Abstract
Aflatrem is a potent tremorgenic mycotoxin produced by the soil fungus Aspergillus flavus and is a member of a large structurally diverse group of secondary metabolites known as indole-diterpenes. By using degenerate primers for conserved domains of fungal geranylgeranyl diphosphate synthases, we cloned two genes, atmG and ggsA (an apparent pseudogene), from A. flavus. Adjacent to atmG are two other genes, atmC and atmM. These three genes have 64 to 70% amino acid sequence similarity and conserved synteny with a cluster of orthologous genes, paxG, paxC, and paxM, from Penicillium paxilli which are required for indole-diterpene biosynthesis. atmG, atmC, and atmM are coordinately expressed, with transcript levels dramatically increasing at the onset of aflatrem biosynthesis. A genomic copy of atmM can complement a paxM deletion mutant of P. paxilli, demonstrating that atmM is a functional homolog of paxM. Thus, atmG, atmC, and atmM are necessary, but not sufficient, for aflatrem biosynthesis by A. flavus. This provides the first genetic evidence for the biosynthetic pathway of aflatrem in A. flavus.
Aflatrem is a potent tremorgenic mycotoxin produced by the soil fungus Aspergillus flavus (17, 18). This compound is a member of a large, structurally diverse group of secondary metabolites known as indole-diterpenes that includes paspaline, paxilline, shearinines, paspalitrems, terpendoles, penitrems, lolitrems, janthitrems, and sulpinines (34). These metabolites all have a common structural core comprised of a cyclic diterpene skeleton derived from geranylgeranyl diphosphate (GGPP) and an indole moiety derived from either tryptophan or a tryptophan precursor. Different patterns of prenylation, hydroxylation, epoxidation, acetylation, and ring stereochemistry around the basic indole-diterpene ring structure define this structural diversity (34). The mechanism by which aflatrem and related indole-diterpenes cause tremorgenicity in mammals is not well defined, but biochemical and clinical studies indicate that these effects are due in part to effects on receptors and to interference with neurotransmitter release in the central and peripheral nervous systems (40).
Very little is known about the pathways of indole-diterpene biosynthesis other than the origins of the indole and diterpene components (7, 10). Biosynthetic schemes based on the chemical identification of likely intermediates have been proposed, but until recently none of these steps had been validated by biochemical or genetic studies (25, 30).
Recently, a cluster of genes for paxilline biosynthesis was cloned from Penicillium paxilli (53). Key genes in this cluster include a GGPP synthase gene (paxG), a FAD-dependent monooxygenase gene (paxM), a prenyl transferase gene (paxC), and two cytochrome P450 monooxygenase genes, paxP and paxQ. The deletion of paxG, paxM, or paxC results in mutants that are defective in paxilline biosynthesis (53; B. Scott, L. McMillan, J. Astin, C. Young, A. Bryant, and E. Parker, unpublished results). PaxM and PaxC may catalyze the addition of indole-3-glycerol phosphate to GGPP and subsequent cyclization to form the first stable indole-diterpene, possibly paspaline (34). paxP and paxQ deletion mutants accumulate paspaline and 13-desoxypaxilline, respectively, suggesting that these compounds are the substrates for the corresponding enzymes (26). Thus, at least five genes are required for the biosynthesis of paxilline in P. paxilli.
Two other fungal gene clusters for diterpene biosynthesis have been reported, namely gibberellin production in Fusarium fujikuroi (44) and aphidicolin synthesis in Phoma betae (42). Like the paxilline biosynthesis cluster in P. paxilli, the gibberellin and aphidicolin gene clusters contain a GGPP synthase gene, named ggs-2 and Pbggs, respectively. P. paxilli and F. fujikuroi also carry a second GGPP synthase gene, suggesting that the presence of two copies of GGPP synthase may be a molecular signature for diterpene biosynthesis (28, 53). Given that many genes for secondary metabolite biosynthesis in fungi are organized in clusters (23), the molecular cloning of GGPP synthases combined with chromosome walking could provide a rapid strategy for cloning new indole-diterpene gene clusters.
The objectives of this study were to (i) isolate GGPP synthases from A. flavus by using degenerate PCR, (ii) determine if one of the GGPP synthases is closely linked to homologs of the P. paxilli pax genes by performing chromosome walking, (iii) determine if the pattern of expression of these genes coincides with the onset of aflatrem biosynthesis, and (iv) test whether one of these genes can functionally complement a P. paxilli pax mutant. Our working hypothesis was that A. flavus contains two GGPP synthase genes, one of which is associated with a cluster of genes for indole-diterpene (aflatrem) biosynthesis. This work broadens our understanding of indole-diterpene metabolite biosynthesis in filamentous fungi and provides the first genetic evidence for the biosynthetic pathway of aflatrem in A. flavus.
MATERIALS AND METHODS
Bacterial strains.
The Escherichia coli strains used for this study, XL-1 Blue (5), KW251 (Promega Corp., Madison, Wis.), and Top 10 (Invitrogen Corp., Carlsbad, Calif.), were grown on Luria-Bertani (LB) agar plates (38) supplemented, when necessary, with either ampicillin (100 μg/ml) or kanamycin (50 μg/ml).
Fungal strains and growth conditions.
A. flavus strain NRRL6541 (47) was obtained from Merck and Co. (Rahway, N.J.) and is maintained at Massey University as strain PN2280. Cultures were maintained on 2.4% potato dextrose agar (Difco, Spark, Md.) plates or as spore suspensions in 10% (vol/vol) glycerol at −80°C. Seed cultures of A. flavus for aflatrem production were started from an inoculum of approximately 5 × 106 spores in a 100-ml Erlenmeyer flask containing 25 ml of YEPGA medium (26) and were grown at 30°C for 2 days with agitation at 200 rpm. Two milliliters of seed culture was added to 50 ml of aflatrem production medium (see below), and the cultures were incubated in the dark without shaking at 30°C until they were harvested. The mycelium (approximately 1 g of fresh weight) was collected by filtration through Miracloth (Calbiochem Corporation, La Jolla, Calif.), washed with water, and blotted dry. Samples of 1 to 3 g (wet weight) were frozen in liquid nitrogen and stored at −80°C for the preparation of RNAs. The remainder of the mycelium was freeze-dried for indole-diterpene analysis. Cultures used for the preparation of genomic DNAs were grown under the same conditions as those used for the seed cultures except that CDYE medium (53) was used.
Aflatrem production medium contained the following ingredients per liter: 80 g of mannitol, 40 g of yeast extract, 10 g of Casamino Acids, 10 g of CaCO3, 10 mg of FeSO4 · 7H2O, 10 mg of ZnSO4 · 7H2O, 2 mg of MnSO4 · 5H2O, 1 mg of CuSO4 · 5H2O, and 0.8 mg of CoCl2 · 6H2O, buffered to pH 6.0 with 10 g of 2-(N-morpholino)ethanesulfonic acid (MES).
P. paxilli cultures were grown and maintained as described previously (21, 26, 53).
Molecular biology.
