Abstract
Human enteric viruses are currently recognized as one of the most important causes of food-borne disease. Implication of enteric viruses in food-borne outbreaks can be difficult to confirm due to the inadequacy of the detection methods available. In this study, a nucleic acid sequence-based amplification (NASBA) method was developed in a multiplex format for the specific, simultaneous, and rapid detection of epidemiologically relevant human enteric viruses. Three previously reported primer sets were used in a single reaction for the amplification of RNA target fragments of 474, 371, and 165 nucleotides for the detection of hepatitis A virus and genogroup I and genogroup II noroviruses, respectively. Amplicons were detected by agarose gel electrophoresis and confirmed by electrochemiluminescence and Northern hybridization. Endpoint detection sensitivity for the multiplex NASBA assay was approximately 10−1 reverse transcription-PCR-detectable units (or PFU, as appropriate) per reaction. When representative ready-to-eat foods (deli sliced turkey and lettuce) were inoculated with various concentrations of each virus and processed for virus detection with the multiplex NASBA method, all three human enteric viruses were simultaneously detected at initial inoculum levels of 100 to 102 reverse transcription-PCR-detectable units (or PFU)/9 cm2 in both food commodities. The multiplex NASBA system provides rapid and simultaneous detection of clinically relevant food-borne viruses in a single reaction tube and may be a promising alternative to reverse transcription-PCR for the detection of viral contamination of foods.
Hepatitis A virus and noroviruses are the two leading causes of viral food-borne illness, with the noroviruses now estimated as the most prevalent agents of food-borne disease (22). Various foods have been implicated in viral outbreaks, including fruits and vegetables, sliced deli meats, shellfish, and hand-prepared foods such as sandwiches and salads (9, 12, 23). Noroviruses, previously referred to as Norwalk-like viruses, are a genetically diverse group of RNA viruses belonging to the Caliciviridae family and consisting of genogroups I and II. The infections associated with noroviruses present as acute vomiting, diarrhea, nausea, and abdominal cramps within 24 to 48 h of exposure (20). Unlike other enteric viruses, hepatitis A virus affects the liver, causing acute infection that can last from one to several weeks, with typical symptoms including fever, nausea, malaise, anorexia, headache, and jaundice (6).
No animal model or cell culture system is available for the isolation or propagation of noroviruses and most wild-type hepatitis A virus strains. Genomic characterization of these viruses has facilitated the development of molecular detection methods, usually based on reverse transcription (RT)-PCR, which were first applied in the clinical realm. While most of these assays are monoplex in nature, a multiplex RT-PCR system was developed by Rosenfield and Jaykus (24) for the simultaneous detection of the human enteroviruses, hepatitis A virus, and Norwalk virus. More recently, Beuret (2) reported a multiplex real-time method to detect genogroup I and genogroup II noroviruses, human enteroviruses, and astroviruses. Both of these studies (2, 24) were developed and tested on cell culture lysates or human stool specimens, as appropriate. Only a limited number of studies have sought to use RT-PCR methods for the detection of enteric viruses in environmental and food samples (25). While shellfish have been the most common food commodity explored, a few studies have reported the RT-PCR detection of viruses in produce and the so-called ready-to-eat foods (10, 18, 27). None of these have been in multiplex format.
Nucleic acid sequence-based amplification (NASBA) is an isothermal nucleic acid amplification method that amplifies an RNA template (7). It is particularly suited for the detection of RNA viruses because there is no need for a separate reverse transcription step. Furthermore, the amplification power of NASBA has been reported to be comparable to or sometimes even better than that of RT-PCR (11, 19). The NASBA approach has been applied to the detection of enteric viruses such as hepatitis A virus (14) and the genogroup I and genogroup II noroviruses (11; www.basickit-support.com, sponsored by bioMerieux, Durham, N.C.). A multiplex format of NASBA has also been reported for the detection of hepatitis A virus and rotavirus in cell culture lysates (15).
The purpose of this study was to develop a multiplex NASBA assay for the simultaneous detection of the most common food-borne viruses, including genogroup I and genogroup II noroviruses and hepatitis A virus. Once developed and optimized, the assay was applied to the detection of these viruses in model ready-to-eat foods artificially seeded to various levels of viral contamination.
MATERIALS AND METHODS
Virus stocks.
The norovirus genogroup I stock was kindly provided by C. L. Moe (Department of International Health, Emory University, Atlanta, Ga.) as pure liquid stool samples from a volunteer experimentally infected with the well-characterized Norwalk 8FIIa prototype strain (17). The norovirus genogroup II stock used in this study was obtained as stool specimens from regional gastroenteritis outbreaks (gift of S. R. Greene, bioMerieux, Durham, N.C.). Three samples of norovirus genogroup II (designated pol064, pol096, and pol108) that were previously screened by monoplex NASBA (16) and demonstrated particularly high NASBA-electrochemiluminescence detection signals (>log10 7.0) were pooled to provide 6 g of a homogeneous stock. All virus stocks were stored at 4°C until use. Assays were done on both the undiluted stocks and stocks serially diluted in RNase-free water (Sigma Chemical Company, Milwaukee, Wis.).
