Abstract
Progressive loss of lung function resulting from the inflammatory response to bacterial colonization is the leading cause of mortality in cystic fibrosis (CF) patients. A greater understanding of these bacterial infections is needed to improve lung disease management. As culture-based diagnoses are associated with fundamental drawbacks, we used terminal restriction fragment (T-RF) length polymorphism profiling and 16S rRNA clone data to characterize, without prior cultivation, the bacterial community in 71 sputa from 34 adult CF patients. Nineteen species from 15 genera were identified in 53 16S rRNA clones from three patients. Of these, 15 species have not previously been reported in CF lung infections and many were species requiring strict anaerobic conditions for growth. The species richness and evenness were determined from the T-RF length and volume for the 71 profiles. Species richness was on average 13.3 ± 7.9 per sample and 13.4 ± 6.7 per patient. On average, the T-RF bands of the lowest and highest volumes represented 0.6 and 59.2% of the total volume in each profile, respectively. The second through fifth most dominant T-RF bands represented 15.3, 7.5, 4.7, and 2.8% of the total profile volume, respectively. On average, the remaining T-RF bands represented 10.2% of the total profile volume. The T-RF band corresponding to Pseudomonas aeruginosa had the highest volume in 61.1% of the samples. However, 18 other T-RF band lengths were dominant in at least one sample. In conclusion, this reveals the enormous complexity of bacteria within the CF lung. Although their significance is yet to be determined, these findings alter our perception of CF lung infections.
The average life expectancy of patients with cystic fibrosis (CF) has increased greatly over the past 50 years (20), with the introduction of antibiotic therapy to treat bacterial lung infections being arguably the most important factor in increasing longevity. Despite these advances, progressive loss of lung function remains the cause of mortality in 90% of CF patients (10, 23, 27, 31). The loss of function in the CF lung stems from damage caused by the intense host inflammatory response to bacteria that colonize the lung over the lifetime of the patient (22, 31). Since patients continue to deteriorate despite seemingly appropriate antibiotic therapy, it is important to determine the nature of the interactions between bacteria and the CF lung. Such understanding will facilitate improvements in therapeutic strategies. The first step in this process is, therefore, to determine accurately what bacteria are present at a given time point in the CF lung.
In traditional diagnostic microbiology, the characterization of the bacteria present within clinical samples requires the prior culture of bacterial species on selective media (37). However, the use of selective media and growth conditions requires assumptions to be made regarding which species will be present. Therefore, CF sputa are assayed routinely for only a limited number of bacterial species considered important, including Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, and Burkholderia cepacia complex (16). All such traditional microbiological techniques are subject to culture bias. Culture media are capable of supporting the in vitro growth of only a small fraction of the total range of bacterial species present (13). The implication of this is that ecological inferences based on the phenotypic characteristics of cultivated bacteria are, by their nature, unrepresentative of the natural populations from which they were obtained (33). In addition, a significant period of time is required for such traditional culture-based approaches to yield data.
Recently, terminal restriction fragment length polymorphism (T-RFLP) profiling has been used to study the bacterial diversity within clinical specimens provided by a relatively small number (n = 14) of CF patients (34). While the use of T-RFLP to characterize the diversity of bacterial communities from many environments has become common (7, 24, 34, 38), it has not been applied widely to the study of human infections. In T-RFLP, nucleic acids are extracted directly from clinical samples. Phylogenetically informative rRNA gene PCR products, specific for the domain Bacteria, are amplified from these nucleic acid extracts. Following digestion with a specific restriction endonuclease, ribosomal gene fragments are separated by gel electrophoresis, forming a T-RFLP profile of the diversity of the bacterial community within the original sample in a single electrophoretic lane. Studies have previously shown T-RFLP to be both reproducible and effective at characterizing bacterial diversity in CF sputum and bronchoalveolar lavage samples (34). It is important to note that expectorated sputum provides an accurate measure of infection and inflammation in the lower airways (15, 36), with insignificant levels of oropharyngeal contamination (15).