Genomic DNAs from A. flavus and P. paxilli were isolated from freeze-dried mycelia by the method of Möller et al. (29), Byrd et al. (6), or Yoder (51). Plasmid and cosmid DNAs were isolated by alkaline lysis (38) and were purified by use of either a Bio-Rad Quantum Prep plasmid miniprep kit (Bio-Rad Laboratories, Hercules, Calif.) or a Qiagen (Hilden, Germany) plasmid mini kit. PCR products amplified with Taq DNA polymerase (Roche, Penzberg, Germany) were routinely cloned into pGEM-T Easy (Promega) and transformed into E. coli XL-1 Blue. Genomic and cosmid digests were transferred to positively charged nylon membranes (Roche) by capillary transfer (41), and DNAs were fixed by UV cross-linking at 254 nm for 2 min. Filters were probed with either [α-32P]dCTP (3,000 Ci/mmol; Amersham, Buckinghamshire, United Kingdom)- or digoxigenin (DIG)-11-dUTP (Roche)-labeled probes. The labeling of DNAs was done by primed synthesis with the Klenow fragment and a High Prime kit (Roche) or by the PCR-mediated incorporation of DIG-11-dUTP (Roche). Hybridizations involving 32P-labeled probes were carried out at 65°C overnight in Denhardt's solution as previously described (52). DIG hybridizations were carried out at 42°C in 50% formamide (DIG system user's guide for filter hybridization; Roche). The membranes were washed, and hybridization signals (phosphorescence) were detected by autoradiography as previously described (52) or by the use of a Fluoro image analyzer (model FLA-5000; Fuji Photo Film Co., Ltd., Tokyo, Japan).
Nested PCR amplification of genomic DNAs.
PCRs were carried out in a final volume of 25 μl containing 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl (pH 8.3), a 50 μM concentration of each deoxynucleoside triphosphate (dNTP), a 200 nM concentration of each primer, 0.5 U of Taq DNA polymerase (Roche), and 5 ng of genomic DNA. The first round of PCR was performed with the degenerate primer pair ggpps27 and ggpps29 (Table 1), followed by a second round with the degenerate primers ggpps27 and ggpps28 and a 1:100 dilution of the reaction mixture from the first round of PCR. The cycling conditions were as follows: 1 cycle of 2 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 45°C, and 1 min at 72°C; and a final extension for 5 min at 72°C. Reactions were carried out in either a PC-960 or FTS-960 thermocycler (Corbett Research, Sydney, Australia). Degenerate primers were designed by using conserved sequences identified from an alignment of Neurospora crassa AL-3 (XP_326920), Gibberella fujikuroi GGS (Q92236), P. paxilli PaxG (AAK11531), Saccharomyces cerevisiae BTS1 (S60921), and G. fujikuroi GGS2 (CAA75568) polypeptide sequences.
TABLE 1.
Primers used for this study
| Primer | Sequence (5′→3′)a |
|---|---|
| ggpps27 | CAYMGIGGTCARGGTATGGA |
| ggpps28 | TTCATRTAGTCGTCICKTATYTG |
| ggpps29 | AACTTTCCYTCIGTSARGTCYTC |
| M13F | CCCAGTCACGACGTTGTAAAACG |
| M13R | AGCGGATAACAATTTCACACAGG |
| T7 | TAATACGACTCACTATAGGG |
| Oligo-dT | GAGAGAATTCGGATCCTCTAGAG(T)20 |
| AF-93 | ACGGATCCAGGAGTGGCGTTATCGTG |
| AF-94 | ACGGATCCAATAGGAGAGGTTAGTCG |
| AF-tub-1 | CTTCTTCATGGTTGGCTTCG |
| AF-tub-2 | CTCTGATCTTCGCAAGTTGG |
| AF-tub-3 | GGTGGAGGACATCTTGAGAC |
| AF-111 | CCTTCTACGTCTCCATCCAG |
| AF-112 | GTGATCTCCTTCTGCATACG |
| atmGF | TAGTCGTCACGGATCTGGAA |
| atmGR | CAAGTTCATTGTCGACCGAG |
| atmMF | GGCATCAATGGCATTATCC |
| atmMR | GGACAATGTATCGTGCAAGG |
| AF-78 | AGGACAGGCTGAACATCATC |
| AF-79 | GTTATCCGCTCTGAGATGTG |
| AF-80 | CTGTCATAGACGTAACCTCC |
| AF-81 | TCTCTTCGAGACATTGCAGC |
| atmCF | TCGGATATTATGTGGCGACC |
| atmCR | GTTGCCGCCTCTGTTGCCTT |
| AF-81 | TCTCTTCGAGACATTGCAGC |
| AF-82 | TTCAACCAGAGGAGCGAGTA |
| AF-106 | CTTAGTCAGCCTCGCTATAC |
| AF-113 | GATTACAAGGTGAGTGGAGC |
| AF-114 | GCTCCACTCACCTTGTAATC |
| AF-115 | AGAGGAAGTCCTACCGCTCA |
| AF-116 | TGAGCGGTAGGACTTCCTCT |
| AF-117 | GGATCAGAGCCGGTGAGACA |
| AF-9 | GAAGACCGAGCACATTGA |
| AF-108 | GTATGTCCTCTTGCATGTCC |
I, inosine; M, A or C; K, G or T; R, A or G; S, C or G; and Y, C or T. Italic bases do not correspond to the A. flavus genomic sequence.
Construction and screening of an A. flavus cosmid library.
Genomic DNA (240 μg) isolated from A. flavus MF6421 (29) was partially digested with MboI (Roche) as described by Frischauf et al. (15) to maximize the yield of 35- to 40-kb DNA fragments. These DNAs were partially end-filled with dATP and dGTP by use of the Klenow fragment of DNA polymerase I (Roche) to generate 5′-GA protruding termini. Aliquots (3.6 μg) of this DNA were ligated in a 9:1 molar ratio of vector to insert with XbaI- and XhoI-digested pMOcosX (6.8 μg) (33). The XbaI termini were dephosphorylated with calf alkaline phosphatase (Roche) (0.5 U/μg of DNA), and the XhoI termini were partially end-filled by the incorporation of dCTP and dTTP to generate protruding 5′-TC ends by use of the Klenow fragment of DNA polymerase I (Roche). Half of the ligated mixture (∼5 μg) was packaged by use of the Packagene system (Promega) according to the manufacturer's instructions (Packagene lambda DNA packaging system technical bulletin no. 005, Promega), the DNAs were transduced into the E. coli host strain KW251, and 2,700 ampicillin-resistant colonies were selected. Individual clones were transferred to 96-well microtiter plates containing LB medium supplemented with ampicillin and were amplified by growing at 37°C overnight. A copy of the cosmid library was maintained at −80°C in LB medium containing 4.4% glycerol (12).
The library was screened by colony PCR with primers ggpps27 and ggpps28. The cycling conditions were as follows: 1 cycle of 2 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C; and a final extension for 5 min at 72°C. Four cosmids, pSZ-17, pSZ-18, pSZ-19, and pSZ-20, contained the atmG gene and one, pSZ-21, contained ggsA.
Subcloning of pSZ-20 and pSZ-21 for sequencing.
The cosmid pSZ-20 was subcloned into the PCR4Blunt-TOPO vector by use of a TOPO shotgun subcloning kit (Invitrogen) using 3.3 μg of cosmid DNA isolated by use of a Qiagen large construct kit. Clones (300) that failed to hybridize to DIG-11-dUTP-labeled pMOcosX DNA were arbitrarily selected for sequencing. Plasmid DNAs were isolated with a Bio-Rad Quantum Prep plasmid miniprep kit, and the ends were sequenced by the use of standard M13 forward and reverse sequencing primers (Table 1). The 7.3-kb BamHI fragment from pSZ-21 was subcloned into pBluescript II KS(+) (Stratagene) to generate pSZ-22, and this plasmid was used as a template to extend the sequence data previously derived from the 218-bp insert of pSZ-2.
RNA isolation and cDNA synthesis and analysis.
