Abstract
Anaerobic nitrogen-fixing consortia consisting of N2-fixing clostridia and diverse nondiazotrophic bacteria were previously isolated from various gramineous plants (K. Minamisawa, K. Nishioka, T. Miyaki, B. Ye, T. Miyamoto, M. You, A. Saito, M. Saito, W. Barraquio, N. Teaumroong, T. Sein, and T. Tadashi, Appl. Environ. Microbiol. 70:3096-3102, 2004). For this work, clostridial populations and their phylogenetic structures in a stand of the grass Miscanthus sinensis in Japan were assessed by a 16S rRNA gene-targeted terminal restriction fragment length polymorphism (TRFLP) analysis combined with most-probable-number (MPN) counts. PCR primers and restriction enzymes were optimized for analyses of the plant clostridia. Clostridia were detected in strongly surface-sterilized leaves, stems, and roots of the plants at approximately 104 to 105 cells/g of fresh weight; they made up a large proportion of N2-fixing bacterial populations, as determined by MPN counts associated with an acetylene reduction assay. Phylogenetic grouping by MPN-TRFLP analysis revealed that the clostridial populations belonged to group II of cluster XIVa and groups IV and V of cluster I; this result was supported by a culture-independent TRFLP analysis using direct DNA extraction from plants. When phylogenetic populations from M. sinensis and the soil around the plants were compared, group II clostridia were found to exist exclusively in M. sinensis.
Several diazotrophs have been isolated and characterized as nitrogen-fixing endophytes from gramineous plants, including Acetobacter (20), Herbaspirillum (5, 8), Klebsiella (2), and Serratia (7). An Azoarcus sp. from Kallar grass abundantly colonizes and expresses nif genes and nitrogenase protein inside its original host as well as in rice roots (11, 18). These diazotrophic endophytes are all gram-negative aerobes and facultative anaerobes. Recently, Minamisawa et al. (16) discovered the existence of anaerobic nitrogen-fixing consortia consisting of N2-fixing clostridia and diverse nondiazotrophic bacteria in nonleguminous plants (16). Clostridia are obligate anaerobic, gram-positive bacteria (1, 4, 9) which have not yet been recognized as bacterial endophytes. A phylogenetic analysis (16) indicated that the plant clostridia fell exclusively into clusters XIVa and I, as defined by Collins et al. (4), and were further subdivided into five groups. In particular, N2 fixation by group II clostridia was induced by metabolites of nondiazotrophs in culture, although N2 fixation in anaerobic nitrogen-fixing consortia is always supported by the elimination of oxygen by the nondiazotrophs (16).
The 16S rRNA gene has been used as a molecular marker that allows the phylogenetic assignment of target organisms in natural environments. Popular methods that rely on 16S rRNA gene analysis include terminal restriction fragment length polymorphism (TRFLP) analysis (14) and denaturing gradient gel electrophoresis (17). Recently, Sessitsch et al. (19) monitored endophytic populations in potato plants by TRFLP and found a wide range of organisms that fell into many distinct phylogenetic groups. Molecular population analyses targeting the 16S rRNA genes of clostridia in the environment have been reported for municipal landfill sites (22), human feces (6), and paddy fields (3, 23).
For this work, we investigated the population levels and phylogenetic structures of clostridia in the grass Miscanthus sinensis by TRFLP analysis targeting the 16S rRNA gene. M. sinensis is a rhizomatous, perennial grass that naturally dominates almost all of the tallgrass-type meadows and wastelands in Japan (13). The plant often grows as a pioneer plant in areas devastated by lahar from volcanic eruptions; in these areas the nitrogen content of the soil is very low. The objective of this work was to determine (i) how the population levels of endophytic clostridia compare with those of culturable diazotrophs and (ii) which are the dominant phylogenetic groups of plant clostridia.
MATERIALS AND METHODS
Bacterial strains, media, and cultivation.
