR is excellent, and it has even been shown to detect differences between yeast mutants (39) and in the metabolic footprint of microbial cells differing in the presence of single genes (25). Our established protocol for FT-IR (10, 35, 43, 47, 48) has the major advantages that it is nondestructive, reproducible, and very rapid both for a single sample (1 to 10 s) and with respect to the automated high throughput of samples in batches of 384.
In this study, the main aim was to investigate the use of FT-IR spectroscopy to characterize 20 coastal sediment isolates putatively identified by morphological and cultural characteristics as Micromonospora. The same isolates were also analyzed by 16S rDNA sequencing and AFLP, and the phylogenetic results of all three methods are compared.
MATERIALS AND METHODS
Isolation.
The sediment samples, 5 to 20 cm under the surface, were collected aseptically during summer at low tide from the coast at North Beach, Aberystwyth, Wales, 52.4000°N, 4.0833°W, United Kingdom. In order to recover as many microorganisms as possible, sediments with fine sand combined with clay were chosen for isolation. One-fourth strength of Ringer's solution was used to dilute the sediment serially. To minimize the number of nonactinomycetes, the sediment was heat treated at 50°C for 10 min, and the isolation media, starch casein agar (SC) and starch casein nitrate agar (28) and humic acid-vitamin agar (HV) (16), were supplemented with 50 μg of cycloheximide per ml, 50 μg of nystatin per ml, and 25 μg of nalidixic acid per ml. Isolation plates were incubated at 25°C for up to 4 weeks. Only starch casein agar and starch casein nitrate agar plates were used for enumeration of actinomycetes (Table 1).
TABLE 1.
Isolation of actinomycetes from marine sediments
| Sediment treatment | Total no. of actinomycetes isolated (CFU/g)
|
|||
|---|---|---|---|---|
| Seawater agar
|
Distilled-water agar
|
|||
| Starch casein | Starch casein nitrate | Starch casein | Starch casein nitrate | |
| None | 2.6 × 105 | 1.7 × 105 | 3.2 × 105 | 3.0 × 105 |
| 45°C for 2 h | 2.4 × 105 | 2.1 × 105 | 2.6 × 105 | 3.2 × 105 |
| 50°C for 10 min | 6.8 × 105 | 5.3 × 105 | 8.7 × 105 | 7.1 × 105 |
| 60°C for 30 min | 2.8 × 105 | 2.2 × 105 | 3.5 × 105 | 2.7 × 105 |
16S rDNA sequencing.
DNA was isolated by the method of Marmur (34). The 16S rDNA was amplified with universal primers 27f (5′-AGAGTTTGATCMTGCCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′) (29). Amplification was carried out with an initial incubation of 5 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 50°C, and 3 min at 72°C, followed by a 10-min final extension at 72°C. The PCR products were purified with a QIAquick PCR extraction kit (Qiagen) and used as templates in two sequencing reactions with primers 27f and 1115r (5′-AGGGTTGCGCTCGTTG-3′) and with 339f (5′-CTCCTACGGGAGGCAGCAG-3′) and 1492r (29). Amplified products were sequenced directly on a 3100 automatic DNA sequencer (Applied Biosystems).
Nearly complete (≈1,400 nucleotides) 16S rDNA sequences of all isolates were aligned with Streptomyces coelicolor A(3)2 (designated Y00411 [which was retrieved from the EMBL database]) and a selection of Micromonospora strains (also retrieved from EMBL) with ClustalW (20). After automatic alignment, the sequences were manually checked. TreeCon version 1.3b software was used to estimate evolutionary distances (Jukes and Cantor, 1969), and to construct phylogenetic trees by the neighbor-joining method (45).
AFLP analysis.
Restriction, ligation, and amplification were performed as described previously (22). Approximately 250 ng of chromosomal DNA was digested with BamHI and MspI (Promega) and subsequently ligated with T4 DNA ligase (Promega) to double-stranded restriction-half-site-specific adaptors (1 μM).
The double-stranded restriction-half-site-specific adaptors were made as follows. For the BamHI adaptor, 100 μl each of the complementary oligonucleotides 5′-TCGTAGACTGCGTACA-3′ and 5′-GATCTGTACGCAGTCTACGA-3′, each at a concentration of 100 μM, were used, and for the MspI adaptor, the complementary oligonucleotides 5′-CGTCAGGACTCATCGTC-3′ and 5′-GACGATGAGTCCTGA-3′ were employed at the same concentration and details as for the BamHI adaptor. The two BamHI adaptor complementary single-stranded oligonucleotides were mixed with 14 μl of 5 M NaCl, heated at 95°C for 3 min, then cooled down at room temperature, and incubated at 25°C overnight. Then 20 μl of 3 M sodium acetate (pH 5.2) and 200 μl of pure 2-isopropanol were added to precipitate the constructed adaptor DNA. TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) was then used to dissolve the above DNA (100 μM). The final concentrations of MspI adaptor and BamHI adaptor for ligation of restriction fragment were 6.6 and 2.3 μM, respectively.
For PCR amplification of the ligated fragments, the BamHI+T primer (5′ modified with fluorescence 7′,8′-benzo-5′-fluoro-2′,4,7-trichloro-5-carboxyfluorescein, 5′-TCGTAGACTGCGTACAGATCCT-3′) and MspI+0 primer (5′-GACGATGAGTCCTGACGG-3′) were used. Amplified fragments were separated on an ABI 3100 DNA sequencer (Applied Biosystems) with a 36-cm array, filter set D, and 400HD (Rox) standard as the internal molecular size marker.