The cytopathic strain HM-175 of hepatitis A virus was propagated in FRhK-4 cells as previously described (8). The viral cell lysate suspension was obtained after two rapid freeze-thaw cycles and purified by extraction with trichlorofluoroethane (1:1, vol/vol) (28). The purified stock solution was stored in 1-ml fractions at −80°C until use.
Estimation of stock virus titer.
Hepatitis A virus strain HM-175 was assayed for infectivity by the plaque technique in FRhK-4 cells as previously described (28) and had a titer of approximately 2 × 106 PFU/ml (data not shown). For the noroviruses, virus concentration was estimated by endpoint titration RT-PCR of RNA extracts (see below) and their serial 10-fold dilutions. Briefly, conventional two-step RT-PCR protocols were done with the RNA PCR Core kit (Roche, Branchburg, N.J.) as described in the manufacturer's instructions. Norovirus genogroup I (SR33, 5′-TGTCACGATCTCATCATCACC-3′; and SR48 5′-GTGAACAGCATAAATCACTGG-3′) and genogroup II (SR33, 5′-TGTCACGATCTCATCATCACC-3′; and SR46 5′-TGGAATTCCATCGCCCACTGG-3′) primers, which produced a 123-bp product between nucleotides 4754 and 4876 within the RNA polymerase gene, were used in these amplifications (1). The reverse transcription reaction volume was increased from 20 to 30 μl to accommodate a 10-μl sample.
RT-PCR was done according to the amplification conditions and parameters described by Ando et al. (1), which included one cycle of reverse transcription at 42°C for 1 h, followed by denaturation at 94°C for 3 min; 40 amplification cycles with denaturation for 60 s at 94°C, annealing for 90 s at 50°C, and extension for 120 s at 60°C; and a final cycle of incubation at 72°C for 7 min. For the genogroup I norovirus suspension, the stock preparation was estimated at 2 × 104 RT-PCR-detectable units/ml; for the genogroup II norovirus suspension, the stock preparation titer was approximately 2 × 105 PCR-detectable units/ml (data not shown). All virus stock suspensions (hepatitis A virus and genogroup I and genogroup II noroviruses) were used consistently in all studies.
Viral contamination and elution from ready-to-eat food.
To simulate the viral contamination of ready-to-eat food, 100 μl of each individual virus suspension or 10-fold serial dilutions thereof was deposited on the center of a leaf of Boston lettuce (3 by 3 cm) previously washed with sterile distilled water or deli sliced turkey samples, both purchased from a local retail source. For multiplex NASBA validation, an experiment was designed in which all three viral suspensions were serially diluted, pooled at different concentrations starting with 2 × 105 PFU, 2 × 103 PCR-detectable units, and 2 × 103 PCR-detectable units/ml for hepatitis A virus and genogroup I and genogroup II noroviruses, respectively, and used as the inoculum to experimentally contaminate the lettuce and turkey samples. Inoculated samples were air dried under a biohazard laminar flow hood for 30 min at room temperature. Viruses were then eluted from the inoculated surfaces with 500 μl of elution buffer (0.5 M glycine, 0.14 NaCl, 0.2% Tween 20, pH 8.5) by repeated pipetting (10 times) of the contaminated area. The elution was repeated a second time with 500 μl of fresh elution buffer. The two eluant solutions were pooled, producing a final 1-ml eluant volume, which was immediately extracted for RNA isolation and purification. All experiments were done in duplicate. Uninoculated control samples were handled exactly as were the experimentally inoculated samples.
Extraction of RNA.
Extraction of RNA was done according to a modification of the guanidine thiocyanate (GuSCN)-silica method of Boom et al. (4) with NucliSens lysis buffer and Isolation reagents (bioMerieux Inc., Durham, N.C.) as suggested by the manufacturer. In brief, 100 μl of stool sample or eluant from food was mixed with 900 μl of lysis buffer and 50 μl of activated silica. The RNA-silica complex was washed twice with 1 ml of wash buffer (10 M GuSCN, 100 mM Tris-HCl [pH 6.4]), twice with 1 ml 70% ethanol, and once with 1 ml of acetone, and dried at 56°C for 10 min. After each wash step, the suspension was centrifuged (10,000 × g) for 1 min. Purified RNA in the silica pellet was eluted in 50 μl of elution buffer at 56°C for 10 min and used immediately in amplification reactions as described below.
Nucleic acid sequence-based amplification primers and oligonucleotide probes.
Oligonucleotides used for NASBA and detection of the different enteric viruses (Table 1) were synthesized and gel-purified by Qiagen-Operon (Alameda, Calif.) and stored at −20°C. All primers and probes are located in highly conserved regions of the viral genomes encoding the RNA polymerase for noroviruses of genogroup I (accession no. M87661) and genogroup II (accession no. X86557) and VP2 for hepatitis A virus (accession no. M14707). The NASBA primers and probes for the detection of the genogroup I noroviruses were previously reported by Greene et al. (11), while those for the amplification of the genogroup II noroviruses were designed in cooperation with bioMerieux, Inc. (www.basickit-support.com). Nucleic acid sequence-based amplification primers and probes for the amplification and detection of hepatitis A virus were initially reported by Jean et al. (14, 15).
TABLE 1.