Sixteen strains of a Clostridium sp. that were previously isolated mainly from M. sinensis were used (16). The strain names (and accession numbers of the sequences) were Sukash-1 (AB114226), Kas203-2 (AB114230), Kas401-4 (AB114231), Kas107-2 (AB114232), Kas203-1 (AB114238), Kas104-4 (AB114239), Kas401-3 (AB114240), Kas107-1 (AB114241), Kas301-1 (AB114242), Kas404-1 (AB114243), Kas303 (AB114244), Kas402-3 (AB114249), Kas202-1 (AB114252), Kas201-1 (AB114258), Kas106-4 (AB114263), and B901-1b (AB114264). The type strains of Clostridium aminovalericum, Clostridium intestinali, Clostridium acetobutylicum, and Clostridium beijerinckii were purchased from culture collections of the Riken Institute (Wako, Japan). Rice extract modified Rennie (RMR) semisolid medium (5) and nutrient agar (NA) (Difco, Detroit, Mich.) were used for the enumeration of bacteria and diazotrophs from plants. Viande-Levure (VL) agar and RMR agar plates were used for the cultivation of clostridia. VL medium contained the following components dissolved in 1 liter of water (pH 7.0): nutrient broth (Difco), 8 g; yeast extract (Difco), 5 g; NaCl, 5 g; glucose, 2 g; cysteine-HCl, 0.3 g. Anaerobic cultivation was carried out with the AnaeroPack system (Mitsubishi Gas Chemical, Tokyo, Japan).
Sample collection and surface sterilization.
Plants were sampled from a stand of naturally vegetated M. sinensis in the town of Kashimadai, in the Miyagi prefecture, Japan, early in summer. Plant materials were carefully washed with tap water and separated into leaf, stem, and root parts. A sample of bulk soil was taken from around the grasses, air dried, and sieved through 2-mm mesh. To ensure the complete surface sterilization of the plant materials, we tested several sterilization conditions (with 1 to 2% NaOCl for 0.5 to 15 min). After surface sterilization, the plant materials were washed several times with sterile distilled water. NaOCl-treated 5-cm-long sections of leaves, stems, or roots or clostridial cell samples were rolled or plated on NA, VL, or RMR agar plates. VL and some RMR plates were anaerobically incubated at 30°C, and NA and other RMR plates were aerobically incubated at 30°C. A similar sterilization experiment was performed with spores of Clostridium sp. strain B901-1b.
Bacterial counts by the MPN method.
The surface-sterilized plant materials were mechanically macerated with 0.8% saline solution and quartz sand and then decimally diluted in 0.8% saline solution. The dilutions were used to seed RMR semisolid medium (five tubes per dilution) for determinations of the most probable numbers (MPN) of total bacteria and N2-fixing bacteria. MPN tubes were monitored for growth in the form of subsurface pellicles 5 days after inoculation. Growth-positive tubes were tested for acetylene reduction. After 24 h of incubation with 10% C2H2, the ethylene formed was measured in a gas chromatograph (GC-7A; Shimadzu, Kyoto, Japan) equipped with a flame ionization detector and a Porapack R column (0.3 mm [inner diameter] by 2 m) at 50°C. MPN counts were calculated at a level of 95% confidence according to the method of Hurley and Roscoe (12).
DNA extraction.
DNAs were isolated by two procedures from semisolid RMR cultures and directly from plant tissues and soil. For DNA isolation from semisolid cultures, a modification of the method of Hiraishi (10) was used as follows. Semisolid cultures (40 μl) from MPN tubes were frozen at −20°C and thawed at room temperature. Lysates were prepared by adding 10 μl of proteinase K (10 mg/ml) and 50 μl of BL buffer (pH 8; 40 mM Tris, 1% Tween 20, 0.5% Nonidet P-40, 1 mM EDTA, 1 N HCl) to the semisolid culture (40 μl), heated at 60°C for 20 min, and then heated at 95°C for 20 min, followed by centrifugation (17,000 × g for 10 min) to remove unbroken cells and large debris. The resultant lysates were used as DNA templates for PCR amplification of the 16S rRNA gene.
DNAs were prepared directly from plants and soil materials by use of a FastDNA kit and a FastDNA Spin kit for soil (BioSystems, Carlsbad, Calif.) according to the manufacturer's instructions. Surface-washed stems of M. sinensis were macerated in a mortar with liquid N2. The powdered tissues (0.1 g) were introduced to tubes containing cell lysis solution for vegetation (BioSystems) and homogenized with a bead beater (BioSpec, Bartlesville, Okla.) with two 1/4-in. cylindrical spheres and a garnet matrix (BioSystems). For DNA extraction from the soil, 0.5 g of air-dried soil was introduced into lysing matrix E (BioSystems) and homogenized as described above.
PCR amplification.