The AFLP results were captured with ABI Prism Genescan 3.7 software (Applied Biosystems), and the fragment data were tabulated by size and fluorescent intensity with the ABI Prism Genotyper 3.7 software (Applied Biosystems). All electropherograms were visually inspected for polymorphous peaks before the final fragment table was produced. Only fragments in the range from 50 to 500 bp were considered further because the resolving capability of the sequencing gel is generally good in this range, when with the Rox internal ladder. Analysis of fluorescent AFLP data was performed with the TreeCon version 1.3b software (45), and an unrooted phylogenetic tree was produced with simple matching and unweighted pair group mean average (UPGMA).
FT-IR spectroscopy.
The 20 Micromonospora isolates were inoculated onto the surface of nylon membranes placed on top of nonsporulation agar medium (NSM; Casamino Acids 2%, soluble starch 2%, yeast extract 0.4%, agar 1.2%, pH 7.2) and incubated at 25°C for 7 days. Each isolate was cultured six times, giving six biological replicates for analysis. After incubation, the biomass was collected carefully and suspended in 0.9% NaCl and then homogenized by 10 s of sonication. Homogenized biomass was loaded into 400-well aluminum IR sample carriers and subsequently dried at 50°C for 30 min. Each biological replicate was analyzed three times (so-called machine replicates).
The FT-IR instrument used was a Bruker IFS28 FT-IR spectrometer (Bruker Spectrospin Ltd., Banner Lane, Coventry, United Kingdom) equipped with an MCT (mercury-cadmium-telluride) detector cooled with liquid N2. The aluminum plate was then loaded onto the motorized stage of a reflectance thin-layer chromatography accessory and analyzed as detailed (10, 35, 43, 47). The IBM-compatible personal computer used to control the IFS28 was also programmed (with Opus version 2.1 software running under IBM O/S2 Warp provided by the manufacturers) to collect spectra over the wave number range 4,000 cm−1 to 500 cm−1. Spectra were acquired at a rate of 20 s−1. The spectral resolution used was 4 cm−1.
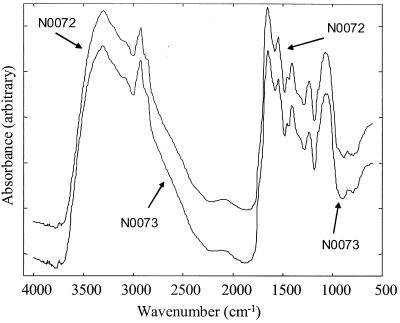
To improve the signal-to-noise ratio, 256 scans were coadded and averaged. All spectra were collected in reflectance mode and displayed in terms of absorbance as calculated from the reflectance-absorbance spectra with the Opus software, which is based on the Kubelka-Munk theory (14). Typical FT-IR spectra are shown in Fig. 1. Ranges of wave numbers are informative and can be associated with special chemical bonds (17, 36); 3,050 to 2,800 cm−1 is the so-called fatty acid region, where peaks show the vibration of CH2 and CH3 groups of fatty acids. The amide section, 1,750 to 1,500 cm−1, is where protein and peptide bands dominate. The range from 1,500 to 1,200 cm−1 is a mixed region containing vibrations of fatty acids, proteins, and polysaccharide. Polysaccharide dominates the region from 1,200 to 900 cm−1. The fingerprint region is from 900 to 700 cm−1. This contains bands which are most characteristic at the species level, but only a few peaks can be assigned to the vibrations of special substances.
FIG. 1.
Typical FT-IR spectra from isolates N0072 and N0073. The spectra are offset on the ordinate for clarity.
Data were exported from the Opus software used to control the FT-IR instrument and imported into Matlab version 6 (The MathWorks, Inc., Natick, Mass), which runs under Microsoft Windows NT on an IBM-compatible personal computer. The FT-IR data were first analyzed by principal component analysis (24). Principal component analysis is a well-known technique for reducing the dimensionality of multivariate data while preserving most of the variance, and this was performed according to the nonlinear iterative partial least squares algorithm (50). Discriminant function analysis (also known as canonical variate analysis) was used to discriminate between groups on the basis of the retained principal components and the a priori knowledge of which spectra were machine replicates, and thus, this process does not bias the analysis in any way (32).
Since the six biological replicates from each isolate clustered on top of one another (data not shown), the means of these were used for dendrogram construction. The Euclidean distance between a priori group centers in discriminant function analysis space from the biological replicates was used to construct a similarity measure, with the Gower general similarity coefficient SG (12), and these distance measures were then processed by an agglomerative clustering algorithm to construct a dendrogram (32).
Nucleotide sequence accession numbers.
The 16S rDNA sequences for the 20 Micromonospora isolates have been deposited in GenBank, and the sequence accession numbers are from AY221484 to AY221503, as detailed in Table 2.
TABLE 2.