Host immune response and antibiotic therapies are among many factors that are important in determining the characteristics of CF lung infections. However, we hypothesize that it is also important to understand bacterial interactions and, more generally, the bacterial ecology of the CF lung. Two key measures used to describe the complexity of communities are species richness and species evenness. Species richness is a measure of the number of separate species present in a community (28). Levels of species richness are influenced both by the number of bacterial species present and by the frequency at which those species are retained. Species evenness describes the relative sizes of species populations within a community (29). As such, while the level of variation in the number of different bacterial species present in samples taken from a group of CF patients is potentially relatively small, the proportion that these different species represent within the community may vary significantly from patient to patient. Therefore, to understand species diversity, the contribution of both species richness and species evenness must be considered (25).
The central aim of this investigation was to determine the composition and diversity of the bacterial communities colonizing the CF lung. This was achieved through the use of T-RFLP analysis, in conjunction with 16S ribosomal DNA (rDNA) clone sequence analysis. T-RFLP profile analysis enabled comparisons of bacterial community composition, richness, and evenness to be made between CF patients.
MATERIALS AND METHODS
Clinical samples and DNA extraction.
Sputum samples were obtained from adult CF patients attending the regional Cystic Fibrosis Centre at Southampton General Hospital. The patient group consisted of 34 individuals, who provided between 1 and 5 sputum samples between 14 September 2001 and 16 July 2002. Over this period of time, 24 patients suffered a period of pulmonary exacerbation and 10 patients remained clinically stable. A variety of oral, intravenous, and nebulized antibiotics were administered to these patients as dictated by the clinicians involved in this study. A total of 71 separate CF sputum samples were collected and processed immediately.
Samples were washed three times in phosphate-buffered saline (Oxoid) prior to DNA extraction. Samples were then treated with Sputasol (Oxoid) in accordance with the manufacturer's instructions, followed by centrifugation for 5 min at 12,000 × g. Pellets were resuspended in 1.5 ml of phosphate-buffered saline and then centrifuged for 5 min at 12,000 × g. This step was repeated three times.
DNA was isolated from clinical specimens by a modification of a procedure described previously (6). Approximately 0.2 ml of each clinical sample was resuspended in 800 μl of 200 mM sodium phosphate buffer (pH 8.0) and 100 μl of guanidinium thiocyanate-EDTA-Sarkosyl. Then, 0.2 g of 0.18-mm-diameter glass beads (B. Braun Biotech International GmbH, Melsungen, Germany) was added, and homogenization was performed for 30 s at 30 Hz in a Mixer Mill 300 (Qiagen, Crawley, United Kingdom). Samples were heated to 70°C for 20 min and then placed on ice for 20 min, and beads and other debris were pelleted by centrifugation at 12,000 × g for 5 min at room temperature. The supernatant was transferred to a fresh microcentrifuge tube, and NaCl (to a final concentration of 0.5 M) and polyethylene glycol (to a final concentration of 15%) were added. Samples were left to precipitate at 4°C for 1 h. DNA was pelleted by centrifugation at 12,000 × g for 10 min at room temperature, and the pellet was resuspended in 300 μl of sterile distilled water. Next, 0.3 ml of Tris-buffered phenol (pH 8.0) was added to each sample before the tubes were vortexed vigorously. After centrifugation at 12,000 × g for 5 min, supernatants were transferred to fresh microcentrifuge tubes. A further 0.3 ml of Tris-buffered phenol (pH 8.0)-chloroform-isoamyl alcohol (25:24:1) was added, and the mixture was vortexed vigorously. After centrifugation at 12,000 × g for 10 min, supernatants were precipitated by using an equal volume of isopropanol and a 1/10 volume of 10 M ammonium acetate for 1 h at −20°C. The pellets formed by centrifugation at 12,000 × g for 10 min at room temperature were washed three times in 70% ethanol. After being dried, pellets were resuspended in 100 μl of sterile distilled water and stored at −20°C. The extracted DNA was verified by Tris-acetate-EDTA-agarose gel electrophoresis.
DNA quantification.
Extracted genomic DNA and restricted PCR products were quantified by use of a CytoFluor series 4000 multiwell plate reader (PerSeptive Biosystems, Foster City, Calif.) and by using the PicoGreen DS DNA quantitation kit (Molecular Probes, Leiden, The Netherlands) according to the manufacturer's instructions.
PCR amplification and restriction endonuclease digestion.