The primers used for amplification of the clostridial 16S rRNA gene are listed in Table 1. A seminested PCR assay was performed to eliminate amplified products from other bacteria. The clostridial 16S rRNA gene from 0.5 μl of a semisolid lysate was amplified in a reaction mixture containing 1.0 μl each of primers C142f and C1090r (20 μM [each]), 0.1 μl (1 U) of Ex Taq polymerase (Takara, Kyoto, Japan), 1.6 μl of 2.5 mM deoxynucleoside triphosphates, 2.0 μl of 10× Ex Taq buffer, and 13.8 μl of sterile distilled water. After a denaturation step of 5 min at 94°C, the amplification reactions were performed, with 30 cycles of denaturation (30 s, 94°C), primer annealing (30 s, 52°C), and primer extension (30 s, 72°C) and a final extension step of 7 min at 72°C. Aliquots (1 μl) of 16S rRNA gene products were used directly for a second PCR amplification of the 16S rRNA gene. The reaction mixture for the second PCR contained 1.5 μl each of primers Cy5-C142f and C796r (20 μM [each]), 0.1 μl (1 U) of Ex Taq polymerase, 4.0 μl of 2.5 mM deoxynucleoside triphosphates, 5.0 μl of 10× Ex Taq buffer, and 37 μl of sterile distilled water. After a denaturation step of 5 min at 94°C, the thermal cycles for the second PCR were identical to those for the first PCR. Aliquots (1 μl) were analyzed by electrophoresis in 1% (wt/vol) agarose gels, followed by staining with ethidium bromide. The bands were visualized by UV excitation. A polyethylene glycol solution (20% [wt/vol] polyethylene glycol, 1.6 M NaCl) was added to the second PCR product at a 0.6 volume and mixed by inversion, and then the mixture was kept at 4°C for 1 h. After centrifugation and discarding of the supernatant, the resulting pellet was resuspended in 150 μl of 70% ethanol and centrifuged at 17,000 × g at 4°C for 10 min. The supernatant was again discarded and the pellet was air dried before the DNA was dissolved in 10 μl of sterile distilled water.
TABLE 1.
Sequences of primers used for seminested PCRs
| Primera | Sequence (5′-3′) | Seminested PCR round |
|---|---|---|
| C142f | AAABGRMKRYTAATACCGCATAA | 1st |
| Cy5-C142f | Cy5-AAABGRMKRYTAATACCGCATAA | 2nd |
| C1090r | TRGCWACTAASRATAAGGG | 1st |
| C796r | CCCNACACCTAGTAYYCATC | 2nd |
TRFLP analysis.
For restriction enzyme digestion, the reagents were prepared as a master mix before being added to the sample. One hundred fifty nanograms of the purified PCR product was digested with either 10 U of HaeIII (Toyobo, Osaka, Japan) or 15 U of MspI (New England BioLabs, Beverly, Mass.) in a total volume of 15 μl at 37°C for 5 h. Next, 2.5 volumes of 99.5% ethanol and 0.1 volume of 3 M sodium acetate were added to the samples before centrifugation at 17,000 × g at 4°C for 15 min. After the supernatant was discarded, the pellet was rinsed with 150 μl of 70% ethanol and centrifuged at 17,000 × g at 4°C for 10 min. The supernatant was again discarded, and the air-dried pellet was resuspended in 2 μl of sterile distilled water. Fluorescently labeled fragments were mixed with 0.5 μl of TE buffer (10 mM Tris-HCl [pH 7.6], 1 mM EDTA) containing 1 fmol each of ALFexpress Sizers 50 and 300 (Amersham Pharmacia Biotech, Piscataway, N.J.) as internal standards and 1.5 μl of loading dye. After gentle agitation, each sample was denatured by heating at 95°C for 2 to 3 min and then was immediately quenched on ice. After electrophoresis in an automated sequencer (ALFexpress II DNA analyzer; Amersham Pharmacia Biotech) and analysis with ALFwin Fragment Analyzer 1.00 software (Amersham Pharmacia Biotech), the sizes (in base pairs) of the terminal restriction fragments (TRFs) were estimated by reference to the internal standard and to the external standard, ALFexpress Sizers 50 to 500. TRFs of clostridial clusters I and XIVa were determined by reference to the sizes of TRFs in mixtures of clostridial groups I, II, IV, and V (Sukash-1, Kas107-2, Kas104-4, and Kas106-4). TRFs with heights of <0.05% for the largest TRF peak were excluded from analyses.
Clostridial counts.
All MPN tubes showing bacterial growth were subjected to TRFLP analysis. The MPNs of group I, II, IV, and V clostridia in the respective MPN tubes were determined by TRFLP analysis. The total number of clostridia was estimated from maximum values of the respective numbers of group I, II, IV, and V clostridia.
Microscopy.