Micromonospora isolates studied and top three 16S rDNA sequence hits against the RDPII database
| Isolate no. | Accession no. | RDPII highest hit (value, species; accession no.) |
|---|---|---|
| N0072 | AY221484 | 0.955, M. fulvoviolaceus; DSM 43905 |
| 0.952, M. yulongensis; DSM 43915 | ||
| 0.949, Micromonospora sp. strain Y10; AF068056 | ||
| N0073 | AY221485 | 0.967, M. yulongensis; DSM 43915 |
| 0.949, M. lacustris; DSM 43908 | ||
| 0.943, M. aurantiaca; AJ245712 | ||
| N0074 | AY221486 | 0.960, M. fulvoviolaceus; DSM 43905 |
| 0.957, M. yulongensis; DSM 43915 | ||
| 0.954, M. lacustris; DSM 43908 | ||
| N0075 | AY221487 | 0.953, M. yulongensis; DSM 43915 |
| 0.950, M. lacustris; DSM 43908 | ||
| 0.958, M. fulvoviolaceus; DSM 43905 | ||
| N0087 | AY221488 | 0.979, M. aurantiaca; AJ245712 |
| 0.972, M. chalceaT; DSM 43026 | ||
| 0.971, M. chalceaT; ATCC12452 | ||
| N0088 | AY221489 | 0.979, M. aurantiaca; AJ245712 |
| 0.972, M. chalceaT; DSM 43026 | ||
| 0.971, M. chalceaT; ATCC12452 | ||
| N0093 | AY221490 | 0.991, M. aurantiaca; AJ245712 |
| 0.989, M. chalceaT; DSM 43026 | ||
| 0.985, M. fulvopurpureus; DSM 43918 | ||
| N0097 | AY221491 | 0.956, M. fulvoviolaceus; DSM 43905 |
| 0.950, M. yulongensis; DSM 43915 | ||
| 0.947, Micromonospora sp. strain Y10; AF068056 | ||
| N0098 | AY221492 | 0.953, M. fulvoviolaceus; DSM 43905 |
| 0.950, M. yulongensis; DSM 43915 | ||
| 0.947, M. lacustris; DSM 43908 | ||
| N0152 | AY221493 | 0.954, M. narashino; DSM 43172 |
| 0.946, M. purpureochromogenesT; DSM 43821 | ||
| 0.944, M. aurantiaca; AJ245712 | ||
| N0153 | AY221494 | 0.991, M. melanospora; DSM 43126 |
| 0.976, M. purpureaT; DSM 43036 | ||
| 0.974, M. echinosporaT; ATCC15837 | ||
| N0155 | AY221495 | 0.990, M. matsumotoenseT; AF152109 |
| 0.943, Micromonospora sp. strain Y10; AF068056 | ||
| 0.933, M. lacustris; DSM 43908 | ||
| N0156 | AY221496 | 0.963, M. fulvoviolaceus; DSM 43905 |
| 0.958, M. yulongensis; DSM 43915 | ||
| 0.955, M. lacustris; DSM 43908 | ||
| N0157 | AY221497 | 0.977, M. aurantiaca; AJ245712 |
| 0.969, M. chalceaT; DSM 43026 | ||
| 0.964, M. chalceaT; ATCC12452 | ||
| N0158 | AY221498 | 0.973, M. matsumotoenseT; AF152109 |
| 0.949, Micromonospora sp. strain Y10; AF068056 | ||
| 0.939, M. lacustris; DSM 43908 | ||
| N0161 | AY221499 | 0.979, M. yulongensis; DSM 43915 |
| 0.936, M. fulvoviolaceus; DSM 43905 | ||
| 0.933, Micromonospora sp. strain Y10; AF068056 | ||
| N0162 | AY221500 | 0.941, M. matsumotoenseT; AF152109 |
| 0.933, M. yulongensis; DSM 43915 | ||
| 0.930, Micromonospora sp. strain Y10; AF068056 | ||
| N0163 | AY221501 | 0.960, M. yulongensis; DSM 43915 |
| 0.950, M. fulvoviolaceus; DSM 43905 | ||
| 0.947, M. matsumotoenseT; AF152109 | ||
| N0168 | AY221502 | 0.963, M. yulongensis; DSM 43915 |
| 0.953, M. lacustris; DSM 43908 | ||
| 0.952, M. fulvoviolaceus; DSM 43905 | ||
| N0179 | AY221503 | 0.956, M. fulvoviolaceus; DSM 43905 |
| 0.950, Micromonospora sp. strain Y10; AF068056 | ||
| 0.950, M. lacustris; DSM 43908 |
RESULTS AND DISCUSSION
Isolation of actinomycetes.
Initial experiments were conducted on several different media types (data not shown) to find the most effective isolation media in terms of maximum CFU (colonies per gram of sediment) of actinomycetes recovered from coastal sediment samples. We found that starch casein agar and humic acid-vitamin agar gave the highest selective yield of actinomycetes. We also investigated the effects on recovery of actinomycetes when the sediments were heat treated at 45°C for 2 h, 50°C for 10 min, or 60°C for 30 min. It was found, as expected, that enumeration of sporeformers increased after heating and that the best heat regimen was 50°C for 10 min (Table 1). Table 1 also shows that there were no differences in recovery of actinomycetes between isolation media based on distilled water or seawater. This result suggests that the isolated actinomycetes were of terrestrial origin or were not obligate halophiles.