Sequences of oligonucleotide primers and probes used for amplification and detection of noroviruses and hepatitis A virus
| Primer or probe | Sequencea |
|---|---|
| NVP1 | 5′-AATTCTAATACGACTCACTATAGGGAGAAGGATCTCATCATCACCATA-3′ |
| NVP2b | 5′-GATGCAAGGTCGCATATGAGATACCACTATGATGCAGATTA-3′ |
| NVP2a | 5′-GATGCAAGGTCGCATATGAGGAATTCCATCGCCCACTGGCT-3′ |
| BB1 | 5′-GATGCAAGGTCGCATATGAGCAGATTGGCTTACTACACA-3′ |
| BB2 | 5′-AATTCTAATACGACTCACTATAGGGAGACATGCAACTCCAAATCTGT-3′ |
| NVG1a | 5′-Biotin-ACAGGCCTATCACCCGATGT-3′ |
| NVG1b | 5′-Biotin-ACTGGCTTATCACCTGATGT-3′ |
| NVG1c | 5′-Biotin-TATCACCTGATGTTATACAATCC-3′ |
| NVG2a | 5′-Biotin-GTCCCCTGACATCATACAGGCT-3′ |
| NVG2b | 5′-Biotin-ACAGGACTAGGCCCCGACAT-3′ |
| NVG2c | 5′-Biotin-TCAGGTCTCTCACCAGAT-3′ |
| BB probe | 5′-Biotin-GATTGATCTGTGCTATGGTTCCTGGTGACC-3′ |
Underlined letters designate T7 promoter sequence (NVP1 and BB2) and the specific sequence for NucliSens ruthenium-linked oligonucleotide detector probe (NVP2a, NVP2b, and BB1).
The forward NASBA primers bore the bacteriophage T7 RNA polymerase promoter sequence (5′-AATTCTAATACGACTCACTATAGGGAGAAGG-3′) at their 5′ end and the backward NASBA primers bore a generic electrochemiluminescence complementary probe sequence (5′-GATGCAAGGTCGCATATGAG-3′). Oligonucleotide probes (Table 1) internal to the amplified regions defined by the different primer pairs were biotinylated during synthesis and subsequently used for the detection of amplified target nucleic acid sequences by solid-phase or liquid-phase hybridizations as described below.
Multiplex and monoplex NASBA.
The monoplex and multiplex procedures were carried out with reagents and protocols as described by Jean et al. (15) with some modifications. The final reaction mixture volume was 25 μl containing 40 mM Tris-HCl (pH 8.5), 50 mM KCl, 12 mM MgCl2, 1 mM each of the deoxyribonucleoside triphosphates, 2 mM each of the ribonucleoside 5′-triphosphates, 10 mM dithiothreitol, and 15% (vol/vol) dimethyl sulfoxide; 5 pmol of each gel-purified primer was used for the multiplex NASBA, and 5 ρmol of the specific primer pair was used for the monoplex NASBA; 5 μl of purified RNA were added to 18 μl of monoplex or multiplex amplification mixture in a 0.6-ml microcentrifuge tube, which was incubated for 5 min at 65°C (in a circulating water bath or thermocycler) in order to disrupt secondary structure in the target RNA. Each tube was immediately cooled to 41 ± 1°C for 5 min, after which 2 μl of an enzyme mixture containing 2.6 μg of bovine serum albumin (in 50% glycerol; Roche Diagnostics Corp., Indianapolis, Ind.), 40 U of T7 RNA polymerase (U.S. Biochemicals, Cleveland, Ohio), 8 U of avian myeloblastosis virus reverse transcriptase (Seikagaku America, Falmouth, Mass.), 0.2 U of RNase H (U.S. Biochemicals), and 12.5 U of RNasin (Promega, Madison, Wis.) were added, followed by incubation at 41 ± 1°C for 120 min. The identity of the amplification products was confirmed by agarose gel electrophoresis or hybridization as described below.
Agarose gel electrophoresis and blotting.
Nucleic acid sequence-based amplification products were separated on a 2% agarose gel by electrophoresis in 1× TAE (0.04 M Tris-acetate and 0.002 M EDTA) and stained with ethidium bromide. After destaining for 15 min, the gel was examined under UV light. For transfer blotting, amplified RNA was transferred from the gel onto a positively charged nylon membrane (Roche Diagnostics) by capillarity in the presence of 10× SSC (1× SSC is 0.15 M NaCl and 15 mM sodium citrate). When dot blot hybridization was used, the NASBA product was diluted 1:1 with 20× SSC, and 3 μl of the diluted product was deposited onto a strip of nylon membrane and allowed to air dry. For both transfer and dot blots, the RNA was cross-linked to the membrane by a 2-min exposure to UV with a UV cross-linker (UVP, Upland, Calif.; model UVG-1) followed by solid-phase hybridization.
Solid-phase hybridization.
