Abstract
We report on a case of cutaneous infection caused by Alternaria infectoria in a cardiac transplant recipient. A rapid molecular diagnosis was obtained by sequence analysis of the internal transcribed spacer domain of the 5.8S ribosomal DNA region amplified from colonies developed on Sabouraud medium. Treatment consisted of a combination of systemic antifungal therapy, first with amphotericin B and then with itraconazole.
Skin diseases in immunocompromised patients are being increasingly encountered, and the range of causative microorganisms is expanding. Alternaria spp., together with other species of the genera Bipolaris, Curvularia, and Exserohilum, are grouped together with dematiaceous fungi because of the formation of grey to black colonies on culture despite the absence of pigmented elements in tissues. All are anamorphs of ascomycetes of the order Pleosporales. They have a worldwide distribution; are commonly isolated from plants, soil, and indoor air environments; and produce large, airborne conidia. Alternaria spp. have been involved in human infections, and their importance as opportunistic pathogens is increasing among immunocompromised patients, especially in transplant recipients (5, 6, 10, 12). Clinically they are more often encountered as traumatic mycoses. It should be noted that cutaneous mycoses caused by such fungi are characterized by brownish hyphal elements in tissue, whereas subcutaneous mycoses often consist of large, hyaline, yeast-like cells. Identification at the species level can be clinically meaningful as long as the possibility of interference with therapy regimens of different degrees of melanization and the occurrence of chlamydospores or meristematic growth has not been excluded. Alternaria alternata and Alternaria infectoria are the most common species found in the clinical setting, but the lack of pigmentation has frequently led to the misidentification of isolates of these species. Molecular identification is a tool that PCR assays have successfully introduced into the armamentarium for the diagnosis of invasive fungal infections. Detection and analysis of the internal transcribed spacer (ITS) domain of the 5.8S ribosomal DNA (rDNA) region are currently proving to be a powerful tool for rapid and precise laboratory diagnoses (2), even if microscopic morphology and a number of additional tests are necessary to confirm the identification.
This paper reports on the usefulness of PCR and DNA typing to identify A. infectoria as the causative agent of diffuse skin infections in a cardiac transplant recipient.
Case report.
A 49-year-old agricultural worker from Verona, Italy, underwent cardiac transplantation for dilated cardiomyopathy. Ten months after the transplant he developed a nodule on his right arm, and several months later nodules also appeared on his left leg. The patient did not recall any local skin trauma occurring since the transplant. A punch biopsy specimen was taken from the leg lesion for histological examination. No dyspnea or fever was present. Routine laboratory tests revealed a leukocyte count of 5,700/mm3 with 86% neutrophils and a hemoglobin level of 127 g/liter. A chest X-ray was normal. The patient was admitted for further evaluation.
On physical examination the lesions were painless erythematous dermal nodules, slightly elevated, with a violet hue. Neither edema nor lymphadenopathy was present. Further physical examination was unremarkable. To exclude the possibility of a systemic infection total-body high-resolution computed tomography was done, with negative results. Bronchoalveolar lavage fluid samples were cultured for bacteria, mycobacteria, yeast, and mould. None of the cultures of bronchoalveolar lavage fluid samples revealed any growth. Histopathological examination of the cutaneous biopsy performed on the left leg showed a few neutrophilic microabscesses scattered in the dermis with a dense nodular granulomatous infiltrate of histiocytic cells along with a number of lymphocytes and plasma cells. Round, yeast-like structures and branched hyphae were found within the granulomas and microabscesses. Periodic acid-Schiff and Grocott's stains confirmed the presence of septate hyphae and spherical bodies.
A sample from the same nodule used for the histological diagnosis was cultured on Sabouraud's dextrose agar at 30 and 35°C. After a 7-day incubation, yellowish-white colonies with an olive-green undersurface developed. Microscopic examination of lactophenol cotton blue-stained smears showed melanized septate hyphae typical of dematiaceous fungi, but no conidia were detected. To enhance sporulation, subcultures on cornmeal agar and water agar were done. Rapid identification was attempted by sequencing the rDNA ITS domain as previously described (2).
Fungal DNA was purified by conventional methods (9) and amplified with primers ITS1 and ITS4 as described by White et al. (17).
The 600-bp amplicon (Fig. 1) was sequenced by AmpliTaq DNA polymerase FS dye terminator cycle sequencing chemistry with ABIPRISM (Perkin-Elmer Applied Biosystems Division, Foster City, Calif.) in accordance with the manufacturer's protocol.
FIG. 1.