Clostridial strains grown in RMR semisolid medium were vortexed and then serially diluted with saline solution. The cell numbers in the solutions were counted in a bacterial counting chamber (0.02 mm [depth] by 0.0025 mm2; Kayagaki Irika Kogyo, Tokyo, Japan) by phase-contrast microscopy (BX50; Olympus, Tokyo, Japan).
RESULTS AND DISCUSSION
Primers for TRFLP analysis of plant clostridia.
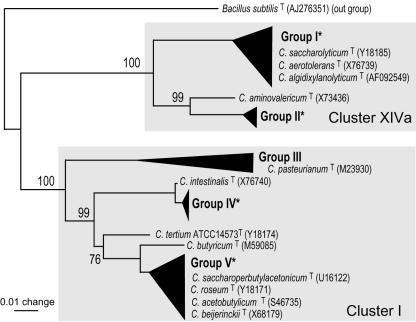
The objective of this work was to assess the densities and phylogenetic populations of endophytic clostridia that had been previously isolated from several gramineous plants (16). Because they were all cluster XIVa or I clostridia according to the system of Collins et al. (4), we aimed to develop a TRFLP detection system specific for these clusters (Fig. 1). The clusters are subdivided into five groups, groups I, II, III, IV, and V (Fig. 1) (16). Group III contains two clostridial isolates that were derived from mature seeds of wild rice but not from fresh materials such as plant shoots and roots (16). Thus, this work focused on group I, II, IV, and V clostridia.
FIG. 1.
Schematic presentation of phylogenetic groups of clostridia isolated from various plant sources, based on 16S rRNA gene sequences. The phylogenetic tree was reconstructed from the results of a previous study (16). Groups denoted by black triangles are phylogenetic groups of clostridia isolated from gramineous plants. The phylogenetic positions of type strains of Clostridium species in and around the groups are indicated by the accession numbers of 16S rRNA gene sequences given in parentheses. Gray regions indicate clostridial clusters as defined by Collins et al. (4). Groups marked with asterisks contain the clostridia isolated from M. sinensis (groups I, II, IV, and V), as shown in Table 3. Group III contains two isolates, B905-1 and B904-4, derived from seeds of wild rice, Oryza officinalis (16). Bar = 0.01 base substitutions per nucleotide.
The PCR primers used for this work are shown in Table 1. In accordance with the 16S rRNA gene sequences of 40 strains of plant clostridia (DDBJ accession numbers AB114225 to -64), primers C142f (Cy5-C142f) and C1090r were designed by the modification of S-*-chis-0150-a-S-23 (6) and S-*-Ccoc-1112-a-A-19 (22), respectively. Preliminary PCR assays suggested that the C142f and C1090r primers produced a PCR product from presumptive enterobacteria other than the plant clostridia from RMR semisolid cultures of M. sinensis (data not shown). Thus, a seminested PCR strategy was adopted with an additional reverse primer, C796r, based on the 16S rRNA gene sequences of the plant clostridia (Table 1; Fig. 2). The Probe Match program (http://rdp.cme.msu.edu/html/) was used to search the matching of the three primers (C142f, C796r, and C1090r) to 16S rRNA gene sequences of bacteria according to the method of Van Dyke et al. (22). The three primers were simultaneously in accord with 16S rRNA gene sequences of 11 clones of cluster XIVa clostridia and 41 clones of cluster I clostridia, although 2 clones of cluster IV clostridia were hit. Therefore, the combinations of these primers permitted PCR amplification of the 16S rRNA genes of cluster XIVa and I clostridia exclusively, including the previous clostridial isolates.
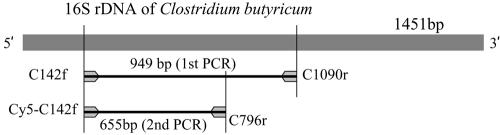
FIG. 2.
Primer pairing and target sites used for seminested PCRs of clostridial 16S rRNA gene. Primer positions were named on the basis of the 16S rRNA gene sequences of Clostridium butyricum ATCC 19398 (DDBJ accession number AB075768).
Restriction enzymes for TRFLP analysis of plant clostridia.