After transfer or dot blotting, nylon membranes were prehybridized for 30 min at 55°C in RNase-free hybridization solution containing 5× SSC, 0.1% (wt/vol) N-laurylsarcosine, 0.02% (wt/vol) sodium dodecyl sulfate, and 1% (wt/vol) protein-blocking reagent (Roche Diagnostics Corp.). Hybridization was carried out for 2 h at 55°C in 5 ml of the same solution containing 50 nM of the specific biotinylated oligoprobe. Unbound probe was removed by washing the membrane twice for 5 min at room temperature in 2× SSC containing 0.1% (wt/vol) sodium dodecyl sulfate and twice for 15 min at 55°C with 0.1× SSC containing 0.1% (wt/vol) sodium dodecyl sulfate. The membrane was incubated for 30 min in blocking solution (0.1 M maleic acid and 0.15 M NaCl, pH 7.5, containing 1% [wt/vol] protein-blocking reagent) prior to detection of the bound probe. The detection procedure consisted of membrane incubation for 30 min at room temperature in the presence of blocking solution containing 0.5 μg of streptavidin-peroxidase (Kirkegaard and Perry Laboratories, Gaithersburg, MD) per ml, followed by washing five times in PBS-T (0.01 mmol of phosphate-buffered saline, pH 7.2, per liter plus 0.85% NaCl and 0.05% Tween 20). Bound peroxidase was assayed with a 3,3′,5,5′-tetramethylbenzidine (Kirkegaard and Perry Laboratories) substrate solution. A positive result was characterized by the formation of a blue precipitate.
Electrochemiluminescent (liquid) hybridization.
The amplicons generated by NASBA were also detected by liquid hybridization with the NucliSens electrochemiluminescence detection kit and reader (bioMerieux). This method uses two oligonucleotides, a 5′-biotinylated specific capture oligonucleotide immobilized onto paramagnetic beads; and a second detector oligonucleotide labeled with ruthenium. The 5′-biotinylated capture probe was complexed to streptavidin-coated magnetic beads in a coupling procedure carried out with the NucliSens electrochemiluminescence detection kit as described by the manufacturer. The detector probe sequence is generic, provided by the NucliSens basic kit, and complementary to the 3′-terminal end of the amplicons transcribed from the stretch of nucleotides identified by the backward primers.
For the liquid hybridization, 5 μl of the NASBA product was diluted in 25 μl of the NucliSens detection diluent, and 5 μl of the diluted NASBA product was then mixed with hybridization mixture containing 10 μl of viral specific capture probe beads and 10 μl of the generic ruthenium-labeled detection probe. This hybridization step was carried out for 30 min at 41°C with vortex intervals of 10 min each and stopped after addition of 300 μl of assay buffer. The RNA target-magnetic bead complexes were immobilized on an electrode by a magnet, and when voltage was applied to the electrode, the generic ruthenium-labeled detection probe could be detected by electrochemiluminscence. The electrochemiluminescence signal generated at 620 nm was detected and recorded by the NucliSens reader.
RESULTS
Feasibility of multiplex NASBA.
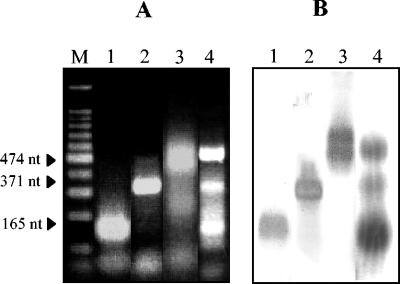
Monoplex reactions for the detection of hepatitis A virus and the genogroup I and genogroup II noroviruses have been described previously (11, 14, 16; www.basickit-support.com). In this study, we sought to develop a multiplex NASBA protocol for the simultaneous detection of these enteric viruses. Based on relatively conserved genomic regions reported for each of these viral genera, three primer pairs were chosen to obtain amplicons of different sizes that could clearly be discriminated from one another by agarose gel electrophoresis. For instance, primers BB1 and BB2, for the specific detection of hepatitis A virus, gave a NASBA product of 474 nucleotides. Primers NVP1 and NVP2b generated a 371-nucleotide amplicon, and NVP1 and NVP2a produced an amplicon of 165 nucleotides for the identification of norovirus genogroup I and genogroup II, respectively. After optimization experiments, all three viruses could be amplified and detected individually and simultaneously with the NASBA primer cocktail, and comparable band intensities were obtained, as visualized by agarose gel electrophoresis (Fig. 1A). Biotinylated oligonucleotide probes corresponding to each virus type were used to confirm amplicon identity in transfer blot hybridization (Fig. 1B).
FIG. 1.
Demonstration of the feasibility of multiplex NASBA for the detection of hepatitis A virus and genogroup I and genogroup II noroviruses, as evaluated by 2% agarose gel electrophoresis (A) and by transfer blot hybridization with specific biotinylated probes (B). Lane M, 100-bp molecular size markers; lane 1, genogroup II norovirus RNA amplified in the multiplex NASBA reaction; lane 2, genogroup I norovirus RNA amplified in the multiplex NASBA reaction; lane 3, hepatitis A virus RNA amplified in the multiplex NASBA reaction; and lane 4, hepatitis A virus and genogroup I and genogroup II norovirus RNAs mixed and amplified in the multiplex NASBA reaction.