RNAs were isolated from frozen mycelia by use of the TRIzol reagent (Invitrogen). mRNAs were isolated by oligo(dT) affinity chromatography with a Sigma (St. Louis, Mo.) GenElute mRNA miniprep kit and were quantitated by measurements of fluorescence with a RiboGreen RNA quantitation kit (Molecular Probes, Inc., Eugene, Oreg.). A denatured sample of mRNA was reverse transcribed by the use of Expand reverse transcriptase (Roche) in the presence of an oligo(dT) primer. Gene-specific amplifications of dilutions (1:10, 1:100, and 1:1,000) of the cDNA were carried out in a reaction volume of 20 μl containing 2.0 μl of 10× reaction buffer (Roche), a 50 μM concentration of each dNTP, a 200 nM concentration of each primer, and 0.5 U of Taq DNA polymerase (Roche). The cycling conditions were as follows: 1 cycle of 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C; and a final extension for 5 min at 72°C. The primer combinations (Table 1) used for cDNA analysis were as follows: atmGF-atmGR (introns 1, 2, and 3) for atmG; AF-79-AF-80 (intron 2), AF-79-atmMR (introns 2 and 3), and AF-80-atmMF (introns 1 and 2) for atmM; and atmCF-AF-82 (intron 2) and atmCR-AF-81 (intron 1) for atmC. These products were cloned into pGEM-T Easy and then sequenced.
For a cDNA analysis of ggsA transcripts, one-step reverse transcription-PCRs (RT-PCRs) were performed with SuperScript III with Platinum Taq (Invitrogen) and DNase I (Invitrogen)-treated mRNAs (5 to 50 ng) purified as described above under the reaction conditions described by the manufacturer. The cycling conditions were as follows: 1 cycle of 30 min at 55°C; 1 cycle of 2 min at 94°C; 40 cycles of 30 s at 94°C, 30 s at 60°C, and 90 s at 72°C; and a final extension for 5 min at 72°C. The primer (Table 1) combinations used were as follows: AF-106-AF-114, AF-113-AF-116 (intron 1), AF-115-AF-117 (intron 2), and AF-9-AF-108 (intron 2). These products were cloned into pGEM-T Easy (Promega Corp.) and then sequenced.
RT-PCR and Northern expression analysis.
RT-PCR expression analysis was performed by use of a SuperScript One-Step RT-PCR with Platinum Taq kit (Invitrogen). PCRs were carried out in a final volume of 25 μl containing 1.2 mM MgCl2, a 200 μM concentration of each dNTP, a 200 nM concentration of each primer, 0.5 μl of reverse transcriptase-Platinum Taq mixture (Invitrogen), and 4 ng of mRNA from each time point. The primer (Table 1) combinations used were AF-tub1-AF-tub3 (spans intron 6) for tub2 (β-tubulin), AF-111-AF-112 for act1 (actin), atmGF-AF-78 (introns 2 and 3) for atmG, AF-79-AF-80 (intron 2) for atmM, and AF-81-atmCR (intron 1) for atmC. The cycling conditions were as follows: 1 cycle of 30 min at 50°C followed by 2 min at 94°C; 27 cycles of 30 s at 94°C, 60 s at 55°C, and 60 s at 72°C; and a final extension for 5 min at 72°C.
For Northern analysis, the total RNA (10 μg) from each time point was mixed with 3 μl of 10× MOPS buffer (38), 15 μl of formamide, 4.8 μl of formaldehyde (37%), 0.12 μl of ethidium bromide (10 mg/ml), and H2O to a final volume of 30 μl. The RNA mixtures were denatured at 65°C for 15 min, loaded into a formaldehyde (6.2%)-agarose (1.2%) gel, and separated by electrophoresis for 4 to 5 h at 120 V. The denatured RNAs were transferred to Hybond N+ (Amersham) membranes by capillary transfer, and the RNAs were fixed by UV cross-linking at 254 nm for 40 s. The filters were probed with [α-32P]dCTP (3,000 Ci/mmol; Amersham)-labeled cDNA probes. Hybridizations were carried out at 68°C overnight in 10× Denhardt's solution. The filters were washed twice with 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate at 68°C. The primer (Table 1) combinations used to generate the cDNA probes were AF-tub2-AF-tub3 for tub2 (β-tubulin), AF-111-AF-112 for act1 (actin), atmGF-atmGR for atmG, AF-79-AF-80 for atmM, and AF-81-atmCR for atmC.
Preparation of complementation construct.
Plasmid pSZ-43 was constructed by ligating a 3,118-bp BamHI fragment containing 1,299 bp of the 5′ sequence, 1,645 bp of the coding sequence, and 174 bp of the 3′ sequence of atmM into pII99 (32). The 3,118-bp BamHI fragment was prepared by digesting a 3,134-bp PCR product that had been amplified by use of the primer set AF-93 and AF-94 with the pSZ-20 cosmid DNA as a template.
P. paxilli transformation and molecular analysis of transformants.
Protoplasts of a paxM deletion mutant (LMM-100) were prepared as previously described (52), except that 10 mg of Glucanex (Chemcolour Industries, Auckland, New Zealand)/ml was used to digest the cell walls, and the mycelium was gently shaken (100 rpm) overnight at 30°C. The protoplasts were transformed with circular pII99 or pSZ-43 by the method of Vollmer and Yanofsky (46) as modified by Itoh et al. (21). Transformants were selected on RG medium (26) containing Geneticin (Invitrogen) at a final plate concentration of 150 μg/ml. Single spores of the resulting transformants were subcultured and maintained on potato dextrose agar supplemented with Geneticin at the same concentration.
Indole-diterpene analysis.
Thin-layer chromatography and reverse-phase high-performance liquid chromatography (HPLC) analyses of A. flavus and P. paxilli indole-diterpenes were performed as described previously (26). For HPLC, eluted products were analyzed by UV detection at either 230 or 280 nm and were compared with the elution times of paxilline and aflatrem standards. Samples were quantitated with respect to known concentrations of paxilline (30) or aflatrem (Merck and Co.). The sensitivity of detection was approximately 50 to 100 ng.
DNA sequencing.
DNA fragments were sequenced by the dideoxynucleotide chain termination method (39) using Big-Dye (version 3) chemistry with oligonucleotide primers (Sigma Genosys) for pBluescript II KS(+), pGEM-T Easy, and PCR4Blunt-TOPO (M13F, M13R, and T7) or for A. flavus sequences. The products were separated in an ABI Prism 377 sequencer (Perkin-Elmer Corp., Foster City, Calif.).
Bioinformatics.
Sequence data were assembled into contigs by the use of Sequencher, version 4.1 (Gene Codes), and were analyzed with the Wisconsin Package, version 9.1 (Genetics Computer Group, Madison, Wis.). Sequence comparisons were performed at the National Center for Biotechnology Information site (http://www.ncbi.nlm.nih.gov/) by using the Brookhaven (PDB), SWISSPROT and GenBank (CDS translation), PIR, and PRF databases, employing algorithms for both local (BLASTX and BLASTP) and global (FASTA) alignments (1, 2, 36). BLASTP analyses were also performed by use of the Whitehead fungal sequence database (http://www-genome.wi.mit.edu/cgi-bin/annotation/fgi/blast_page.cgi).
The pSZ-20 and ggsA sequence contigs were annotated and analyzed with MacVector 7.2 (Accelrys Inc., San Diego, Calif.).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the ggsA and pSZ-20 sequences are AY559848 and AY559849, respectively.