Many colonies with different morphological and cultural characteristics were picked from the 1- to 4-week-old isolation plates and transferred to starch casein agar medium for purification and morphology observation. Most of the isolates were tentatively assigned as belonging to the genera Streptomyces and Micromonospora according to their macro- and micromorphological and cultural characteristics. Streptomyces isolates had rich aerial mycelia and matured to form short, long, spiral, or straight spore chains. Most of their colonies showed fuzzy surface because of sporulation. Compared with the Streptomyces isolates, Micromonospora spp. have smaller colonies with yellow, pink, or purple vegetative mycelium and sporulate after longer incubation. Some rare actinomycetes, such as Nocardia spp. with branched substrate mycelium, were also tentatively identified.
While an investigation of the exact community diversity and structure of this marine sediment would be useful, this was not our current objective. It would not be practical to pursue this in the present study, since only cultured organisms are considered and sampling errors will be manifest in taking such a small subsample from such a large beach area. We therefore considered whether FT-IR and AFLP can be used as complementary tools to 16S rDNA sequencing for the characterization of a subset of these actinomycetes.
Twenty Micromonospora isolates were selected for analysis by a polyphasic taxonomic approach. These were coded as N0072, N0073, N0074, N0075, N0087, N0088, N0093, N0097, N0098, N0152, N0153, N0155, N0156, N0157, N0158, N0161, N0162, N0163, N0168, and N0179. All 20 Micromonospora isolates gave rise to relatively small colonies on starch casein agar, and the substrate mycelia were orange or purplish. On further incubation, the colony surface of some of them became black as they started to sporulate. A typical scanning electron micrograph of one of the isolates is shown in Fig. 2, where it can be seen that spores were generated singly from the substrate mycelium.
FIG. 2.
Scanning electron micrograph of isolate N0072 grown on starch casein agar for 3 weeks at 25°C. Single spores were produced from the substrate mycelium.
16S rDNA sequencing.
In contemporary actinomycete systematics, 16S rDNA sequencing is routinely used to identify freshly isolated organisms by inferring evolutionary relationships at the genus and species levels with known described organisms in an rDNA sequence database (either held in-house or one that is publicly available). For the description of new genera and species which do not appear in the published literature, the isolates are then evaluated and refined with other taxonomic criteria, notably from chemosystematic and DNA-DNA relatedness studies.
Nearly complete 16S rDNA sequences from the 20 isolates were amplified and sequenced. The sequences were compared with all the 16S rDNA data stored in the Ribosomal Database Project II (RDP II) with a fast Sequence Match search program (31). All 20 isolates were placed in the genus Micromonospora according to the hit lists of similar sequences found in RDP II (Table 2).
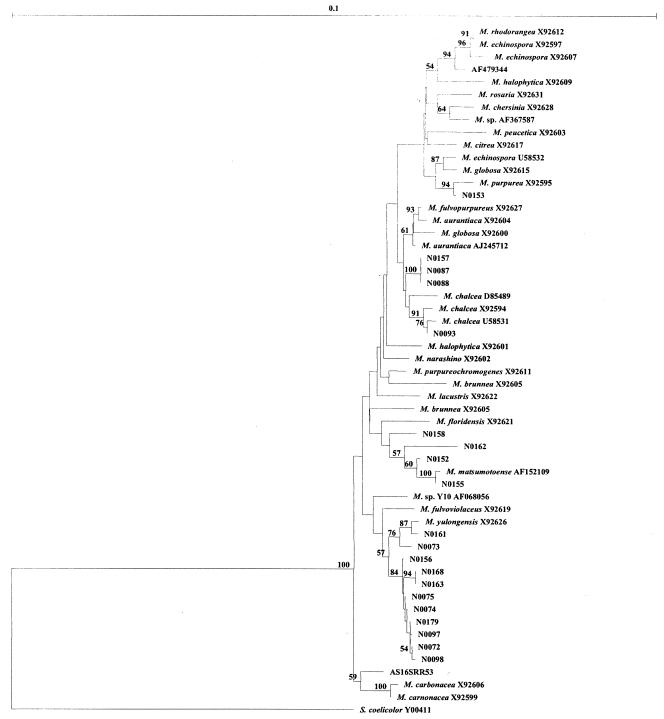
A selection of Micromonospora spp. from RDP II were downloaded and aligned with the 20 isolates from this study along with the 16S rDNA sequence from S. coelicolor A(3)2 Y00411 (the outgroup). All of these Micromonospora strains exhibited a very high level of 16S rDNA sequence similarity (>97%, data not shown). A phylogenetic tree was generated with the neighbor-joining method, and the results are shown in Fig. 3.
FIG. 3.
Neighbor-joining phylogenetic tree constructed with 20 Micromonospora isolates from this study (denoted N072 to N0179) plus Micromonospora from the RDPII based on their 16S rDNA sequences. S. coelicolor A(3)2 Y00411 was used as the outgroup. Bootstrap values greater than 50% are indicated at the nodes (100 replications). The bar stands for 0.1 substitution per nucleotide position.
From a closer inspection of Fig. 3, it is likely that isolates N0161, N0073, N0156, N0168, N0163, N0075, N0074, N0179, N0097, N0072, and N0098, which are loosely associated with M. yulongensis, might constitute a novel species of Micromonospora. Since we isolated 11 organisms in this clade, this will be an area of future study with chemotaxonomic methods and DNA homology. Likewise, N00157, N0087, and N0088 may also be a novel species; however, since the isolate numbers are lower and all three have identical 16S rDNA sequences, it is probably unjustified to investigate these further. The same can be said of N00158 and N0162, which are certainly taxonomically distinct from M. matsumotoense.