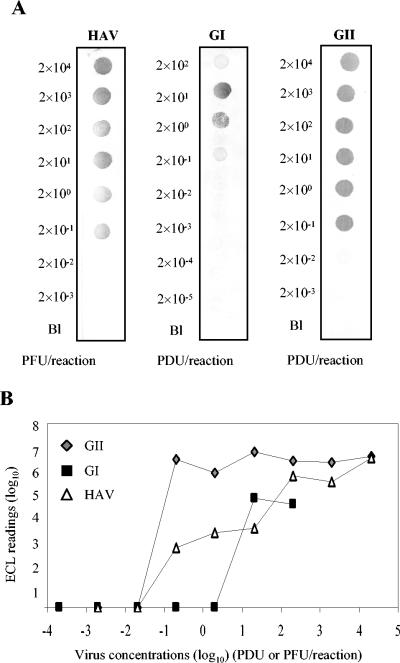
Subsequent experiments focused on elucidating the detection limits for hepatitis A virus and the noroviruses when the RNA extracted from each individual virus was amplified in the presence of the multiplex NASBA primer cocktail. For hepatitis A virus, a detection limit of 2 × 10−1 PFU/reaction was possible after dot blot hybridization (Fig. 2A). Similarly, we were able to detect as few as 2 × 10−1 RT-PCR-detectable units/reaction for the genogroup I and genogroup II noroviruses by dot blot hybridization (Fig. 2A). Similar detection limits were obtained for the genogroup II norovirus and hepatitis A virus when electrochemiluminescence was used as an alternative to dot blot hybridization. However, the detection limit obtained by electrochemiluminescence hybridization was 10-fold less sensitive than that for dot blot hybridization when applied to the detection of genogroup I norovirus amplicons (Fig. 2B).
FIG. 2.
Detection limits for hepatitis A virus and the noroviruses when serial 10-fold dilutions of the RNA extracted from each individual virus were amplified in the presence of the multiplex NASBA primer cocktail and detected by dot blot (A) and electrochemiluminescence (B) hybridizations. Electrochemiluminescence readings reflect the mean and standard deviation of duplicate samples. The detection limit is inferred as the minimum concentration of enteric viruses giving a detectable signal by the hybridization method.
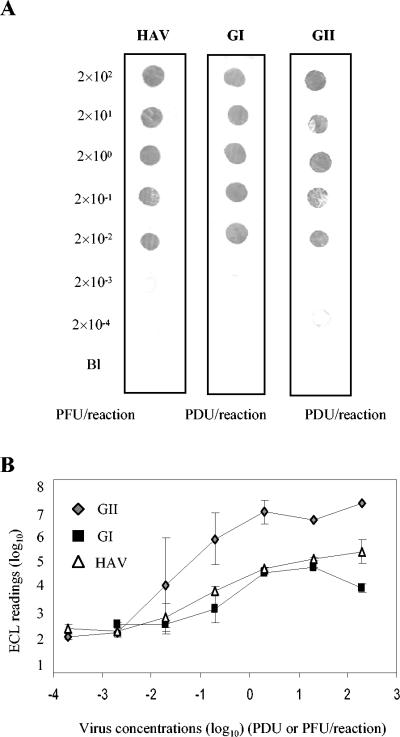
In order to systematically compare the sensitivity of the multiplex NASBA system to the monoplex system, each individual virus suspension was extracted for RNA isolation and 10-fold serial dilutions were subjected to monoplex amplification. In general, the detection limit of the monoplex system was 10-fold (2 × 10−2 PCR-detectable units or PFU/reaction) better than that of the multiplex system (Fig. 3A). While both hybridization systems effectively detected amplicons, electrochemiluminescence hybridization was 10-fold less sensitive than dot blot when applied to the detection of genogroup I norovirus and hepatitis A virus amplicons (Fig. 3B).
FIG. 3.
Detection limits for hepatitis A virus and the noroviruses when serial 10-fold dilutions of the RNA extracted from each individual virus were amplified with the monoplex NASBA system and detected by dot blot (A) and electrochemiluminescence (B) hybridizations. Electrochemiluminescence readings reflect the mean and standard deviation of duplicate samples. The detection limit is inferred as the minimum concentration of enteric viruses giving a detectable signal by the hybridization method.
Application of multiplex NASBA to the detection of viruses on artificially contaminated ready-to-eat foods.
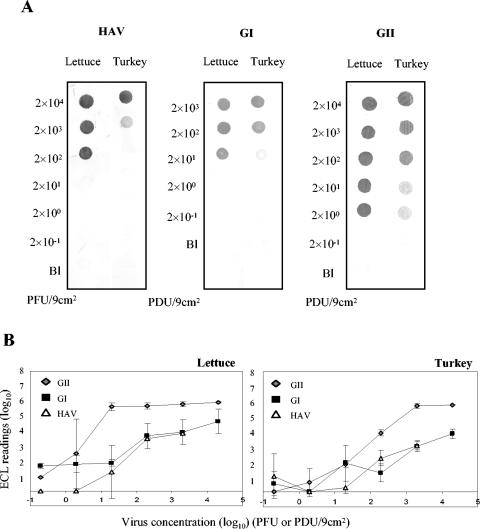
To further demonstrate the applicability of the multiplex NASBA reaction for the detection of viral contamination in complex sample matrices, two at-risk food items (lettuce and sliced deli turkey) were surface contaminated with various concentrations of hepatitis A virus and genogroup I or genogroup II noroviruses, followed by elution of the virus, RNA extraction, NASBA amplification, and amplicon confirmation by hybridization. After contamination of the foods with one virus type alone followed by detection with the multiplex NASBA cocktail, the minimum level of detection was 2 × 102 PFU of hepatitis A virus/9 cm2 for lettuce and 2 × 103 PFU of hepatitis A virus/9 cm2 for turkey, as confirmed by dot blot hybridization (Fig. 4A). When similar experiments were done with the noroviruses artificially inoculated onto the surface of lettuce and turkey, detection limits of 2 × 101 and 2 × 100 PCR-detectable units/9 cm2 were obtained for the genogroup I and genogroup II noroviruses, respectively, regardless of product type (Fig. 4A). As in previous instances, electrochemiluminescence hybridization was not quite as sensitive as dot blot hybridization for confirmation of amplicon identity (Fig. 4B).