The oligonucleotide primers used to amplify a region of the 16S rRNA gene for members of the domain Bacteria, 8f700 and 926r (5′-CCG TCA ATT CAT TTG AGT TT-3′), were described previously (24). Primer 8f700 was labeled at the 5′ end with IRD700; primer 926r was unlabeled. PCR mixtures comprised 1× PCR buffer, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, each primer at a concentration of 0.2 mM, and 1 U of REDTaq DNA polymerase (Sigma-Aldrich) in a final volume of 50 μl. An initial denaturation step of 94°C for 2 min was followed by 32 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 2 min, with a final extension step at 72°C for 10 min. Amplification was carried out by using a GeneAmp PCR System 2400 (Perkin-Elmer), with PCR products for T-RFLP analysis stored at −20°C after verification on Tris-acetate-EDTA-agarose gels as described above.
PCR products (ca. 20 ng) were digested by using the restriction endonuclease CfoI (Roche, Lewes, United Kingdom) for 3 h at 37°C with the reaction buffer supplied by the manufacturer. All restriction endonuclease digestions were carried out to complete digestion as shown by comparing PCR products after various digestion incubation times (data not shown). The restriction endonuclease was inactivated by heating at 90°C for 20 min. An approximately 0.7-μg portion of T-RFLP PCR products was separated by length by using a 25-cm SequagelXR denaturing acrylamide gel (National Diagnostics) prepared in accordance with the manufacturer's instructions with the addition of 8.3 M urea and 10% (final concentration) formamide and by using an IR2 automated DNA sequencer (LI-COR Biosciences) at 55°C and 1,200 V.
T-RFLP profile analysis.
Gels were analyzed by using Phoretix one-dimensional advanced software, version 5.10 (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom). The sizes of the bands resolved by T-RFLP were determined by comparing their relative positions with two sets of size markers, one set that formed bands at 75, 100, 150, 200, 250, 300, 350, 400, 450, 500, 600, 700, 800, 900, and 1,000 bases (microSTEP 15a [700 nm]) and one set that formed bands at 155, 209, 214, 238, and 364 bases (microSTEP custom GR [700 nm]). Both sets of size markers were obtained from Microzone (Lewes, United Kingdom). In addition, this software was also used to determine the volume of each band (with band volume being the product of the area over which a band was detected and the intensity of signal recorded over that area). Band volume was expressed as a percentage of the total volume of bands detected in a given electrophoretic profile. T-RFLP bands were resolved over the region between 50 and 958 bases. No bands shorter than 50 bases in length were recorded, as they were in the region susceptible to high levels of signal stemming from the IR tag on either unattached or nonused primer 8f700IR. In this study, the threshold used to detect bands was 0.01% of the total signal between the 50- and 958-base region.
In silico T-RFLP band size determination.
Published bacterial 16S rRNA gene sequence data stored at GenBank (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db_Nucleotide) were retrieved. MapSort (Wisconsin Package, version 10.3; Accelrys) was used to predict T-RFLP band sizes by locating the position of a specific restriction endonuclease recognition motif, here CfoI, in a given sequence. T-RFLP band lengths (in bases) were therefore obtained for the region from the 5′ end of primer 8f700IR to the first cleavage position of the restriction endonuclease CfoI in each 16S rRNA gene sequence. A total of 853 sequences were analyzed in this way with the data stored at http://www.kcl.ac.uk/kis/schools/life_sciences/life_sci/TRFLP.html. Of the 853 sequences analyzed, approximately 10% matched an empirically derived T-RF band length (Table 1).
TABLE 1.