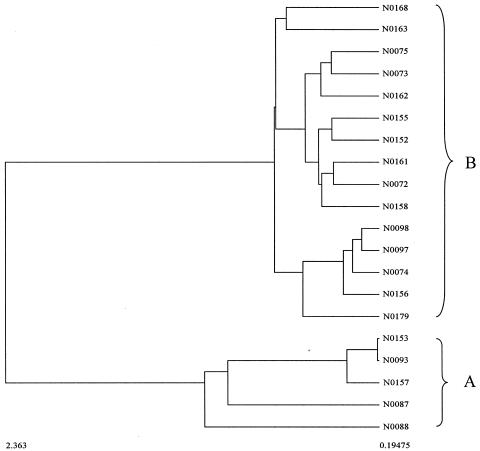
In order to simplify the comparison of this phylogeny based on 16S rDNA sequences with those generated from AFLP and FT-IR spectroscopy, phylogenetic analysis of just the 20 isolates, with Y00411 as the outgroup, was undertaken. This led to the production of the tree shown in Fig. 4, which clearly shows that the strains fall into two subgroups (labeled A and B).
FIG. 4.
Neighbor-joining phylogenetic tree constructed with 20 Micromonospora isolates based on their 16S rDNA sequences. Streptomyces coelicolor A(3)2 Y00411 was used as the outgroup. Bootstrap values greater than 50% are indicated at the nodes (100 replications). The bar stands for 0.1 substitution per nucleotide position.
Fluorescent AFLP analysis.
Chromosomal DNA was digested with two restriction enzymes, the hexacutter BamHI and the tetracutter MspI. Fluorescent AFLP patterns of the 20 Micromonospora spp. isolates were generated and analyzed automatically with a DNA sequencer as detailed above. Typically, the fluorescent AFLP barcodes contained ≈100 fragments which ranged from 50 to 500 bp and the fragments were compiled into a data matrix in Microsoft Excel for visual inspection prior to cluster analysis.
The frequency of fluorescent AFLP bands shared between the 20 Micromonospora isolates analyzed was assessed, and the results are shown in Table 3. It can be seen in this table that no single band was present in all 20 isolates, and moreover, the majority of the isolates had few common bands. That many of the isolates had unique fluorescent AFLP barcodes with few shared polymorphisms will have implications for phylogenetic analyses (i.e., the branches on the trees will be long and the bootstrap values are likely to be low).
TABLE 3.
Frequency of fluorescent AFLP bands shared among the 20 Micromonospora isolates analyzed
| No. of shared bands | Frequency (%) | No. of shared bands | Frequency (%) |
|---|---|---|---|
| 0 | 87 | 11 | 13 |
| 1 | 32 | 12 | 8 |
| 2 | 32 | 13 | 10 |
| 3 | 24 | 14 | 5 |
| 4 | 19 | 15 | 5 |
| 5 | 17 | 16 | 4 |
| 6 | 22 | 17 | 0 |
| 7 | 21 | 18 | 1 |
| 8 | 22 | 19 | 0 |
| 9 | 12 | 20 | 0 |
| 10 | 17 |
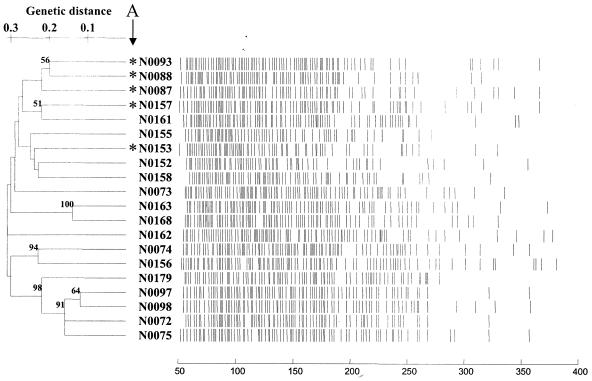
Analysis of the fluorescent AFLP bands was performed with TreeCon with simple matching and the UPGMA clustering method, and the unrooted tree is shown in Fig. 5. As expected, the branches of this tree were long and few nodes had significant bootstrap values. Isolates N0093, N0088, N0087, N0157, and N0153 (labeled in Fig. 5 with an asterisk) from cluster A in the 16S rDNA tree (Fig. 4) were not recovered together in a single group. N0093, N0088, and N0087 did cluster together, and these isolates were closely related to N0157 and N0161. While N0153 was recovered away from these organisms, it can be seen on closer inspection of the 16S rDNA tree (Fig. 4) that N0153 was the most distantly related organism in cluster A, as evident by its long connection branch to N0093.
FIG. 5.
Cluster analysis of the fluorescent AFLP data from the 20 Micromonospora isolates with simple matching and UPGMA. Also shown are the AFLP profiles for these isolates.
FT-IR spectroscopy.
Typical FT-IR spectra obtained from the analyses of these Micromonospora isolates are shown in Fig. 1. These and indeed all of the spectra collected showed broad similarities, and as for the DNA analyses outlined above, cluster analysis was necessary in order to reveal how similar they were to one another.