RESULTS
GGPP synthase sequences in A. flavus.
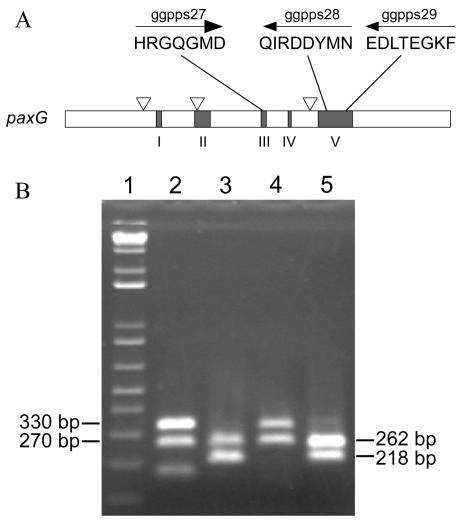
An alignment of fungal GGPP synthase polypeptide sequences identified three highly conserved motifs within domains III and V that flanked an intron in P. paxilli paxG, but not ggs1, and that were suitable for the design of degenerate primers (Fig. 1A). Nested PCR amplification of genomic DNAs from P. paxilli and A. flavus with primers ggpps27 and ggpps29 and then with primers ggpps27 and ggpps28 resulted in two products from each reaction (Fig. 1B). Products of 330 and 270 bp (lanes 2 and 4) were obtained after the first round of amplification, and products of 262 and 218 bp (lanes 3 and 5) were observed after the nested amplification. The P. paxilli products corresponded to sequences of the GGPP synthase genes ggs1 (the smaller band) and paxG (the larger band) (53). The difference in size was due to an intron in paxG. The two A. flavus PCR products (lane 5) each displayed approximately 50% amino acid sequence identity to the P. paxilli GGPP synthase genes. Southern blot analysis confirmed that each sequence was unique and present as a single copy in the genome (Fig. 2). The larger, 262-bp PCR product was predicted to contain an intron and, by analogy with the sequences from P. paxilli, was proposed to be the copy required for aflatrem biosynthesis and was designated atmG (aflatrem GGPP synthase). The smaller, 218-bp product was designated ggsA (A. flavus GGPP synthase) and, as described below, appears to be a pseudogene.
FIG. 1.
A. flavus contains two GGPP synthases. (A) P. paxilli paxG gene map showing conserved domains (I to V), intron positions (inverted triangles), and consensus polypeptide sequences used to design degenerate primers to amplify the region between domains III and V. (B) PCR products amplified with degenerate primers ggpps27 and ggpps29 (lanes 2 and 4) and ggpps27 and ggpps28 (lanes 3 and 5) from genomic DNAs of P. paxilli (lanes 2 and 3) and A. flavus (lanes 4 and 5). The products shown in lanes 3 and 5 were from nested PCR amplifications of 1/100 dilutions of the products shown in lanes 2 and 4, respectively. Lane 1 contains a 1-kb DNA ladder (Invitrogen).
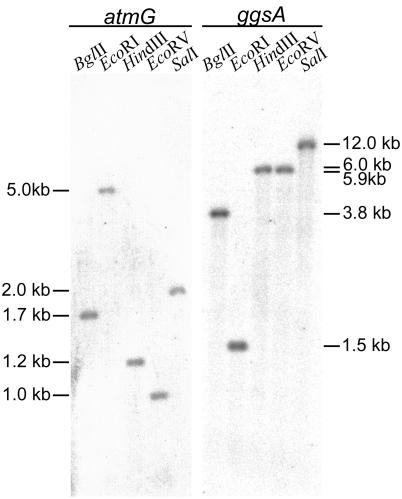
FIG. 2.
Southern analysis of A. flavus genomic DNA probed with atmG and ggsA. The autoradiograph shows Southern blots of A. flavus genomic DNA (1 μg) digested with five different restriction enzymes and probed with 32P-labeled 262-bp atmG and 218-bp ggsA probes. The numbers on the left and right correspond to the sizes of the restriction fragments that hybridized to the probes.
From an A. flavus cosmid library constructed in pMOcosX (33), 2,700 clones were screened by PCR, and five of them, pSZ-17 to pSZ-21, carried atmG or ggs1. Cosmids pSZ-17 to pSZ-20 contained overlapping fragments, but cosmid pSZ-21 had a unique restriction pattern (data not shown). The 262-bp atmG probe hybridized to 2.4-kb BamHI and 5.0-kb EcoRI fragments of cosmids pSZ-17 to pSZ-20 but did not hybridize with pSZ-21. Conversely, the 218-bp A. flavus ggsA product hybridized to a 7.3-kb BamHI fragment of pSZ-21 but did not hybridize with the other four cosmids (data not shown).
Organization of genes for indole-diterpene biosynthesis in A. flavus.
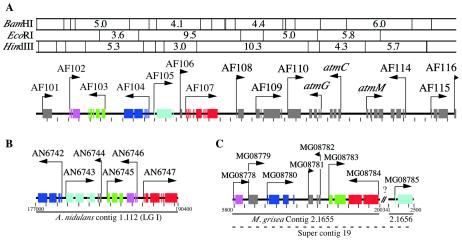
We sequenced the atmG-containing cosmid pSZ-20 and identified a potential gene cluster (Fig. 3) with high sequence similarity to the P. paxilli pax genes, which are required for the biosynthesis of the indole-diterpene paxilline. These genes included atmG (AF111), atmC (AF112), and atmM (AF113), which are orthologs of paxG, encoding a GGPP synthase, paxC, encoding a prenyltransferase (cyclase), and paxM, encoding a FAD-dependent monooxygenase (Table 2), respectively. Overall, 16 putative genes, denoted AF101 to AF116, were identified within this 38.9-kb sequence (Fig. 3).
FIG. 3.
Physical gene map of A. flavus atm locus and conserved synteny with A. nidulans and M. grisea. (A) Physical map of A. flavus insert DNA from cosmid pSZ-20 showing restriction enzyme sites for BamHI, EcoRI, and HindIII and the sizes of fragments of >3 kb. The 16 putative genes identified within this 38,895-bp sequence are labeled AF101 to AF116. Orthologs of P. paxilli indole-diterpene biosynthetic genes are labeled as atm (aflatrem biosynthesis) genes in accordance with A. nidulans nomenclature, as described by Bennett and Lasure (4). Exons of each gene are indicated by blocks, and the arrows indicate the direction of transcription. Tick marks are at 1-kb intervals. Syntenous regions for the A. flavus atm locus are shown for A. nidulans (B) and M. grisea (C). Exons with the same color are orthologous.
TABLE 2.