Detection frequencies and species correlations of resolved T-RF bandsa
| T-RF band length (bases) | % of patients with band | Bacterial species assignation |
|---|---|---|
| 59 | 62 | Craurococcus roseus |
| 61 | 44 | Rhizobium loti, Ochrobactrum anthropi, Peptostreptococcus anaerobius |
| 68 | 21 | Ralstonia gilardii |
| 92 | 9 | Bilophila wadsworthia |
| 93 | 3 | Desulfovibrio desulfuricans, Tistrella mobilis |
| 96 | 3 | Wolinella succinogenes, Campylobacter rectus, Bacteroides forsythus |
| 98 | 6 | Bacteroides gracilis |
| 102 | 57 | Prevotella loescheii, P. salivae, P. buccae, P. oris, Porphyromonas gingivalis |
| 104 | 71 | Porphyromonas endodontalis, Prevotella denticola, P. melaninogenica, P. nigrescens, P. veroralis, P. intermedia |
| 155 | 88 | Pseudomonas aeruginosa |
| 197 | 3 | Acinetobacter junii |
| 201 | 9 | Mycobacterium tuberculosis |
| 207 | 15 | Acinetobacter johnsonii |
| 208 | 3 | Pandoraea pulmonicola |
| 209 | 15 | Burkholderia cepacia complexb |
| 210 | 9 | Burkholderia gladioli |
| 211 | 15 | Abiotrophia paraadiacens |
| 213 | 12 | Actinomyces viscosus, A. naeslundii, Xanthomonas campestris, Morganella morganii, Neisseria species (N. meningitidis, N. gonorrhoeae), Xanthomonas hyacinthi, Pseudomonas aureofaciens, Legionella pneumophila |
| 214 | 9 | Stenotrophomonas maltophilia, Fusobacterium gonidiaformans |
| 215 | 6 | Aeromonas caviae, Abiotrophia defectiva |
| 225 | 6 | Flavobacterium indologenes |
| 238 | 18 | Staphylococcus aureus, S. epidermidis, S. cohnii, S. hominis |
| 256 | 3 | Lactobacillus murinus |
| 266 | 6 | Staphylococcus sciuri |
| 279 | 3 | Rhizobium radiobacter |
| 339 | 3 | Methylobacterium radiotolerans |
| 363 | 6 | Mycobacterium chelonae |
| 364 | 9 | Haemophilus paraphrophilus, H. influenzae, Actinobacillus actinomycetemcomitans |
| 367 | 3 | Mycobacterium flavescens |
| 372 | 15 | Salmonella enterica serovar typhimurium, Bifidobacterium pseudocatenulatum, Proteus mirabilis |
| 445 | 3 | Bordetella hinzii |
| 562 | 6 | Eubacterium saburreum |
| 563 | 6 | Halomonas variabilis |
| 566 | 18 | Comamonas testosteroni |
| 569 | 26 | Paracoccus halodenitrificans |
| 571 | 3 | Pseudomonas huttiensis |
| 574 | 35 | Ralstonia pickettii, Oligella urethralis |
| 576 | 6 | Streptococcus pneumoniae, S. salivarius, S. pyogenes, S. macedonicus, S. sanguinis |
| 585 | 32 | Selenomonas sputigena, Streptococcus intermedius |
| 587 | 6 | Peptococcus-like species oral clone JM048 |
| 588 | 9 | Veillonella parvula |
| 589 | 3 | Capnocytophaga sputigena |
| 590 | 3 | Streptococcus anginosus, Veillonella atypica, V. ratti |
| 843 | 3 | Alcaligenes xylosoxidans |
| 846 | 9 | Treponema pallidum |
T-RF bands were detected in the sample set, and corresponding species band sizes were predicted (as determined by software-driven analysis of published sequence data). The percentages of patients in the sample set in which each band was detected and the names of the species with which they are consistent are listed.
Sequences from genomovars B. cepacia, B. multivorans, B. stabilis, B. vietnamensis, B. ambifaria, and B. anthina of the B. cepacia complex were all subjected to in silico analysis. However, there was insufficient sequence data to derive in silico T-RFLP band size lengths for genomovars B. dolosa, B. pyrrocinia, B. ubonensisa, or B. cenocepacia.
Preparation and analysis of 16S rDNA clones libraries.
PCR products for 16S rRNA clone analysis were amplified with the universal bacterial primers 8f700 (as described above) and 338r (5′-GCT GCC TCC CGT AGG AGT-3′) (32) obtained from MWG-Biotech (Milton Keynes, United Kingdom). The PCR mixture composition, amplification program parameters, and equipment were all as described above. Samples of the PCR products generated were cloned with a pGEM-T Easy Vector system (Promega, Southampton, United Kingdom) according to the manufacturer's instructions. DNA was extracted from clones as follows: a single white colony was resuspended in 200 μl of sterile distilled water and then boiled for 10 min, with the cell debris pelleted by centrifugation at 12,000 × g for 5 min. Then, 15 μl of the supernatant was added to a 50-μl (final volume) PCR mixture, comprising 1× PCR buffer, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, each primer at a concentration of 0.2 μM, and 1 U of REDTaq DNA polymerase (Sigma-Aldrich) in a final volume of 50 μl. Amplification was carried out with the primers gemsp6 (5′-GCT GCG ACT TCA CTA GTG AT-3′) and gemt7 (5′-GTG GCA GCG GGA ATT CGA T-3′) as previously described (34). The PCR mixture composition, amplification program parameters, and equipment were all as described above.