To determine appropriate restriction enzymes for TRFLP analysis, we calculated the lengths of TRFs derived from the 655-bp PCR product of the 16S rRNA gene sequence (Fig. 2) by the use of several restriction enzymes (Table 2). Two enzymes, HaeIII and MspI, were selected to discriminate TRF sizes in a group-specific manner (Table 2). HaeIII digestion generated the following expected TRFs: 152 or 108 bp for group I, 109 bp for group II, 76 bp for group IV, and 154 bp for group V. MspI digestion generated the following expected TRFs: 58 bp for group I, 134 bp for group II, and 375 bp for groups IV and V. Thus, the combination of TRF sizes for products that were independently digested with HaeIII and MspI could be used to specify the groups of clostridia. From the strains for which we estimated TRF sizes, the 16 strains of M. sinensis-derived clostridia were examined (Table 2). Seminested PCR assays were performed with their lysates, which were subsequently subjected to a TRFLP analysis using HaeIII and MspI. The observed sizes of the TRFs were almost identical to the predicted TRF sizes, except for group I clostridia (Table 2). Two strains (Kas203-2 and Kas401-4) in group I showed both a 152-kb and a 108-bp TRF upon HaeIII digestion. Three strains in group I showed both a 152-kb and a 58-bp TRF upon MspI digestion. These differences were probably caused because different rRNA operons within group I clostridia have slightly different sequences. For group II, IV, and V clostridia, the observed TRF sizes were identical to the predicted sizes.
TABLE 2.
Predicted and observed lengths of the 5′ TRF 16S rRNA genes of plant clostridia and their neighbors digested with various restriction enzymesa
| Cluster and group | Strain | Predicted TRF size (bp)
|
Observed size (bp)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HaeIII | MspI | TaqI | HhaI | RsaI | Tsp5091 | AluI | MboI | HaeIII | MspI | ||
| Cluster XIVa | |||||||||||
| Group I | Sukash-1 | 152 | 58 | 581 | 24 | 129 | 493 | 275 | 124 | 152 | 58, 152 |
| Kas203-2 | 108 | 58 | 581 | 24 | 303 | 493 | 467 | 124 | 108, 152 | 58, 152 | |
| Kas401-4 | 108 | 58 | 581 | 24 | 303 | 493 | 467 | 124 | 108, 152 | 58, 152 | |
| C. saccharolyticum (Y18185) | 108 | 58 | 581 | 24 | 303 | 493 | 275 | 124 | |||
| C. aerotolerans (X76739) | 108 | 58 | 580 | 24 | 303 | 492 | 275 | 124 | |||
| C. algidixylanolyticum (AF092549) | 108 | 58 | 581 | 24 | 303 | 493 | 467 | 124 | |||
| Group II | Kas107-2 | 109 | 134 | 584 | 24 | 306 | 496 | 90 | 125 | 109 | 134 |
| OkiF101 | 109 | 134 | 584 | 24 | 306 | 496 | 90 | 125 | 109 | 134 | |
| OkiN108 | 109 | 134 | 584 | 24 | 306 | 496 | 90 | 125 | 109 | 134 | |
| C. aminovalericum (X73436) | 172 | 153 | 601 | None | 41 | 446 | 108 | 144 | 172 | 153 | |
| Cluster I | |||||||||||
| Group IV | Kas104-4 | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | 76 | 375 |
| Kas107-1 | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | 76 | 375 | |
| Kas203-1 | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | 76 | 375 | |
| Kas301-1 | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | 76 | 375 | |
| Kas303 | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | 76 | 375 | |
| Kas401-3 | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | 76 | 375 | |
| Kas404-1 | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | 76 | 375 | |
| UsIt102-1 | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | |||
| C. intestinalis (X76740) | 76 | 375 | 583 | 84 | 305 | 495 | 91 | 150 | 76 | 375 | |
| Group V | Kas202-1 | 154 | 375 | 583 | None | 31 | 48 | 91 | 150 | 154 | 375 |
| UsIt102-2 | 154 | 375 | 583 | None | 31 | 48 | 91 | 150 | |||
| UsIt101-1 | 154 | 375 | 583 | None | 31 | 48 | 91 | 150 | |||
| UsS102-1 | 154 | 375 | 583 | None | 31 | 48 | 91 | 150 | |||
| UsS101-2 | 154 | 375 | 583 | None | 31 | 48 | 91 | 150 | |||
| Kas106-4 | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | 154 | 375 | |
| Kas201-1 | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | 154 | 375 | |
| Kas402-3 | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | 154 | 375 | |
| Usu-S-R3 | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | |||
| UsIt102-3 | 154 | 375 | 492 | None | 305 | 48 | 91 | 150 | |||
| UsS101-1 | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | |||
| B901-1b | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | 154 | 375 | |
| C. saccharoperbutylacetonicum (U16122) | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | |||
| C. roseum (Y18171) | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | |||
| C. acetobutylicum (S46735) | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | 154 | 375 | |
| C. beijerinckii (X68179) | 154 | 375 | 583 | None | 305 | 48 | 91 | 150 | 154 | 375 | |
| C. butyricum (M59085) | 154 | 375 | 583 | 397 | 305 | 48 | 91 | 150 | |||
| C. tertium (Y18174) | 76 | 375 | 583 | 84 | 305 | 48 | 91 | 150 | |||
Clusters XIVa and I are as defined by Collins et al. (4). The groups are based on phylogenetic analyses of the 16S rRNA genes of plant clostridia (16). None, no restriction sites were found in the region of the amplified PCR product (655 bp). All clostridial strains with species names are type strains.