FT-IR spectroscopy was performed on the six biological replicates from each of the 20 isolates. Therefore, 120 samples were cultivated, and each of these was analyzed three times (the machine replicates). The cluster analysis of these data by the discriminant function analysis algorithm used a priori group knowledge encoded at the machine replicate level; that is to say, 120 groups were used rather than 20, allowing an unbiased analysis and allowing us to ascertain how reproducible growth and sample preparation were. Considering that 360 objects were clustered in this discriminant function analysis plot (data not shown), it is necessarily cluttered; however, each of the six biological replicates clustered synonymously for each of the 20 Micromonospora isolates. Therefore, we were satisfied that the growth and preparation of these actinomycetes were conducted in a reproducible manner.
The dendrogram was therefore constructed with the group means in the discriminant function analysis plot from the biological replicates, as detailed above, and is shown in Fig. 6. The clustering of the Micromonospora isolates from their FT-IR spectra (Fig. 6) was in good agreement with 16S rDNA sequencing analysis. The two main groups, A and B, were the same, although the subclustering within these two groups was slightly different. Isolates N0163 and N0168 have 100% 16S rDNA similarity and are clustered together based on FT-IR spectral information. This shows that they are duplicates, at least on the species level. The nearly complete (≈1,400 nt) 16S rDNA sequence similarity between N0073 and N0075, N0152 and N0155, N0072 and N0161, N0097 and N0098, and N0093 and N0153 is 99.6%, 99.6%, 99.4%, 99.9%, 99.4%, respectively, and they are grouped together in FT-IR clustering analysis. Isolates N0087, N0088, and N0157 have identical 16S rDNA sequences and N0153 has 99% similarity with N0087 and N0157, which means that they are the same species. The FT-IR dendrogram also support this grouping, that those five isolates are in the same group, A. All the isolates from the subgroups in groups A and B have more than 98.2% 16S rDNA similarity. That means that FT-IR can detect duplicates on the species level. This result is highly encouraging when one considers that FT-IR is a phenotypic typing method and measures the total biochemical makeup of the sample rather than being based on a single gene.
FIG. 6.
Dendrogram constructed from the FT-IR spectra from the 20 Micromonospora isolates with hierarchical cluster analysis as detailed in the text.
Previous studies show that FT-IR spectra are influenced by many factors such as plating methods, growth temperature, incubation time, and type of medium (27). Thus, standard sample preparation should be adopted for reproducible results. In our study, those 20 isolates were inoculated on nonsporulation agar and 1-week-old cultures were analyzed. All the samples were from the same batch culture to reduce experimental error.
Concluding remarks.
FT-IR spectroscopy is becoming established for the rapid identification of microorganisms (for a review see reference 33). The interest in FT-IR for the characterization of actinomycetes is growing, and strains from several genera have been analyzed, including Streptomyces and Micromonospora (15) and Corynebacterium and Rhodococcus (37). One of the exciting prospects is the generation of validated comprehensive FT-IR spectral reference databases, and this has been demonstrated in food-borne yeasts (27) and some selected coryneform bacteria and related taxa from the suborders Micrococcineae and Corynebacterineae (Actinomycetales and Actinobacteria) (38).
Oberrueuter and colleagues (37) compared their FT-IR results with 16S rDNA sequence analysis to assay the intraspecific diversity of three different actinomycetes species, Brevibacterium linens, Corynebacterium glutamicum, and Rhodococcus erythropolis. No correlation was found between 16S rDNA sequence similarity and FT-IR spectral distance for the three species below the species level. In our study, the characterization of a group of 20 marine sediment isolates belonging to Micromonospora by FT-IR was compared to two molecular methods, the established 16S rDNA sequence analysis and AFLP, which is itself a relatively new molecular typing approach. All methods gave very similar dendrograms, and FT-IR and 16S rDNA were found to be the most congruent, at least on the species level. We also studied 15 Streptomyces isolates with both the FT-IR and 16S rDNA sequence analysis methods (unpublished data), which showed that the FT-IR spectral dendrogram correlated to the phylogeny tree very well. Thus, FT-IR could be used as preliminary dereplication for screening novel isolates.
All three methods in this study characterize bacteria in different ways. 16S rDNA analysis focuses on a small highly conserved region of the genome, which, while useful for inferring deep phylogenies, suffers in that only a small part of the genome is analyzed. By contrast, AFLP analyzes DNA fragments from restriction analysis of genomic DNA and so more of the genotype is interrogated. FT-IR spectroscopy also effectively analyzes more of the organism, although at the phenotypic level, by measuring the vibrational properties of molecules.
FT-IR is a very rapid method, typically taking 1 to 10 s per sample, and samples can be loaded in batches of 96 or 384. It is very easy to use and inexpensive with respect to laboratory consumables (although an initial capital outlay for the equipment is needed). By contrast, both 16S rDNA sequencing and AFLP are slower (taking many hours if not days), require skilled operators, and are expensive at both the consumables and instrument levels. It is worth noting that a DNA sequencer is equivalent in price to an FT-IR instrument. Since FT-IR spectroscopy gives a measure of the organism's phenotype, standard growth parameters such as medium, temperature, and incubation time need to be controlled stringently in order to get reproducible results, especially for construction and comparison of spectral databases.
In conclusion, although currently FT-IR cannot be used alone for microbial classification because extensive microbial databases do not currently exist, we believe that FT-IR will become a very valuable first step in microbial screening programs. In particular, it can be used as a reliable method for the dereplication of environmental isolates prior to more expensive and time-consuming characterization of organisms producing potentially important and novel pharmacophores.
Acknowledgments
We thank the UK's Natural Environment Research Council for supporting this work.