Sequence and bioinformatic analysis of genes within the A. flavus pSZ-20 cosmid insert
| Gene/ORF or ORF | Size (aa) | Putative Function | Top functionally characterized BLASTP match
|
|||
|---|---|---|---|---|---|---|
| Protein | E-value | Organism | Reference or accession no. | |||
| AF101 | 315 | Unknown | ||||
| AF102 | 299 | Dioxygenase | DodA | 4e−06 | Portulaca grandiflora | CAE45178 |
| AF103 | 472 | Dehydrogenase | Sim15 | 2e−06 | Streptomyces antibioticus | 19 |
| AF104 | 694 | Monooxygenase | SalA | 6e−22 | Acinetobacter sp. strain ADP1 | 22 |
| AF105 | 591 | Acyl esterase | GACAAP | 7e−17 | Bacillus laterosporus | 3 |
| AF106 | 87 | Unknown | BinA | 6e−13 | Aspergillus nidulans | AJ011295 |
| AF107 | 819 | Transcription factor | RegA | 6e−79 | Aspergillus fumigatus | CAE46958 |
| AF108 | 214 | Dehydrogenase | P22 | 2e−05 | Gallus gallus | 54 |
| AF109 | 785 | Zn2Cys6 transcription factor | Hap1 | 0.002 | Saccharomyces cerevisiae | 37 |
| AF110 | 408 | Monooxygenase | SalA | 1e−15 | Acinetobacter sp. strain ADP1 | 22 |
| atmG | 340 | Geranylgeranyl diphosphate synthase | PaxG | 2e−97 | P. paxilli | 53 |
| atmC | 334 | Prenyltransferase | PaxC | 1e−109 | P. paxilli | 53 |
| atmM | 479 | Monooxygenase | PaxM | 1e−139 | P. paxilli | 53 |
| AF114 | 378 | Polytopic membrane protein | Pth | 3e−27 | Blumeria graminis | AF329397 |
| AF115 | 508 | Cytochrome P450 | Cyp8b1 | 3e−13 | Mus musculus | 16 |
| AF116a | 75 | Acyl transferase | ||||
AF116 is truncated at the 3′ end in pSZ-20, and therefore only a partial sequence was studied.
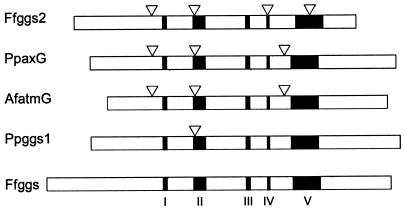
atmG has three introns whose positions and phase are the same as those found in P. paxilli paxG. The first two are also shared with F. fujikuroi ggs-2 (Fig. 4). atmG is predicted to encode a polypeptide of 340 amino acids with an unmodified molecular mass of 38.5 kDa (Table 2) that has 49% identity (64% similarity) to PaxG from P. paxilli. AtmG contains the five conserved domains found in all prenyl diphosphate synthases (8), including the highly conserved aspartate-rich motifs, DDXXD and DDXXN/D, of domains II and V that are the proposed binding sites for the isopentenyl diphosphate and allyl isoprenoid substrates (Fig. 4).
FIG. 4.
Comparison of gene structure of A. flavus GGPP synthase gene with those of other fungal GGPP synthase genes. Gene structures are given for F. fujikuroi primary (Ffggs) and secondary (Ffggs2), P. paxilli primary (Ppggs1) and secondary (PppaxG), and A. flavus atmG (AfatmG) GGPP synthases. The positions of introns (inverted triangles) and conserved domains I to V (shading) are identified.
atmC contains two introns whose positions and phase are the same as those found in paxC. atmC is predicted to encode a polypeptide of 334 amino acids with an unmodified molecular mass of 38.1 kDa (Table 2). This polypeptide contains the five conserved domains found in other prenyl diphosphate synthases (8) and has 55% identity (70% similarity) to PaxC.
atmM has three introns and is predicted to encode a polypeptide of 479 amino acids with an unmodified molecular mass of 53.9 kDa (Table 2). AtmM has 52% identity (67% similarity) to PaxM from P. paxilli. AtmM and PaxM both contain four highly conserved motifs, the dinucleotide binding domain (48), an ATG motif (45), a GD motif (11), and a G helix. These motifs suggest a modified Rossman fold, which is used by many flavoproteins to bind FAD.
To the right of atmM are three genes that encode a putative polytopic membrane protein (AF114), a cytochrome P450 monooxygenase (AF115), and a partial sequence for an acyltransferase (AF116). AF114 is predicted to be an integral membrane protein with seven transmembrane domains. Adjacent to the AF114 gene is a gene encoding a putative cytochrome P450 monooxygenase, AF115, that contains the classical signature motifs of cytochrome P450 enzymes, including a heme-binding domain (FGSSPHICPGRHFA) (20), but that does not appear to be an ortholog of either PaxP (E value of 2e−16) or PaxQ (E value of 7e−10), which are cytochrome P450 enzymes that are required for paxilline biosynthesis by P. paxilli (26). The best match (1e−111) is to a hypothetical protein, AN4117 from Aspergillus nidulans. AF115 (partial sequence) has 41 to 43% similarity to trichothecene 3-O-acetyltransferases from Gibberella and Fusarium spp. as its most closely related polypeptides.
Immediately to the left of atmG are three genes that are predicted to encode a dehydrogenase (AF108), a transcription factor (AF109), and a FAD-dependent monooxygenase (AF110). AF109 appears to be a transcription factor of the classical Zn(II)2Cys6 binuclear cluster type, with its best match being a hypothetical protein from Magnaporthe grisea (MG02408; E value, 5e−49). AF110 also has the conserved motifs that are indicative of a modified Rossman fold, but it does not appear to have an ortholog in the pax gene cluster.
Further to the left of these three genes is a set of six genes (AF102 to -107) with synteny to orthologs from A. nidulans and M. grisea, although the gene order and proposed transcriptional orientation vary among species (Fig. 3). The A. nidulans (AN6746) and M. grisea (MG08778) orthologs of AF102 share 71 and 69% identity, respectively, with AF102. AF103 is predicted to be a dehydrogenase and has orthologs in the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus (19).
AF104 is a third FAD-dependent monooxygenase found in the atm region (Table 2), with four introns and E values of 0 and 2e−38 for the A. nidulans (AN6742) and M. grisea (MG08780) orthologs, respectively. AF104 does not appear to have an ortholog in the pax gene cluster. Although AF106 has a very small open reading frame (encoding 87 amino acids), the predicted polypeptide shares significant similarity with the C terminus of A. nidulans AN6743 and with A. nidulans BinA, with the latter being defined as functional by the isolation of an expressed sequence tag (27). Thus, A. nidulans AN6743 appears to be split into two genes in A. flavus, corresponding to AF105 and AF106.
AF107 belongs to a class of transcriptional activators, including the proteins encoded by regA from Aspergillus fumigatus (accession number CAE46958), CMR1 from Colletotrichum lagenarium (43), and PIG1 from M. grisea (43), that contain both Cys2His2-type zinc finger and Zn(II)2Cys6 binuclear cluster DNA binding motifs. Both motifs are present in the N-terminal region of AF107, with the Cys2His2 zinc finger motif being present in two copies.
Molecular analysis of ggsA.
A 7.3-kb BamHI fragment of pSZ-21 containing ggsA was subcloned into pBluescript II KS(+) to generate pSZ-22, which was used as a template to extend the sequence data previously derived from the 218-bp insert in pSZ-2. This sequence (ggsA) contained the five conserved domains found in other prenyl diphosphate synthase genes (8) and had significant identity to paxG (E value of 5e−40). RT-PCR confirmed that ggsA is transcribed and has three exons and two introns. The position of intron 1 is identical to that found in P. paxilli paxG and F. fujikuroi ggs-2 (Fig. 4). Intron 2 is in the same position as intron 4 from F. fujikuroi ggs-2. However, a detailed analysis of the ggsA genomic and cDNA sequences failed to identify a putative methionine start codon. Furthermore, a frameshift mutation resulting in a premature translation stop codon was also identified, indicating that ggsA is a nonfunctional gene.
atm gene expression and aflatrem biosynthesis.