Sensitivity and independence of TRFs.
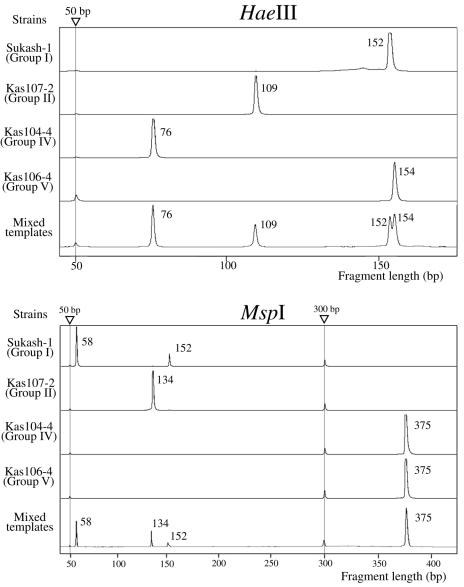
To examine the sensitivity of TRFLP for semisolid RMR cultures, we independently cultivated Sukash-1 (group I), Kas104-4 (group IV), and Kas106-4 (group V) anaerobically in this medium. After the cell densities had been measured by microscopy, cell lysates were prepared from 40 μl of each culture. By using a dilution series of lysates, we performed seminested PCR amplification with the primers shown in Table 1. As a result, the detection limit for these clostridia was <100 cells/ml of culture for the second PCR (data not shown). When the templates (lysates) of representative strains of groups I, II, IV, and V were mixed, all peaks of TRFs derived from the different groups were equally detected. This indicates that there was no PCR bias in the TRFLP system (Fig. 3).
FIG. 3.
Electrograms of the 5′ TRFLPs of HaeIII- and MspI-digested 16S rRNA genes amplified from representatives of different groups of clostridia and their mixed templates. Triangles indicate the positions of internal standards (50 and 300 bp). Numbers with peaks are sizes in base pairs.
Surface sterilization.
To estimate the endophytic populations of clostridia by a combined MPN-TRFLP system, we determined the optimum surface sterilization conditions for the exclusion of epiphytic bacteria. One week after the anaerobic or aerobic incubation of RMR, VL, and RMR plates, no bacterial growth was observed under the following sterilization conditions: 1% NaOCl for 0.5 to 15 min (leaves), 1 to 2% NaOCl for 10 to 15 min (stems), and 2% NaOCl for 15 min (roots). When spores of Clostridium sp. strain B901-1b, which is a close relative of C. beijerinckii, were exposed to a 1% NaOCl solution for 0.5 min, the spores completely lost their viability on VL and RMR plates. Based on these results, the surface sterilization conditions were fixed as follows: 1% NaOCl for 0.5 min for leaves of M. sinensis and 2% NaOCl for 15 min for stems and roots.
MPN counts of N2-fixing bacteria and clostridia.
MPN counts of N2-fixing bacteria and total numbers of bacteria were assessed by an acetylene reduction assay and bacterial growth in MPN tubes containing RMR semisolid medium. Diazotrophic bacteria were detected at levels from 104 to 105 cells/g of fresh weight in surface-sterilized leaves, stems, and roots of M. sinensis. Subsequently, we prepared cell lysates from the cultures in all MPN tubes showing cell growth in RMR semisolid medium and performed TRFLP analyses under conditions established as described above. For culture lysates from the respective MPN tubes, we observed clear peaks without noise; the peaks corresponded to those of group II, IV, and V clostridia, as was the case for the electrograms (Fig. 3) (data not shown). The populations of group II, IV, and V clostridia were separately estimated by the MPN method (Table 3). The populations of group II, IV, and V clostridia ranged from 1 × 103 to 4 × 104 cells/g of fresh weight for all tissues of M. sinensis, but group I clostridia were not detected at all. This indicates that clostridial cells of groups II, IV, and V resided in all parts of the plants, including the aerial parts. The total clostridial population was estimated from the maximum values of the respective groups (Table 3). When the population of clostridia was compared with that of diazotrophs, clostridial cells made up a large proportion of the diazotrophs in the leaves, stems, and roots of M. sinensis (C/D ratio in Table 3).