FIG. 4.
Detection of single virus suspensions on artificially contaminated foods with the multiplex NASBA system. Serial 10-fold serial dilutions of hepatitis A virus and genogroup I and genogroup II noroviruses were individually spotted onto lettuce and deli sliced turkey, followed by elution and RNA extraction. Samples were subjected to NASBA with the multiplex cocktail and detected by dot blot (A) and electrochemiluminescence (B) hybridizations. Electrochemiluminescence readings reflect the mean and standard deviation of duplicate samples. The detection limit is inferred as the minimum concentration of each enteric virus giving a detectable signal by the hybridization method.
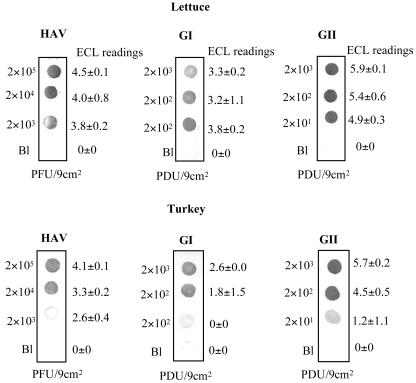
In an effort to systematically evaluate the ability of the multiplex NASBA method to simultaneously detect enteric viruses in a single reaction, an experiment was designed in which the candidate food commodities were artificially contaminated with a mixture of hepatitis A virus and genogroup II norovirus. Interestingly, similar detection limits (e.g., 2 × 100 and 2 × 101 PCR-detectable units/9 cm2 on lettuce and turkey, respectively, for the genogroup II noroviruses and 2 × 102 and 2 × 103 PFU/9 cm2 on lettuce and turkey, respectively, for hepatitis A virus) were obtained when the two viruses were simultaneously subjected to multiplex NASBA (data not shown). Figure 5 shows the results obtained with multiplex NASBA in the presence of the three viruses simultaneously. Detection limits were as low as 2 × 103 PFU/9 cm2 for hepatitis A virus, 2 × 102 PCR-detectable units/9 cm2 for the genogroup I noroviruses, and 2 × 101 PCR-detectable units/9 cm2 for the genogroup II noroviruses and did not differ by commodity. Electrochemiluminescence hybridization again was approximately one log10 less sensitive at amplicon detection than dot blot hybridization.
FIG. 5.
Simultaneous detection of hepatitis A virus and genogroup I and genogroup II noroviruses by multiplex NASBA on artificially contaminated foods. Serial 10-fold dilutions of a three-virus cocktail were spotted onto lettuce and deli sliced turkey, followed by elution and RNA extraction. Samples were subjected to NASBA with the multiplex cocktail and detected by dot blot hybridization. The initial inoculum level is displayed to the left of each blot; the corresponding electrochemiluminescence reading (mean and standard deviation) is displayed to the right of each blot.
DISCUSSION
In spite of the considerable progress in using molecular techniques for the detection of human enteric viruses, few studies have focused their efforts on harnessing these methods to the detection of viral contamination of foods. In this study, we report the development of a multiplex NASBA assay for the simultaneous detection of epidemiologically significant enteric viruses, with particular application to their detection in representative ready-to-eat foods. In experiments done on viral stock cultures alone, monoplex NASBA detection limits were similar to those reported for more established amplification methods such as RT-PCR and were also consistent with the genogroup I norovirus NASBA detection limits reported recently by Greene et al. (11).
In the multiplex format, we found an approximately 10-fold decrease in detection sensitivity, and this, too, is consistent with the reports of other investigators. For instance, Rosenfield and Jaykus (24), who developed an RT-PCR system for the simultaneous detection of the human enteroviruses hepatitis A virus and Norwalk virus, demonstrated a one-log10 decrease in detection sensitivity in comparing their system to monoplex RT-PCR. Similar results were obtained by Tsai et al. (29) in comparing the detection of poliovirus with a monoplex versus a triplex RT-PCR system. However, even with reduced test sensitivity with our multiplex NASBA, detection limits remained <1 PCR-detectable unit or PFU per amplification reaction, which should be sufficient for routine detection of enteric viruses in clinical samples and perhaps even in naturally contaminated foods. Most recently, Beuret (2), using a real-time Sybr Green multiplex RT-PCR protocol for the simultaneous detection of genogroup I and genogroup II noroviruses, human enteroviruses, and rotaviruses, found that real-time amplification actually improved detection limits by approximately 10-fold. These investigators also reported negligible loss in detection sensitivity when combining all four primer pairs compared to a monoplex amplification platform. Indeed, as investigators increasingly use real-time detection approaches, the decreases in amplification efficiency described for combined systems may begin to disappear.