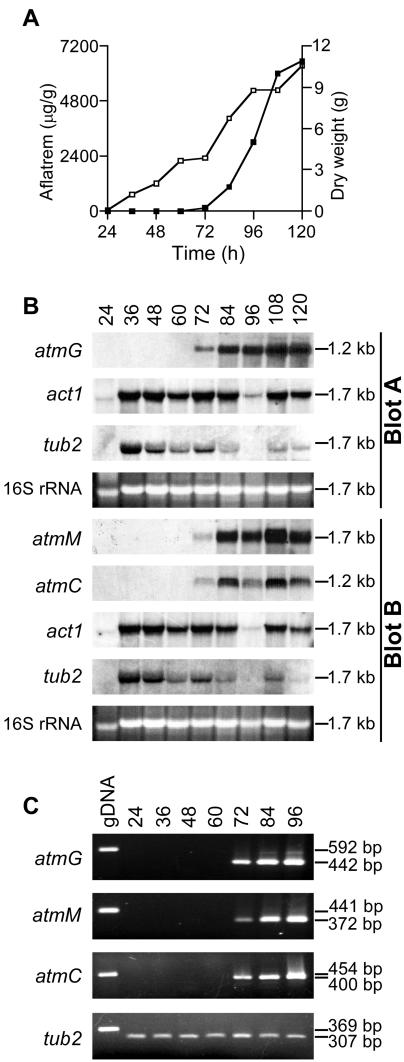
Total RNAs were isolated from mycelia harvested at different times during the growth of cultures (Fig. 5). The steady-state levels of mRNA for atmG, atmC, and atmM increased dramatically with the onset of aflatrem biosynthesis, at approximately 72 h postinoculation (Fig. 5). In contrast, the steady-state levels of tub2 (β-tubulin) and act1 (actin) were relatively constant. The same expression pattern was found when the samples were analyzed by semiquantitative RT-PCR. Thus, an increased expression of atmG, atmC, and atmM is coincident with the onset of aflatrem biosynthesis.
FIG. 5.
Gene expression analysis. (A) Growth of A. flavus showing changes in dry weight (□) and aflatrem production (▪). (B) Autoradiographs of Northern blots of A. flavus total RNAs (10 μg) probed with 32P-labeled 568-bp atmG, 400-bp atmC, 372-bp atmM, 562-bp act1, and 358-bp tub2 cDNAs. The ethidium bromide-stained 16S rRNA region of each gel is shown for comparison. The numbers on the right correspond to the sizes of the hybridized transcripts by comparison to a set of RNA standards (Promega). (C) RT-PCR analysis from 24 to 96 h, with mRNA as the template together with the genomic product (gDNA). Fragment sizes are indicated in base pairs.
Expression of A. flavus atmM in P. paxilli.
Plasmid pSZ-43, containing a wild-type copy of atmM, was introduced into protoplasts of a paxM deletion derivative of P. paxilli, strain LMM-100. Twenty arbitrarily selected Genr and Hygr transformants were purified by subculturing of single spores and then were analyzed by PCR to confirm the correct genetic background. With a primer set specific for paxM, 3.0-kb products were amplified from LMM-100, LMM-100/pII99, and six LMM-100/pSZ-43 transformants, and a 2.3-kb fragment was amplified from the wild type (results not shown). Thus, all six transformants had an LMM-100 background. With a primer set specific for atmM, 3.1-kb products were amplified from pSZ-43, an A. flavus wild-type strain, and six transformants, but no product was obtained from the P. paxilli wild-type, LMM-100, or LMM-100/pII99 (results not shown).
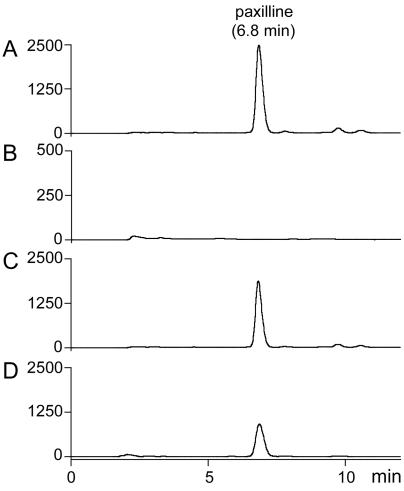
Nine of 10 LMM-100/pSZ-43 transformants produced a metabolite with the same Rf as authentic paxilline (results not shown). No paxilline was found in extracts from LMM-100 and LMM-100/pII99 cells by either thin-layer chromatography or reverse-phase HPLC analysis (Fig. 6). Extracts of LMM-100/pSZ-43 (Fig. 6C) accumulated an indole-diterpene that eluted at the same retention time (6.8 min) as authentic paxilline (Fig. 6D). No indole-diterpenes were detected in extracts of LMM-100 (Fig. 6B). Thus, the A. flavus atmM gene can functionally replace the P. paxilli paxM gene.
FIG. 6.
HPLC analysis of indole-diterpenes from extracts of a P. paxilli paxM mutant containing the A. flavus atmM gene. (A) Wild type. (B) LMM-100 (paxM mutant). (C) LMM-100 containing atmM (pSZ-43). (D) Paxilline standard (5 μg). The units on the y axis are milli-absorbance units at 230 nm, and the retention time on the x axis is given in minutes.
DISCUSSION
The putative indole-diterpene gene cluster from A. flavus that we have cloned may be responsible for aflatrem biosynthesis. This gene cluster contains three genes, atmG, atmM, and atmC, that are up-regulated at the onset of aflatrem biosynthesis and that have functional orthologs in the paxilline biosynthesis gene cluster of P. paxilli (53), although the arrangement of the genes on the chromosome is not conserved across species. The A. flavus cluster is the second indole-diterpene gene cluster to be characterized for a filamentous fungus.
atmG probably encodes a GGPP synthase and is presumed to catalyze the first step in the biosynthesis of aflatrem, i.e., the synthesis of GGPP. A second coding sequence for a GGPP synthase in A. flavus appears to be nonfunctional. Thus, AtmG presumably provides GGPP for aflatrem biosynthesis as well for other GGPP metabolic requirements, such as the C-20 prenylation of proteins (13, 24, 50).
Deletions of paxG, paxM, and paxC in P. paxilli result in mutants with a paxilline-negative phenotype. No identifiable indole-diterpene intermediates have been identified in these strains, suggesting that paxG, paxM, and paxC are involved in very early steps in the pathway. We have been unable to make targeted replacements of any of the atm genes due to a lack of sufficient numbers of protoplasts from A. flavus strain NRRL6541. Procedures that we have used successfully with P. paxilli (21) and Epichloë endophytes (31) as well as previously described protocols for A. flavus (35, 49) were all unsuccessful (unpublished results). Strains of A. flavus with either a tan or white genotype have been successfully induced to form protoplasts (35, 49), suggesting that the pigments produced by strain NRRL6541 may inhibit protoplast formation. A hyaline mutant was also needed to obtain protoplasts from Venturia inaequalis (14). Agrobacterium-mediated transformation (9) has been successful for some filamentous fungi and might circumvent the present difficulties.