TABLE 3.
Cell densities of total bacteria, diazotrophs, clostridia, and phylogenetic populations of clostridia in M. sinensis by the MPN methoda
| Tissue | Total no. (cells g−1 of fresh wt)
|
C/D ratiob (%) | No. of cells (cells g−1 of fresh wt) in phylogenetic populationc
|
|||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Diazotrophs | Clostridia | Cluster XIVa
|
Cluster I
|
||||
| Group I | Group II | Group IV | Group V | |||||
| Leaf | 2.3 × 105 | 4.2 × 104 | 4.2 × 104 | 100 | ND | 4.2 × 104 | 2.6 × 103 | 1.7 × 104 |
| Stem | 2.5 × 106 | 1.5 × 105 | 3.5 × 104 | 23 | ND | 3.5 × 104 | 3.5 × 104 | 1.3 × 103 |
| Root | 7.9 × 106 | 2.9 × 104 | 1.9 × 104 | 66 | ND | 1.9 × 104 | 6.4 × 103 | 4.0 × 103 |
Surface sterilization was carried out with 1% NaOCl for 0.5 min for leaves of M. sinensis and 2% NaOCl for 15 min for stems and roots. MPN counts of diazotrophic bacteria and total bacteria were assessed by an acetylene reduction assay and by bacterial growth in MPN tubes containing RMR semisolid medium. Thus, the total number of bacteria indicates the counts of bacteria that can grow only in this medium. TRFLP analysis was performed with cultures that showed cell growth in the MPN tubes. The total number of clostridia was assessed by the maximum population level among individual clostridial groups. The MPN results were single determinations for 150 test tubes.
C/D ratio, the ratio of the total number of clostridia to that of diazotrophs.
The conditions used for the surface sterilization of leaves and stems by NaOCl were effective enough to completely avoid contamination by epiphytic bacteria. We may have underestimated the population levels of endophytic clostridia by MPN-TRFLP analysis because NaOCl is able to penetrate the plant tissues and cause partial sterilization of endophytic clostridia. Thus, the observed population levels were probably minimum values for endophytic clostridia in M. sinensis. Endophytic bacteria are ubiquitous in most plant species. However, the known diazotrophic endophytes (2, 5, 7, 8, 11, 18, 20) and nondiazotrophic endophytes (15, 21) are all aerobes and facultative anaerobes. To our knowledge, this is thus the first report that substantial populations of obligate anaerobic clostridia actually reside in plants (Table 3).
TFRLP analysis of directly extracted DNA.
TRFLP is generally used for the detection and phylogenetic assignment of target organisms in natural environments without cultivation (3, 14, 19). When DNAs directly extracted from M. sinensis stems were used as a PCR template for the clostridium-specific TRFLP method, group I, II, IV, and V clostridia were detected (Fig. 4A). After a 102 dilution of the PCR template with water, signature peaks of groups II, IV, and V remained (Fig. 4A), although no signature peaks were detected at a 104 dilution for any group. This result agreed with the analysis of clostridial populations by MPN-TRFLP (Table 3) in that group II, IV, and V clostridia were dominant in M. sinensis stems. When a bulk sample of soil from around the M. sinensis stand was analyzed, peaks specific for group I, IV, and V clostridia were observed (Fig. 4B, asterisk), but no peaks for group II were detected (Fig. 4B, arrowhead).
FIG. 4.
TRFLP profiles of HaeIII- and MspI-digested 16S rRNA genes amplified from DNA extracts of M. sinensis stems (A) and from soil near the stand of plants (B). Clostridial groups (I, II, IV, and V) were identified by the TRF sizes shown in Table 3. M50 and M300 indicate size markers of 50 and 300 bp, respectively. Asterisks show peaks specific to clostridial groups. Arrowheads indicate the positions of group II-specific clostridia. Note that the TRF signal of group II clostridia was not detected in the soil (B) but was detected in plant stems (A).
Group II clostridia do not include currently known species of the genus Clostridium (Fig. 1). It was empirically difficult to isolate them because they showed very weak growth on VL and RMR agar media (16). This is why only small numbers of group II clostridia were isolated (Table 3) (16). Moreover, they showed community-dependent N2 fixation. It is therefore surprising that group II clostridia dominated the populations of diazotrophs and clostridia (Table 3). Group II clostridia may function as diazotrophic endophytes in M. sinensis regardless of their poor culturability.