We are grateful to Steve Wade for scanning electron microscopic analysis.
REFERENCES
- 1.Anzai, Y., T. Okuda, and J. Watanabe. 1994. Application of the random amplified polymorphic DNA using polymerase chain reaction for efficient elimination of duplicate strains in microbiological screening. J. Antibiot. 47:183-193. [DOI] [PubMed] [Google Scholar]
- 2.Atalan, E., G. P. Manfio, A. C. Ward, R. M. Kroppenstedt, and M. Goodfellow. 2000. Biosystematic studies on novel streptomycetes from soil. Annu. Rev. Microbiol. 77:337-353. [DOI] [PubMed] [Google Scholar]
- 3.Brandão, P. F. B., M. Torimura, R. Kurane, and A. T. Bull. 2002. Dereplication for biotechnology screening: PyMS analysis and PCR-RFLP-SSCP(PRS) profiling of 16S rRNA genes of marine and terrestrial actinomycetes. Appl. Microbiol. Biotechnol. 58:77-83. [DOI] [PubMed] [Google Scholar]
- 4.Chun, J., E. Atalan, S. B. Kim, H. J. Kim, M. E. Hamid, M. E. Trujillo, J. G. Magee, G. P. Manfio, A. C. Ward, and M. Goodfellow. 1993. Rapid identification of Streptomycetes by artificial neural network analysis of pyrolysis mass spectra. FEMS Microbiol. Lett. 114:115-119. [DOI] [PubMed] [Google Scholar]
- 5.Coenye, T., L. M. Schouls, J. R. W. Govan, K. Kersters, and P. Vandamme. 1999. Identification of Burkholderia species and genomovars from cystic fibrosis patients by AFLP fingerprinting. Int. J. Syst. Bacteriol. 49:1657-1666. [DOI] [PubMed] [Google Scholar]
- 6.Crueger, W., and A. Crueger. 1989. Biotechnology: a textbook of industrial microbiology. Sinauer Associates, Sunderland, Mass.
- 7.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demain, A. L. 1999. Pharmaceutically active secondary metabolites of microorganisms. Appl. Microbiol. Biotechnol. 52:455-463. [DOI] [PubMed] [Google Scholar]
- 9.Drucker, D. B. 1981. Microbiological applications of gas chromatography. Cambridge University Press, Cambridge, United Kingdom.
- 10.Goodacre, R., E. M. Timmins, R. Burton, N. Karderbhai, A. M. Woodward, D. B. Kell, and P. J. Rooney. 1998. Rapid identification of urinary tract infection bacteria using hyperspectral, whole organism fingerprinting and artificial neural networks. Microbiology 144:1157-1170. [DOI] [PubMed] [Google Scholar]
- 11.Goodfellow, M., K. Isik, and E. Yates. 1999. Actinomycete systematics: an unfinished synthesis. Nova Acta Leopoldina NF 80 312:47-82. [Google Scholar]
- 12.Gower, J. C. 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325-338. [Google Scholar]
- 13.Grady, R., D. Blanc, P. Hauser, and J. Stanley. 2001. Genotyping of European isolates of methicillin-resistant Staphylococcus aureus by fluorescent amplified-fragment length polymorphism analysis (fluorescent AFLP) and pulsed-field gel electrophoresis (PFGE) typing. J. Med. Microbiol. 50:588-593. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths, P. R., and J. A. de Haseth. 1986. Fourier transform infrared spectrometry. John Wiley, New York, N.Y. [DOI] [PubMed]
- 15.Haag, H., H.-U. Gremlich, R. Bergmann, and J. J. Sanglier. 1996. Characterization and identification of actinomycetes by FT-IR spectroscopy. J. Microbiol. Methods 27:157-163. [Google Scholar]
- 16.Hayakawa, M., and H. Nonomura. 1987. Humic acid-Vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65:501-509. [Google Scholar]
- 17.Helm, D., H. Labischinski, G. Schallehn, and D. Naumann. 1991. Classification and identification of bacteria by Fourier transform infrared spectroscopy. J. Gen. Microbiol. 137:69-79. [DOI] [PubMed] [Google Scholar]
- 18.Helm, D., H. Labischinski, G. Schallehn, and D. Naumann. 1991. Classification and identification of bacteria by Fourier transform infrared spectroscopy. J. Gen. Microbiol. 137:69-79. [DOI] [PubMed] [Google Scholar]
- 19.Heyrman, J., J. Mergaert, R. Denys, and J. Swings. 1999. The use of fatty acid methyl ester analysis (FAME) for the identification of heterotrophic bacteria present on three mural paintings showing severe damage by microorganisms. FEMS Microbiol. Lett. 181:55-62. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huys, G., R. Coopman, P. Janssen, and K. Kersters. 1996. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int. J. Syst. Bacteriol. 46:572-580. [DOI] [PubMed] [Google Scholar]
- 22.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kersters. 1996. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 23.Jendrossek, D., G. Tomasi, and R. M. Kroppenstedt. 1997. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol. Lett. 1:179-188. [DOI] [PubMed] [Google Scholar]
- 24.Jolliffe, I. T. 1986. Principal component analysis. Springer-Verlag, New York, N.Y.