Our working model for paxilline biosynthesis in P. paxilli is that PaxM and PaxC are required for the epoxidation and cyclization of GGPP and the addition of indole-3-glycerol phosphate to form the first stable indole-diterpene, possibly paspaline (34). By analogy, we propose that AtmM and AtmC catalyze the same early reactions in aflatrem biosynthesis, since atmM is a functional homolog of paxM and can complement a paxM mutant of P. paxilli. paxP and paxQ, which encode cytochrome P450 enzymes, are required for paxilline biosynthesis, and similar modifications to the paspaline skeleton are predicted to be required for the generation of aflatrem. However, only a single P450 gene, AF115, is known to be closely linked to the core atmG, atmC, and atmM genes, and it does not appear to be a functional ortholog of either paxP or paxQ. Orthologs of paxP and paxQ may be further to the right of AF116. Other modifications to the paspaline skeleton in the biosynthetic pathway to aflatrem include oxidation and acetal formation at C-7 and prenylation of the C-20 carbon of the indole group. Candidate genes for the former are AF104 and AF110, both of which encode putative FAD-dependent monooxygenases of a type predicted to be capable of making this type of biochemical modification. If the break in conserved synteny with A. nidulans and M. grisea between AF107 and AF108 defines the left-hand boundary of the atm cluster, then AF110 is the best candidate for this function.
In summary, we predict that at least eight genes are required for the biosynthesis of aflatrem, including atmG, atmC, atmM, two genes encoding cytochrome P450 enzymes, one FAD-dependent monooxygenase gene, a dehydrogenase gene, and a prenyltransferase gene. Further genetic analyses of the genes identified here and of their adjacent genes will help us to elucidate the pathway for aflatrem biosynthesis. Comparisons with the steps required for paxilline biosynthesis in P. paxilli should elucidate the basic biochemistry and genetics of this important group of secondary metabolites.
ADDENDUM IN PROOF
A recent analysis of A. flavus EST sequences (53a) identified a GGPP synthase sequence (accession number CO146826) which is neither atmG nor ggsB, suggesting the presence of three GGPP synthase genes in A. flavus. Further analysis of our data showed that the 262-bp atmG and 218-bp ggsA nested products (Fig. 1B, lane 5) originate from the upper 330-bp product of the first-round PCR (Fig. 1B, lane 4). Sequence analysis of the lower 270-bp product (Fig. 1B, lane 4) identified a GGPP synthase sequence that corresponds to CO146826. This gene, designated ggsB, is proposed to be the GGPP synthase for primary metabolism.
Acknowledgments
This research was supported by a contract from Merck & Co. (Rahway, N.J.) and by a grant, MAU010, from the Royal Society of New Zealand Marsden Fund.
We thank Andrea Bryant for technical assistance, Sanjay Saikia, Carolyn Young, and Michelle Bryant for technical advice, Emily Parker for discussions of the chemistry of aflatrem biosynthesis, and Emily Parker and Rosie Bradshaw for comments on the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aramori, I., M. Fukagawa, M. Tsumura, M. Iwami, H. Ono, H. Kojo, M. Kohsaka, Y. Ueda, and H. Imanaka. 1991. Cloning and nucleotide sequencing of a novel 7-beta-(4-carboxybutanamido) cephalosporanic acid acylase gene of Bacillus laterosporus and its expression in Escherichia coli and Bacillus subtilis. J. Bacteriol. 173:7848-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, J. W., and L. L. Lasure. 1985. Conventions for gene symbols, p. 537-544. In J. W. Bennett and L. L. Lasure (ed.), Gene manipulation in fungi. Academic Press, London, United Kingdom.
- 5.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-378. [Google Scholar]
- 6.Byrd, A. D., C. L. Schardl, P. J. Songlin, K. L. Mogen, and M. R. Siegel. 1990. The β-tubulin gene of Epichloë typhina from perennial ryegrass (Lolium perenne). Curr. Genet. 18:347-354. [DOI] [PubMed] [Google Scholar]
- 7.Byrne, K. M., S. K. Smith, and J. G. Ondeyka. 2002. Biosynthesis of nodulisporic acid A: precursor studies. J. Am. Chem. Soc. 124:7055-7060. [DOI] [PubMed] [Google Scholar]
- 8.Chen, A. P., P. A. Kroon, and C. D. Poulter. 1994. Isoprenyl diphosphate synthases: protein sequence comparisons, a phylogenetic tree, and predictions of secondary structure. Protein Sci. 3:600-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Groot, M. J. A., P. Bundock, P. J. J. Hooykaas, and A. Beijersbergen. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16:839-842. [DOI] [PubMed] [Google Scholar]
- 10.de Jesus, A. E., C. P. Gorst-Allman, P. S. Steyn, F. R. van Heerden, R. Vleggar, P. L. Wessels, and W. E. Hull. 1983. Tremorgenic mycotoxins from Penicillium crustosum. Biosynthesis of penitrem A. J. Chem. Soc. Perkin Trans. 1983:1863-1868. [Google Scholar]
- 11.Eggink, G., H. Engel, G. Vriend, P. Terpstra, and B. Witholt. 1990. Rubredoxin reductase of Pseudomonas oleovorans: structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J. Mol. Biol. 212:135-142. [DOI] [PubMed] [Google Scholar]
- 12.Farman, M. L. 2001. Analysis of filamentous fungal genomes. In N. J. Talbot (ed.), Molecular and cell biology of filamentous fungi: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 13.Farnsworth, C. C., M. Kawata, Y. Yoshida, Y. Takai, M. H. Gelb, and J. A. Glomset. 1991. C terminus of the small GTP-binding protein smg p25A contains two geranylgeranylated cysteine residues and a methyl ester. Proc. Natl. Acad. Sci. USA 88:6196-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald, A. M., A. M. Mudge, A. P. Gleave, and K. M. Plummer. 2003. Agrobacterium and PEG-mediated transformation of the phytopathogen Venturia inaequalis. Mycol. Res. 207:803-810. [DOI] [PubMed] [Google Scholar]
- 15.Frischauf, A. M., H. Lehrach, A. Poustka, and N. Murray. 1983. Lambda replacement vectors carrying polylinker sequences. J. Mol. Biol. 170:827-842. [DOI] [PubMed] [Google Scholar]
- 16.Gafvels, M., M. Olin, B. P. Chowdhary, T. Raudsepp, U. Andersson, B. Persson, M. Jansson, I. Bjorkhem, and G. Eggertsen. 1999. Structure and chromosomal assignment of the sterol 12 alpha-hydroxylase gene (CYP8B1) in human and mouse: eukaryotic cytochrome P-450 gene devoid of introns. Genomics 56:184-196. [DOI] [PubMed] [Google Scholar]
- 17.Gallagher, R. T., J. Clardy, and B. J. Wilson. 1980. Aflatrem, a tremorgenic toxin from Aspergillus flavus. Tetrahedron Lett. 21:239-242. [Google Scholar]
- 18.Gallagher, R. T., and B. J. Wilson. 1978. Aflatrem, a tremorgenic toxin from Aspergillus flavus. Mycopathology 66:183-185. [DOI] [PubMed] [Google Scholar]
- 19.Galm, U., J. Schimana, H.-P. Fiedler, J. Schmidt, S.-M. Li, and L. Heide. 2003. Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tu 6040. Arch. Microbiol. 178:102-114. [DOI] [PubMed] [Google Scholar]
- 20.Graham-Lorence, S. E., and J. A. Peterson. 1996. Structural alignments of P450s and extrapolations to the unknown. Methods Enzymol. 272:315-326. [DOI] [PubMed] [Google Scholar]