The sequencing of individual clones was carried out by the Genetics Core Facility, Hammersmith Hospital, London, United Kingdom. Sequences were analyzed by BLAST (18) and by using the Ribosomal Database Project II (19). BLAST matches described below all had an e-value greater than e−142. Novel sequence types obtained in this study were stored in GenBank under accession numbers AJ555128 to AJ555147.
RESULTS
Species identification and relative prevalence.
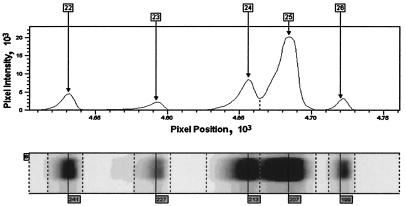
T-RFLP profiles were generated for each of the 71 sputum samples studied. Figure 1 shows a region of the electrophoretic output of a typical lane and the band size data contained within it being extracted. By such comparisons, a total of 248 distinct T-RF band sizes were detected over the sizeable range of the T-RFLP gel.
FIG. 1.
Gel image and band extraction. The figure shows both the trace from a region of an electrophoretic lane that forms the basis of the band assignment and signal intensity measurement process (upper panel) and the corresponding image from which this trace is derived as it appears on the automated sequencer output (lower panel).
The frequency at which each of these 248 different T-RF bands was detected in the sample set was determined. Detection frequencies ranged from 1.4% (representing a T-RF band that was present in a single sample) to 87.5% of samples (the T-RF band of 155 bases in length). The frequency at which the T-RF bands were detected in each individual patient was also determined. Little difference was found between these detection frequencies and those for the sample set as a whole, with detection rates ranging from 3% (present in a single patient) to 88%. However, certain T-RF band sizes were confined to a relatively small subset of the patient group. For example, the T-RF band 64 bases in length was detected in 32% of samples overall but in only 17% of patients (data not shown).
Where possible, individual T-RF bands were assigned to bacterial species by comparison of band size with in silico band size predictions made for the range of bacteria, as described in Materials and Methods. Through this process, 45 (18%) of the 248 different T-RF band sizes resolved in this set of patients were found to match the band length predicted for one or more bacterial species in silico (Table 1). Of these 45 matches, 29 corresponded to a single bacterial species from our in silico data set. Sixteen T-RF band sizes corresponded to several species belonging to more than one genus, and four corresponded to more than one species from within a single genus (Table 1). It should be noted that the resolution of a band of the length predicted by in silico analysis for a particular bacterial species does not indicate the presence of that species but is merely consistent with the presence of that species.
Species prevalence and identification—16S rRNA gene clone sequence analysis.
Samples from three patients were selected at random for sequence analysis of 16S rRNA gene PCR product clone libraries. Clone libraries of 16S rRNA regions amplified from nucleic acids extracted directly from sputum samples from these patients were generated. A total of 53 cloned 16S rRNA gene regions were sequenced (20 clones from each of two patients and 13 clones from the third patient). 16S rRNA gene sequences generated here were compared to published 16S rRNA sequence data to enable bacterial species identification. A total of 19 bacterial species were identified within these 53 cloned 16S rRNA regions (Table 2).
TABLE 2.
Bacterial species identified in the sample set through 16S rDNA clone sequence analysisa
| Bacterial species | No. of clones detected in patient:
|
Detected by T-RFLP in:
|
Detected previously in CF | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | Sample cohort | Same sample | ||
| Abiotrophia adiacens | 1 | + | + | |||
| Abiotrophia paraadiacens | 1 | + | ||||
| Bacteroides gracilis | 1 | + | + | |||
| Burkholderia cepacia complex | 2 | + | + | |||
| Eubacterium brachy | 1 | |||||
| Lactobacillus crispatus/L. gasseri | 2 | + | ||||
| Mycoplasma salivarium | 2 | |||||
| Porphyromonas sp. | 1 | + | + | |||
| Prevotella melaninogenica | 1 | 4 | + | |||
| Prevotella salivae | 1 | + | + | |||
| Prevotella sp. oral clone | 2 | + | + | |||
| Pseudomonas aeruginosa | 1 | + | + | + | ||
| Ralstonia taiwanensis | 1 | |||||
| Rothia mucilaginosa | 3 | |||||
| Staphylococcus hominis | 1 | + | + | |||
| Streptococcus anginosus | 18 | + | ||||
| Streptococcus pneumoniae/mitis | 3 | 4 | + | +/− | + | |
| Treponema vincentii | 2 | |||||
| Veillonella atypica | 1 | + | ||||
Bacterial species detected by 16S rDNA clone analysis. The number of clones of each species detected is shown. All of the identifications were made based on matches to published sequence data (see Materials and Methods).