- 24a.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 25.Kaderbhai, N. N., D. I. Broadhurst, D. I. Ellis, R. Goodacre, and D. B. Kell. 2003. Functional genomics via metabolic footprinting: Monitoring metabolite secretion by Escherichia coli tryptophan metabolism mutants using FT-IR and direct injection electrospray mass spectrometry. Comp. Func. Genomics 4:376-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieser, T., M. Bibb, J., M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces Genetics. The John Innes Foundation, Norwich, United Kingdom.
- 27.Kümmerle, M., S. Scherer, and H. Seiler. 1998. Rapid and reliable identification of food-borne yeast by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 64:2207-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Küster, E., and S. T. Williams. 1964. Selection of media for isolation of streptomycetes. Nature 22:928-929. [DOI] [PubMed] [Google Scholar]
- 29.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, England.
- 30.Luedemann, G. M., and B. C. Brodsky. 1964. Taxonomy of gentamicin-producing Micromonospora. Antimicrob. Agents Chemother. 63:116-124. [PubMed] [Google Scholar]
- 31.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiejie. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acid Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manly, B. F. J. 1994. Multivariate statistical methods: a primer. Chapman & Hall, London, United Kingdom.
- 33.Maquelin, K., C. Kirschner, L.-P. Choo-Smith, N. van den Braak, H. P. Endtz, D. Naumann, and G. J. Puppels. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255-271. [DOI] [PubMed] [Google Scholar]
- 34.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 35.McGovern, A. C., D. Broadhurst, J. Taylor, N. Kaderbhai, M. K. Winson, D. A. Small, J. J. Rowland, D. B. Kell, and R. Goodacre. 2002. Monitoring of complex industrial bioprocesses for metabolite concentrations using modern spectroscopies and machine learning: application to gibberellic acid production. Biotechnol. Bioeng. 78:527-538. [DOI] [PubMed] [Google Scholar]
- 36.Naumann, D., V. Fijala, H. Labischinski, and P. Giesbrecht. 1988. The rapid differentiation and identification of pathogenic bacteria using Fourier-transform infrared spectroscopic and multivariate statistical analysis. J. Mol. Struct. 1988:165-170. [Google Scholar]
- 37.Oberrueuter, H., J. Charzinski, and S. Scherer. 2002. Intraspecific diversity of Brevibacterium linens, Corynebacterium glutamicum and Rhodococcus erythropolis based on partial 16S rDNA sequence analysis and Fourier-transform infrared (FT-IR) spectroscopy. Microbiology 148:1523-1532. [DOI] [PubMed] [Google Scholar]
- 38.Oberrueuter, H., H. Seiler, and S. Scherer. 2002. Identification of coryneform bacteria and related taxa by Fourier-transform infrared (FT-IR) spectroscopy. Int. J. Syst. Environ. Microbiol. 52:91-100. [DOI] [PubMed] [Google Scholar]
- 39.Oliver, S. G., M. K. Winson, D. B. Kell, and F. Baganz. 1998. Syst. functional analysis of the yeast genome. Trends Biotechnol. 16:373-378. [DOI] [PubMed] [Google Scholar]
- 40.Sanglier, J. J., H. Haag, T. A. Huck, and T. Fehr. 1993. Novel bioactive compounds from actinomycetes: a short review (1988-1992). Res. Microbiol. 144:633-642. [DOI] [PubMed] [Google Scholar]
- 41.Sanglier, J. J., D. Whitehead, G. S. Saddler, E. V. Ferguson, and M. Goodfellow. 1992. Pyrolysis mass spectrometry as a method for the classification, identification and selection of actinomycetes. Gene 115:235-242. [DOI] [PubMed] [Google Scholar]
- 42.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 43.Timmins, E. M., S. A. Howell, B. K. Alsberg, W. C. Noble, and R. Goodacre. 1998. Rapid differentiation of closely related Candida species and strains by pyrolysis mass spectrometry and Fourier transform infrared spectroscopy. J. Clin. Microbiol. 36:367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandamme, P., B. Pot, M. Gillis, P. DeVos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van de Peer, Y., and R. De Wachter. 1996. Construction of evolutionary distance trees with TREECON for windows: accounting for variation in nucleiotide substitution rate among sites. Comput. Appl. Biosci. 13:227-230. [DOI] [PubMed] [Google Scholar]
- 46.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winder, C. L., E. Carr, R. Goodacre, and R. Seviour. 2004. The rapid identification of Acinetobacter species using Fourier transform infrared spectroscopy. J. Appl. Microbiol. 96:328-339. [DOI] [PubMed] [Google Scholar]
- 48.Winson, M. K., R. Goodacre, É. M. Timmins, A. Jones, B. K. Alsberg, A. M. Woodward, J. J. Rowland, and D. B. Kell. 1997. Diffuse reflectance absorbance spectroscopy taking in chemometrics (DRASTIC). A hyperspectral FT-IR-based approach to rapid screening for metabolite overproduction. Anal. Chim. Acta 348:273-282. [Google Scholar]
- 49.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wold, H. 1966. Estimation of principal components and related models by iterative least squares, p. 391-420. In K. R. Krishnaiah (ed.), Multivariate analysis. Academic Press, New York, N.Y.
- 51.Woods, C. R. J., V. J., T. Koeuth, and J. R. Lupski. 1992. Analysis of relationships among isolates of Citrobacter diversus by using DNA fingerprints generated by repetitive sequence-based primers in the polymerase chain reaction. J. Clin. Microbiol. 30:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]