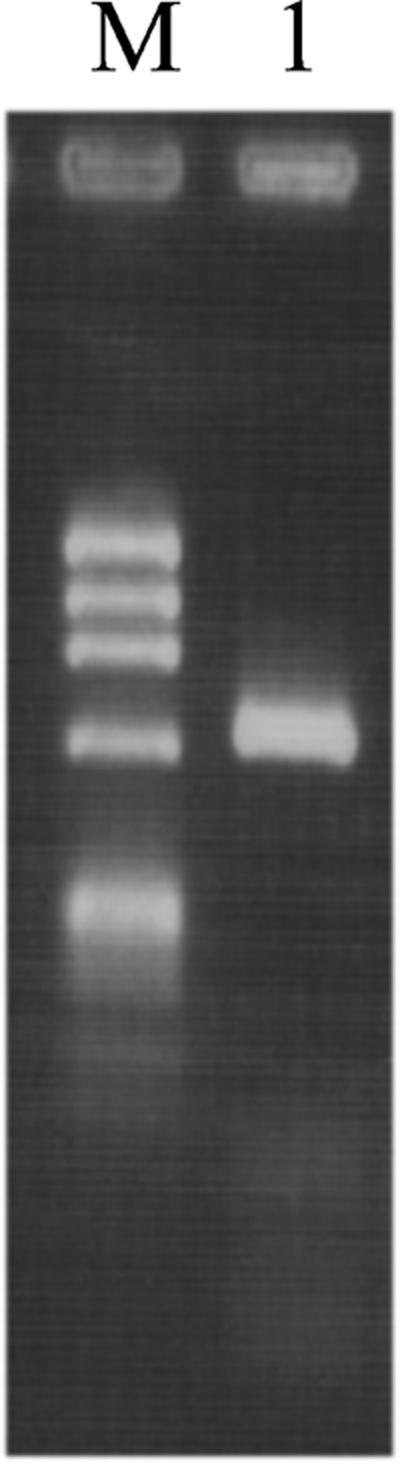
PCR amplification of Alternaria DNA. Lanes: M, molecular size marker consisting of HaeIII-digested X174 replicative-form DNA (1,353, 1,078, 872, 603, 310, 234, 194, 118, and 72 bp); 1, A. infectoria.
The sequence was aligned with sequences in the EMBL GenBank database with the Fasta 2.0 program and found to be 100% identical to that of Lewia infectoria (teleomorph of A. infectoria) (EMBL GenBank fungal library accession no. AY154688). To exclude any misidentification, the sequence of our clinical strain was compared with the sequences of six previously published strains of A. infectoria (accession no. AF229458, AF229480, AJ2760558, Y17066, and Y17067) (2). The homologies of the 5.8S rDNA ITS domain sequences were 99.4, 99.6, and 99.8%, respectively. The molecular identification was subsequently confirmed after repeated transfer on water agar; conidiophores were dark, septate, and mostly unbranched. Conidia were ovoid and smooth walled with one or more transverse and sometimes longitudinal septa. The conidia, often in chains, showed a short apical beak that in some conidia extends into a secondary conidiophore.
An antifungal susceptibility test was performed in accordance with the NCCLS M38-A reference method (13). The MICs were 0.125 mg/liter for amphotericin B, 64 mg/liter for fluconazole, 0.125 mg/liter for itraconazole, 0.5 mg/liter for ketoconazole, 0.5 mg/liter for voriconazole, and >64 mg/liter for flucytosine.
Intravenous amphotericin B (3 mg/kg/day) was administered for 14 days, followed by in-house treatment with 400 mg of itraconazole daily for 4 weeks. Given the patient's underlying immunosuppression, under continuous treatment with tacrolimus, attention to the possibility of interaction with itraconazole was recommended. The lesions healed completely, and after 15 months, no relapse has occurred.
More than 100 species belonging to at least 57 genera are known to be agents of phaeohyphomycosis (15). A review of the literature shows just four cases—before our report—of human infections caused by A. infectoria (4, 5, 7, 8), and in most of them PCR-based methods yielded a rapid, specific diagnosis. Our study strengthens the usefulness of the detection and sequence analysis of the ITS domain of the 5.8S rDNA region in the identification of dematiaceous mycetes.
Alternaria is a common fungus found in the environment. Although the genus comprises a large number of saprophytic and plant pathogenic species, only a few of these are implicated in human diseases, namely, A. alternata, A. infectoria, A. tenuissima, and A. chartarum (3). Most of the clinical presentations were localized skin infections resulting from direct, traumatic inoculation, even if systemic spread is possible in a compromised host. Phaeohyphomycosis caused by Alternaria may be difficult to recognize because lesions are variable in size and aspect, ranging from crusted lesions to erythematous macules or subcutaneous nodules. Histopathologically, Alternaria infection is sometimes misdiagnosed as a yeast infection (10) or blastomycosis, and distinguishing between species in vitro may be problematic because clinical isolates may remain degenerate and sterile (3). Additionally, differentiation to the species level is recommended because species may differ in virulence or resistance to antimycotic therapy because of the occurrence of chlamydospores (3, 14). Moreover, blastomycosis, sporotrichosis, cryptococcosis, and other subcutaneous mycoses that may present similar clinical features require different therapeutic approaches.
In the patient described here, the diagnosis of phaeohyphomycosis was confirmed by both molecular and cultural evidence. The presence of two lesions at noncontiguous body sites (the right arm and left leg) suggests a possible disseminated disease, which was disproven by further investigations such as chest X-ray and high-resolution computed tomography.