The MPN approach to comparing relative population sizes of different clostridial groups requires that the groups have similar levels of culturability. Thus, culturing bias is one potential factor affecting the reliability of the population size estimates by MPN-TRFLP analysis. On the other hand, TRFLP analysis using directly extracted DNAs is a rapid and convenient method to survey clostridial populations in plants without a culturing bias.
Acknowledgments
We are grateful for a grant from the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) to support this work.
REFERENCES
- 1.Cato, E. P., W. L. George, and S. M. Finegold. 1986. Genus Clostridium, p. 1141-1200. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md.
- 2.Chelius, M., and E. W. Triplett. 2000. Immunolocalization of dinitrogenase reductase produced by Klebsiella pneumoniae in association with Zea mays L. Appl. Environ. Microbiol. 66:783-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin, K., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. E. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 5.Elbeltagy, A., K. Nishioka, T. Sato, H. Suzuki, B. Ye, T. Hamada, T. Isawa, H. Mitsui, and K. Minamisawa. 2001. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67:5285-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franks, A. H., H. J. M. Harmsen, G. C. Raags, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gyaneshwar, P., E. K. James, N. Mathan, P. M. Reddy, B. Reinhold-Hurek, and J. K. Ladha. 2001. Endophytic colonization of rice by a diazotrophic strain of Serratia marcescens. J. Bacteriol. 183:2634-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyaneshwar, P., E. K. James, P. M. Reddy, and J. K. Ladha. 2002. Herbaspirillum colonization increases growth and nitrogen accumulation in aluminium-tolerant rice varieties. New Physiol. 154:131-145. [Google Scholar]
- 9.Hippe, H., J. R. Andreesen, and G. Gottschalk. 1992. The genus Clostridium—nonmedical, p. 1800-1866. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, Heidelberg, Germany.
- 10.Hiraishi, A. 1992. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial culture without DNA purification. Lett. Appl. Microbiol. 15:210-213. [DOI] [PubMed] [Google Scholar]
- 11.Hurek, T., L. L. Handley, B. Reinhold-Hurek, and Y. Piche. 2001. Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Mol. Plant-Microbe Interact. 15:233-242. [DOI] [PubMed] [Google Scholar]
- 12.Hurley, M. A., and M. E. Roscoe. 1983. Automated statistical analysis of microbial enumeration by dilution series. J. Appl. Bacteriol. 55:159-164. [Google Scholar]
- 13.Kobayashi, K., and Y. Yokoi. 2003. Spatiotemporal patterns of shoots within an isolated Miscanthus sinensis patch in the warm-temperature region of Japan. Ecol. Res. 18:41-51. [Google Scholar]
- 14.Liu, W., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McInroy, J. A., and J. W. Kloepper. 1995. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil 173:337-342. [Google Scholar]
- 16.Minamisawa, K., K. Nishioka, T. Miyaki, B. Ye, T. Miyamoto, M. You, A. Saito, M. Saito, W. Barraquio, N. Teaumroong, T. Sein, and T. Tadashi. 2004. Anaerobic nitrogen-fixing consortia consisting of clostridia isolated from gramineous plants. Appl. Environ. Microbiol. 70:3096-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis of polymerase chain reaction-amplified genes encoding 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinhold-Hurek, B., and T. Hurek. 1998. Life in grasses: diazotrophic endophytes. Trends Microbiol. 6:139-144. [DOI] [PubMed] [Google Scholar]
- 19.Sessitsch, A., B. Reiter, U. Pfeifer, and E. Wilhelm. 2002. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39:23-32. [DOI] [PubMed] [Google Scholar]
- 20.Sevilla, M., R. H. Burris, N. Gunapala, and C. Kennedy. 2001. Comparison of benefit to sugarcane plant growth and 15N2 incorporation following inoculation of sterile plants with Acetobacter diazotrophicus wild-type and nif− mutant strains. Mol. Plant-Microbe Interact. 14:359-366. [DOI] [PubMed] [Google Scholar]
- 21.Sturz, A. V., B. R. Christie, B. G. Matheson, and J. Nowak. 1997. Biodiversity of endophytic bacteria which colonize red clover nodules, roots, stems and foliage and their influence on host growth. Biol. Fertil. Soils 25:13-19. [Google Scholar]
- 22.Van Dyke, M. I., and A. J. McCarthy. 2002. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Appl. Environ. Microbiol. 68:2049-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber, S., S. Stubner, and R. Conrad. 2001. Bacterial populations colonizing and degrading rice straw in anoxic paddy soil. Appl. Environ. Microbiol. 67:1318-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]