- 21.Itoh, Y., R. Johnson, and B. Scott. 1994. Integrative transformation of the mycotoxin-producing fungus, Penicillium paxilli. Curr. Genet. 25:508-513. [DOI] [PubMed] [Google Scholar]
- 22.Jones, R. M., L. S. Collier, E. L. Neidle, and P. A. Williams. 1999. areABC genes determine the catabolism of aryl esters in Acinetobacter sp. strain ADP1. J. Bacteriol. 181:4568-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 24.Khosravi-Far, R., R. J. Lutz, A. D. Cox, L. Conroy, J. R. Bourne, M. Sinensky, W. E. Balch, J. E. Buss, and C. J. Der. 1991. Isoprenoid modification of rab proteins terminating in CC or CXC motifs. Proc. Natl. Acad. Sci. USA 88:6264-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantle, P. G., and C. M. Weedon. 1994. Biosynthesis and transformation of tremorgenic indole-diterpenoids by Penicillium paxilli and Acremonium lolii. Phytochemistry 36:1209-1217. [Google Scholar]
- 26.McMillan, L. K., R. L. Carr, C. A. Young, J. W. Astin, R. G. T. Lowe, E. J. Parker, G. B. Jameson, S. C. Finch, C. O. Miles, O. B. McManus, W. A. Schmalhofer, M. L. Garcia, G. J. Kaczorowski, M. A. Goetz, J. S. Tkacz, and B. Scott. 2003. Molecular analysis of two cytochrome P450 monooxygenase genes required for paxilline biosynthesis in Penicillium paxilli and effects of paxilline intermediates on mammalian maxi-K ion channels. Mol. Gen. Genomics 270:9-23. [DOI] [PubMed] [Google Scholar]
- 27.Melin, P., J. Schnürer, and E. G. H. Wagner. 1999. Changes in Aspergillus nidulans gene expression induced by bafilomycin, a Streptomyces-produced antibiotic. Microbiology 145:1115-1122. [DOI] [PubMed] [Google Scholar]
- 28.Mende, K., V. Homann, and B. Tudzynski. 1997. The geranylgeranyl diphosphate synthase gene of Gibberella fujikuroi: isolation and expression. Mol. Gen. Genet. 255:96-105. [DOI] [PubMed] [Google Scholar]
- 29.Möller, E. M., G. Bahnweg, H. Sandermann, and H. H. Geiger. 1992. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 20:6115-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munday-Finch, S. C., A. L. Wilkins, and C. O. Miles. 1996. Isolation of paspaline B, an indole-diterpenoid from Penicillium paxilli. Phytochemistry 41:327-332. [Google Scholar]
- 31.Murray, F. R., G. C. M. Latch, and D. B. Scott. 1992. Surrogate transformation of perennial ryegrass, Lolium perenne, using genetically modified Acremonium endophyte. Mol. Gen. Genet. 233:1-9. [DOI] [PubMed] [Google Scholar]
- 32.Namiki, F., M. Matsunaga, M. Okuda, I. Inoue, K. Nishi, Y. Fujita, and T. Tsuge. 2001. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant-Microbe Interact. 14:580-584. [DOI] [PubMed] [Google Scholar]
- 33.Orbach, M. J. 1994. A cosmid with a HygR marker for fungal library construction and screening. Gene 150:159-162. [DOI] [PubMed] [Google Scholar]
- 34.Parker, E. J., and D. B. Scott. 2004. Indole-diterpene biosynthesis in ascomycetous fungi, p. 405-426. In Z. An (ed.), Handbook of industrial mycology. Marcel Dekker, New York, N.Y.
- 35.Payne, G. A., and C. P. Woloshuk. 1989. Transformation of Aspergillus flavus to study aflatoxin biosynthesis. Mycopathologia 107:139-144. [DOI] [PubMed] [Google Scholar]
- 36.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeifer, K., K.-S. Kim, S. Kogan, and L. Guarente. 1989. Functional dissection and sequence of yeast HAP1 activator. Cell 56:291-301. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selala, M. I., G. M. Laekeman, B. Loenders, A. Masuka, A. G. Herman, and P. Schepens. 1991. In vitro effects of tremorgenic mycotoxins. J. Nat. Products 54:207-212. [DOI] [PubMed] [Google Scholar]
- 41.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 42.Toyomasu, T., K. Nakaminami, H. Toshima, T. Mie, K. Watanabe, H. Ito, H. Matsui, W. Mitsuhashi, T. Sassa, and H. Oikawa. 2004. Cloning of a gene cluster responsible for the biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase alpha. Biosci. Biotechnol. Biochem. 68:146-152. [DOI] [PubMed] [Google Scholar]
- 43.Tsuji, G., Y. Kenmochi, Y. Takano, J. Sweigard, L. Farrall, I. Furusawa, O. Horino, and Y. Kubo. 2000. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 38:940-954. [DOI] [PubMed] [Google Scholar]
- 44.Tudzynski, B., and K. Hölter. 1998. Gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet. Biol. 25:157-170. [DOI] [PubMed] [Google Scholar]
- 45.Vallon, O. 2000. New sequence motifs in flavoproteins: evidence for common ancestry and tools to predict structure. Proteins 38:95-114. [DOI] [PubMed] [Google Scholar]
- 46.Vollmer, S. J., and C. Yanofsky. 1986. Efficient cloning of genes of Neurospora crassa. Proc. Natl. Acad. Sci. USA 83:4869-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wicklow, D. T., P. F. Dowd, M. R. TePaske, and J. B. Gloer. 1988. Sclerotial metabolites of Aspergillus flavus toxic to a detritivorous maize insect (Carpophilus hemipterus, Nitidulidae). Trans. Br. Mycol. Soc. 91:433-438. [Google Scholar]
- 48.Wierenga, R. K., P. Terpstra, and W. G. J. Hol. 1986. Predictions of the occurrence of the ADP-binding βαβ-fold in proteins using an amino acid sequence fingerprint. J. Mol. Biol. 187:101-107. [DOI] [PubMed] [Google Scholar]
- 49.Woloshuk, C. P., E. R. Seip, G. A. Payne, and C. R. Adkins. 1989. Genetic transformation system for the aflatoxin-producing fungus Aspergillus flavus. Appl. Environ. Microbiol. 55:86-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamane, H. K., C. C. Farnsworth, H. Xie, T. Evans, W. N. Howald, M. H. Gelb, J. A. Glomset, S. F. Clarke, and B. K. Fung. 1991. Membrane-binding domain of the small G protein G25K contains an S-(all-trans-geranylgeranyl) cysteine methyl ester at its carboxyl terminus. Proc. Natl. Acad. Sci. USA 88:286-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoder, O. C. 1988. Cochliobolus heterostrophus, cause of southern corn leaf blight. Adv. Plant Pathol. 6:93-112. [Google Scholar]
- 52.Young, C., Y. Itoh, R. Johnson, I. Garthwaite, C. O. Miles, S. C. Munday-Finch, and B. Scott. 1998. Paxilline-negative mutants of Penicillium paxilli generated by heterologous and homologous plasmid integration. Curr. Genet. 33:368-377. [DOI] [PubMed] [Google Scholar]
- 53.Young, C. A., L. McMillan, E. Telfer, and B. Scott. 2001. Molecular cloning and genetic analysis of an indole-diterpene gene cluster from Penicillium paxilli. Mol. Microbiol. 39:754-764. [DOI] [PubMed] [Google Scholar]
- 53a.Yu, J., C. A. Whitelaw, W. C. Nierman, D. Bhatnagar, and T. E Cleveland. 2004. Aspergillus flavus expressed sequence tags for identification of genes with putative roles in aflatoxin contamination of crops. FEMS Microbiol. Lett. 237:333-340. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, J., S. E. Bloom, E. Lazarides, and C. Woods. 1995. Identification of a novel Ca2+-regulated protein that is associated with the marginal band and centrosomes of chicken erythrocytes. J. Cell Sci. 108:685-698. [DOI] [PubMed] [Google Scholar]