Table 2 shows the bacterial species that provided the closest matches, in terms of sequence similarity, to database entries. The 19 different bacterial species identified were held within 15 different genera. The phylogenetic diversity of these cloned sequences was wide, with genus members within the Bacteroidetes/Chlorobi group (genera Bacteroides, Porphyromonas, and Prevotella), the Proteobacteria (genera Burkholderia β, Ralstonia β, and Pseudomonas γ), the Firmicutes (genera Abiotrophia, Eubacterium, Lactobacillus, Mycoplasma, Staphylococcus, Streptococcus, and Veillonella), the Actinobacteria (genus Mycobacterium), and the Spirochaetes (genus Treponema).
Species richness and evenness. (i) Richness.
The number of separate T-RF bands resolved in the samples studied ranged from 2 to 37. A mean of 13.3 (±7.9 [standard deviation]) distinct T-RF bands were detected for each clinical sample analyzed. Twenty of the 34 patients provided samples on more than one occasion. The mean number of T-RF bands detected in samples from each patient was 13.4 (±6.7). On average, the interval between the highest and lowest levels of species richness in samples provided by the same person was 9.9 (±8.0).
(ii) Evenness.
Band volume refers to the area over which each T-RF band was detected (above an automatically determined background level) multiplied by the intensity of the signal at every given point within it. The volume of every band detected in the T-RFLP profiles was determined and expressed as a percentage of the total volume of a given profile (data not shown). On average, the smallest T-RF band volume detected in a profile was 0.6%, although in one instance it was as low as 0.012%.
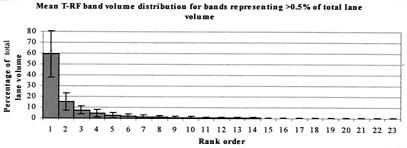
The bands detected in each profile were placed in rank order of descending volume. This order of descending band volume was referred to as the order of dominance. Mean volumes for bands occupying each position in the order were then determined for the sample set as a whole. These mean volumes are represented in Fig. 2. It was found that the volume of the most dominant band was, on average, approximately four times as great as that of the second most dominant band (59.2 and 15.3%, respectively). The second most dominant band was found to be approximately twice the volume of the third (15.3% compared with 7.5%, respectively), with the fourth and fifth most dominant T-RF mean band volumes representing 4.7 and 2.8%, respectively. The majority of T-RF bands represented less than 1% of the total band volume. Outside of the five most dominant T-RF bands, the subtotal of mean band volumes together represented only 10.2% of the overall lane volume.
FIG. 2.
Graph of mean percentage of total band volume represented by rank-ordered T-RF bands.
The number of times a T-RF band occupies the dominant, second, third, fourth, or fifth position in a profile was determined (data not shown). Further, the percentage of samples in which the band was detected at this position in the dominance hierarchy and the percentage of samples in which it was detected were determined. The T-RF band size of 155 bases, corresponding to that predicted for P. aeruginosa, had the highest band volume in the majority of samples (61.1%). However, when not the dominant T-RF band, the 155-base fragment was relatively rarely detected in one of the other top five ranked positions (Table 3). In total, 18 non-155 T-RF band sizes were the highest-volume band in at least one of the 71 profiles examined. A total of 79 different T-RF band sizes occupied one of the five highest-volume bands in at least one profile.
TABLE 3.
Dominance of key CF pathogens in profiles generated from data seta
| Species | % of samples in which species had dominance rank of:
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| P. aeruginosa | 61.1 | 0 | 1.4 | 1.0 | 0 |
| S. aureus | 5.6 | 1.4 | 1.4 | 0 | 1.0 |
| B. cepacia complex | 1.4 | 1.4 | 1.4 | 0 | 2.0 |
| S. maltophilia | 4.2 | 0 | 2.8 | 0 | 0 |
| H. influenzae | 1.4 | 7.0 | 1.4 | 3.0 | 2.0 |
| Other | 26.3 | 90.2 | 91.6 | 96 | 95 |
Percentages of samples in which a T-RF band consistent with each of the five key recognized CF pathogens was detected at a given point in the dominance hierarchy. In addition, the percentage of samples where a T-RF band of a length not consistent with any of these species occupies a top five dominance position is shown.