Few studies have described methods for the detection of enteric viruses in foods other than bivalve molluscan shellfish, and many of these do not definitively describe detection limits. Schwab et al. (27) developed a method to detect enteric virus contamination of delicatessen foods with a guanidinium-phenol-based surface wash and reported detection limits of 10 to 100 RT-PCR-detectable units of hepatitis A virus and Norwalk virus in 20 to 40 g of food. Bidawid et al. (3) used a combination of filtration, immunomagnetic separation, and RT-PCR to detect hepatitis A virus in experimentally contaminated lettuce and strawberries, reporting detection limits as low as 10 PFU per piece of lettuce or strawberry (approximate surface area, 50 cm2). Leggitt and Jaykus (18) were able to consistently detect viral RNA at initial inoculum levels of 102 to 103 units/50 g of food sample (lettuce and hamburger) for both hepatitis A virus and Norwalk virus with their concentration/RT-PCR approach. More recently, by refining RNA extraction and purification methods, Sair et al. (26) were able to improve these detection limits to 100 to 102 units/6 g of food sample. In our study, we report multiplex NASBA detection limits in the range of 100 to 103 RT-PCR-detectable units (or PFU)/9 cm2 in both food commodities, a level approaching that which might be anticipated in naturally contaminated food products.
Nucleic acid sequence-based amplification requires sample preparations similar to those employed for RT-PCR, particularly with respect to removal of matrix-associated inhibitory substances and reduction in overall sample volume prior to the initiation of amplification. In our study, although the three viruses were detected in both food items studied, the detection limits for lettuce were generally better than those for deli sliced turkey. Visible turbidity in the eluants recovered from deli turkey, perhaps consisting of inhibitory substances such as lipids (27), was a likely cause of this reduced detection sensitivity. Also, due to the surface characteristics of turkey, it is possible that virus recovery after elution was less than optimal for this commodity, although this issue was not systematically investigated in our study. Consistent with what was reported in the early optimization experiments, detection in foods was more sensitive with the monoplex than with the multiplex NASBA system. Indeed, some investigators have found that the amplification efficiency of one target may decrease by 1 to 3 log10 in the presence of excess competitive target (13, 21). The reduced detection sensitivity of electrochemiluminescence compared to dot blot hybridization may be explained by differences in temperature and time used in the hybridization protocols or differences in the mechanisms and chemistries upon which detection of positive signals was based.
Nucleic acid sequence-based amplification has several important advantages over RT-PCR. For the detection of enteric viruses, NASBA may be especially well suited because it is an RNA amplification method, allowing the investigator to use the initial single-stranded RNA genome as the template in the reaction. Furthermore, NASBA generates the same number of copies in a shorter period of time than RT-PCR because it results in an exponential increase every cycle due to the simultaneous action of the three enzymes, whereas PCR progresses in a binary fashion (5). Furthermore, there is no denaturation step in NASBA, so contaminating DNA is not coamplified. Unfortunately, NASBA is not a panacea, as the low incubation temperature (41°C) in the NASBA reaction may increase the risk of nonspecific amplification. Also, since RNA is the final amplification product, the possibilities for postamplification analyses such as sequencing or fingerprinting may be limited.
In summary, this study describes the development of a multiplex NASBA assay for the simultaneous detection of hepatitis A virus and genogroup I and genogroup II noroviruses and the application of this assay to the detection of viral contamination in model ready-to-eat foods. Excellent detection limits were obtained for all viruses when evaluated from virus stock suspensions and from food samples artificially contaminated with a single virus type. When all three viruses were present in the food matrix simultaneously, they could be detected in a single amplification reaction at detection limits ranging from 2 × 101 to 2 × 103 PCR-detectable units or PFU/9 cm2. Future efforts will focus on applying the assay to other food matrices and to naturally contaminated food samples and on the development of real-time NASBA methods.
Acknowledgments
This work was supported in part by a grant from the USDA National Research Initiative, Competitive Grants Program, contract 2002-35201-11610. Julie Jean was the recipient of a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada. We also acknowledge bioMérieux, Inc., for providing technical support for this study.
The use of trade names in this paper does not imply endorsement by the North Carolina Agricultural Research Service or criticism of similar ones not mentioned.