Alternaria skin infections have been successfully treated with itraconazole (1), amphotericin B, ketoconazole, and fluconazole, even if data on susceptibility to antifungals are limited and variable (3, 11, 14, 16). Furthermore, no data on the susceptibility of A. infectoria to antifungals have been published. For our isolate, the MICs of amphotericin B, ketoconazole, itraconazole, fluconazole, and voriconazole can be considered to be equivalent to those published for Alternaria spp. and A. alternata (3, 11, 16), suggesting a therapeutic approach with amphotericin B and itraconazole although no breakpoint has been recommended by the NCCLS or the European Committee on Antimicrobial Susceptibility Testing for antifungals and filamentous fungi. The duration of treatment is controversial because relapses may occur after clinical resolution (12). The patient described here was successfully treated with amphotericin B (250 mg daily) for 14 days, followed by itraconazole (400 mg daily) for 1 month, with no surgical intervention.
Today, in the era of modern medicine, critical use of molecular methods is essential in a microbiology laboratory. Application of DNA typing in this case yielded a rapid diagnosis and resulted in the administration of specific, effective therapy. Moreover, it confirmed the role of A. infectoria in the differential diagnosis of phaeohyphomycosis, especially in transplant patients.
REFERENCES
- 1.Acland, K. M., R. J. Hay, and R. Groves. 1998. Cutaneous infection with Alternaria alternata complicating immunosuppression: successful treatment with itraconazole. Br. J. Dermatol. 138:354-356. [DOI] [PubMed] [Google Scholar]
- 2.De Hoog, G. S., and R. Horré. 2001. Molecular taxonomy of the Alternaria and Ulocladium species from humans and their identification in the routine laboratory. Mycoses 45:259-276. [DOI] [PubMed] [Google Scholar]
- 3.De Hoog, R. D., and J. Guarro. 1995. Atlas of clinical fungi. Centraalbureau voor Schimmelcultures, Baarn, The Netherlands.
- 4.Ferrer, C., J. Montero, J. L. Aliò, J. L. Abad, J. M. Ruiz-Moreno, and F. Colom. 2003. Rapid molecular diagnosis of posttraumatic keratitis and endophthalmitis caused by Alternaria infectoria. J. Clin. Microbiol. 41:3358-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdsen, R., M. Uerlich, G. S. de Hoog, T. Bieber, and R. Horré. 2001. Sporotrichoid phaeohyphomycosis due to Alternaria infectoria. Br. J. Dermatol. 145:484-486. [DOI] [PubMed] [Google Scholar]
- 6.Gilmour, T. K., E. Rytina, P. B. O'Connell, and J. C. Sterling. 2001. Cutaneous alternariosis in a cardiac transplant recipient. Aust. J. Dermatol. 42:46-49. [DOI] [PubMed] [Google Scholar]
- 7.Halaby, T., H. Boots, A. Vermeulen, A. van der Ven, H. Beguin, H. van Hooff, and J. Jacobs. 2001. Phaeohyphomycosis caused by Alternaria infectoria in a renal transplant recipient. J. Clin. Microbiol. 39:1952-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laumaillé, C., F. Le Gall, B. Degeilh, E. Gueho, and M. Huerre. 1998. Cutaneous Alternaria infectoria infection after liver transplantation. Ann. Pathol. 18:192-194. [PubMed] [Google Scholar]
- 9.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 10.Mayser, P., M. Nilles, and G. S. de Hoog. 2002. Case report. Cutaneous phaeohyphomycosis due to Alternaria alternata. Mycoses 45:338-340. [DOI] [PubMed] [Google Scholar]
- 11.McGinnis, M. R., and L. Pasarell. 1998. In vitro testing of susceptibilities of filamentous ascomycetes to voriconazole, itraconazole, and amphotericin B, with consideration of phylogenetic implications. J. Clin. Microbiol. 36:2353-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merino, E., J. Banulus, V. Boix, A. Franco, J. Guijarro, J. Portilla, and I. Betlloch. 2003. Relapsing cutaneous alternariosis in a kidney transplant recipient cured with liposomal amphotericin B. Eur. J. Clin. Microbiol. Infect. Dis. 22:51-53. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi: approved standard. Document M38-A. National Committee for Clinical Laboratory Standards., Wayne, Pa.
- 14.Pujol, I., C. Aguilar, J. Gené, and J. Guarro. 2000. In vitro antifungal susceptibility of Alternaria spp. and Ulocladium spp. J. Antimicrob. Chemother. 46:323-342. [DOI] [PubMed] [Google Scholar]
- 15.Rinaldi, M. G. 1996. Phaeohyphomycosis. Dermatol. Clin. 14:147-153. [DOI] [PubMed] [Google Scholar]
- 16.Sutton, D. A., A. W. Fothergill, and M. G. Rinaldi. 1998. Guide to clinically significant fungi. Williams & Wilkins, Baltimore, Md.
- 17.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In N. Innis, J. Gelfand, and T. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, N.Y.