Footnotes
This work represents paper number FSR 04-11 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Gliss. 1995. Detection and differentiation of antigenically distinct small round structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beuret, C. 2004. Simultaneous detection of enteric viruses by multiplex real-time RT-PCR. J. Virol. Methods 115:1-8. [DOI] [PubMed] [Google Scholar]
- 3.Bidawid, S., J. M. Farber, and S. A. Sattar. 2000. Rapid concentration and detection of hepatitis A virus from lettuce and strawberries. J. Virol. Methods 88:175-185. [DOI] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M.,Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, A. B., and J. D. Fox. 1999. Nucleic acid sequence-based amplification and other transcription-based amplification methods for research and diagnostic microbiology. Rev. Med. Microbiol. 10:185-196. [Google Scholar]
- 6.Ciocca, M. 2000. Clinical course and consequences of hepatitis A infection. Vaccine 18(Suppl. 1):S71-S74. [DOI] [PubMed] [Google Scholar]
- 7.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 350:91-92. [DOI] [PubMed] [Google Scholar]
- 8.Cromeans, T., M. D. Sobsey, and H. A. Fields. 1987. Development of a plaque assays for a cytopathic, rapidly replicated isolate of hepatitis A virus. J. Med. Virol. 22:45-56. [DOI] [PubMed] [Google Scholar]
- 9.Daniels, N. A., D. A. Bergmire-Sweat, et al. 2000. A food-borne outbreak of gastroenteritis associated with Norwalk-like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J. Infect. Dis. 181:1467-1470. [DOI] [PubMed] [Google Scholar]
- 10.Gouvea, V., N. Santos, Carmo-Timenetsky, M., and M. K. Estes. 1994. Identification of Norwalk virus in artificially seeded shellfish and selected foods. J. Virol. Methods 48:177-187. [DOI] [PubMed] [Google Scholar]
- 11.Greene, S. R., C. Moe, L.-A. Jaykus, M. Cronin, L. Grosso, and P. van Aarle. 2003. Evaluation of the NucliSens Basic kit assay for detection of Norwalk virus RNA in stool specimens. J. Virol. Methods 108:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutin, Y. J., V. Pool, E. H. Cramer, et al. 1999. A multistate, food-borne outbreak of hepatitis A. National Hepatitis A Investigation Team. N. Engl. J. Med. 34:595-602. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, R., D. J. Morris, R. J. Cooper, A. S. Bailey, P. E. Klapper, G. M. Cleator, and A. B. Tullo. 1996. Multiplex polymerase chain reaction for adenovirus and herpes simplex virus in eye swabs. J. Virol. Methods 56:41-48. [DOI] [PubMed] [Google Scholar]
- 14.Jean, J., B. Blais, A. Darveau, and I. Fliss. 2001. Detection of hepatitis A virus by the nucleic acid sequence-based amplification (NASBA) technique and comparison with RT-PCR. Appl. Environ. Microbiol. 67:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jean, J., B. Blais, A. Darveau, and I. Fliss. 2002. Simultaneous detection and identification of hepatitis A virus and rotavirus by multiplex nucleic acid sequence-based amplification (NASBA) and microtiter plate hybridization system. J. Virol. Methods 105:123-132. [DOI] [PubMed] [Google Scholar]
- 16.Jean, J., D. H. D'Souza, and L.-A. Jaykus. 2003. Transcriptional enhancement of RT-PCR for rapid and simultaneous detection of noroviruses. FEMS Microbiol. 226:339-345. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, X., D. Y. Graham, K. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 18.Leggitt, P. R., and L.-A. Jaykus. 2000. Detection methods for human enteric viruses in representative foods. J. Food Prot. 63:1738-1744. [DOI] [PubMed] [Google Scholar]
- 19.Lunel, F., P. Cresta, D. Vitour, et al. 1999. Comparative evaluation of hepatitis C virus RNA quantitation by branched DNA NASBA, and monitor assays. Hepatology 29:528-535. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy, M., M. K. Estes, and K. C. Hyams. 2000. Norwalk-like virus infections in military forces: epidemic potential, sporadic disease, and the future direction of prevention and control efforts. J. Infect. Dis. 181:S387-S391. [DOI] [PubMed] [Google Scholar]
- 21.McElhinney, L. M., R. J. Cooper, and D. J. Morris. 1995. Multiplex polymerase chain reaction for human herpesvirus-6, human cytomegalovirus, and human B-globin DNA. J. Virol. Methods 53:223-233. [DOI] [PubMed] [Google Scholar]
- 22.Mead, P. S. L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M., Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenblum, L. S., I. R. Mirkin, D. T. Allen, D. Safford, and S. C. Hadler. 1990. A multifocal outbreak of hepatitis A traced to commercially distributed lettuce. Am. J. Public Health 80:1075-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenfield, S. I., and L.-A. Jaykus. 1999. A multiplex reverse transcription polymerase chain reaction method for the detection of food-borne viruses. J. Food Prot. 62:1210-1214. [DOI] [PubMed] [Google Scholar]
- 25.Sair, A. I., D. H. D'Souza, and L.-A. Jaykus. 2002. Hum. enteric viruses as causes of food-borne disease. Comprehensive Rev. Food Sci. Food Safety. 1:73-89. [DOI] [PubMed] [Google Scholar]
- 26.Sair, A. I., D. H. D'Souza, C. L. Moe, and L.-A. Jaykus. 2002. Improved detection of human enteric viruses in foods by RT-PCR. J. Virol. Methods 100:57-69. [DOI] [PubMed] [Google Scholar]
- 27.Schwab, K. J., F. H. Neill, R. L. Fankhauser, N. A. Daniels, S. S. Monroe, Bergmire- D. A. Sweat, M. K. Estes, and R. L. Atmar. 2000. Development of methods to detect “Norwalk-like viruses” (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLVs outbreak. Appl. Environ. Microbiol. 66:213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobsey, M. D., R. J. Carrick, and H. R. Jensen. 1978. Improved methods for detecting enteric viruses in oysters. Appl. Environ. Microbiol. 36:121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai, Y.-L., B. Tran, L. R. Sangermano, and C. J. Palmer. 1994. Detection of poliovirus, hepatitis A virus, and rotavirus from sewage and ocean water by triplex reverse transcriptase PCR. Appl. Environ. Microbiol. 60:2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]