The dominance of T-RF band sizes corresponding to four other bacterial species (B. cepacia complex at 209 bases, Stenotrophomonas maltophilia at 214 bases, S. aureus at 238 bases, and H. influenzae at 364 bases) traditionally considered to be key CF pathogens were also assessed (Table 3). It should be noted that, in the case of S. aureus and the B. cepacia complex, several species from the same genus produce T-RF bands of the same length, and in the case of H. influenzae and S. maltophilia, species from other genera produce identical band lengths (34). Therefore, it is possible that the dominance attributed to these species may be contributed to by other, in some cases unrelated, species. Further, in the case of B. cepacia complex, the species B. cepacia, Burkholderia multivorans, Burkholderia stabilis, Burkholderia vietnamensis, Burkholderia ambifaria, and Burkholderia anthina all have identical in silico T-RFLP band sizes (209 bases); however, there is currently insufficient sequence data to derive in silico T-RFLP band size lengths for the species Burkholderia dolosa, Burkholderia pyrrocinia, Burkholderia ubonensisa, or Burkholderia cenocepacia.
No T-RF band representing a non-key species was detected in any top five dominance position in more than 8.3% of the profiles. On average, such bands were detected in 2.3% of the profiles. A number of species were present in a large proportion of profiles but only as bands of low relative volume. These included bands of 585, 268, 203, and 580 bases in length detected in 33, 30, 27, and 27% of patients, respectively. The assignment of a bacterial species on the basis of in silico length prediction was possible in the case of only one of these band sizes, namely the T-RF band of 585 bases in size to either Streptococcus intermedius or Selenomonas sputigena.
DISCUSSION
The findings of this study change our understanding of the diversity of bacteria present in the respiratory tract of CF patients. This knowledge is crucial if the management of respiratory infections, a major determinant of prognosis, is to be improved. Two related approaches were used to characterize the diversity of the bacteria in sputum samples from a group of 34 patients attending a regional adult CF clinic in Southampton, England. Sequence analysis of cloned 16S rRNA gene PCR products enabled the identification of bacteria from a subset of these patients. However, such analysis of clone library data allows the characterization of only a very small fraction of the bacteria present in such samples. T-RFLP profiling was therefore used to examine the diversity of the dominant bacterial community members in each of these CF patients. The resulting data provided information on the levels of bacterial species richness and evenness in these samples.
Prior to this study, there was little appreciation of the number of bacterial species present in CF sputum. This study has shown that, typically, individual CF patients were colonized by many different species. These species spanned a wide range of bacterial growth strategies with respect to oxygen, with aerobes, facultative anaerobes, and obligate anaerobes all detected in this group of patients. Further, many of the species identified here have not previously been reported in CF lung infections. These findings are highly significant because they reveal the complexity of CF lung bacterial ecology. Moreover, they demonstrate the inadequacy of community analysis carried out with traditional culture-based techniques. This study highlights a gap in our understanding of the mechanisms involved in CF lung infections and underlines the importance of the application of molecular microbial ecology to the analysis of complex bacterial infections in general.
Sputum samples from CF patients were carefully washed to remove saliva and bacteria that adhered to the bolus during its passage through the upper airways and mouth. Despite these careful attempts, it is important to acknowledge the possibility that oropharyngeal contamination could contribute to the diversity of the species identified in this study. Certain species were detected that are associated typically as flora found in the oral cavity or upper respiratory tract. However, these T-RF bands may represent species that have genuinely colonized the CF lung, as there is both a very low level of saliva carryover from CF sputa and a high bacterial load in the sputum itself. The analysis of cloned 16S rRNA sequences is an important tool in identifying the bacterial species present in a given sample. Data derived through this process confirmed the presence of five bacterial species whose corresponding T-RF bands were detected in the sample group (B. cepacia complex, B. multivorans, Rhodotorula mucilaginosa, P. aeruginosa, Streptococcus pneumoniae, and Streptococcus mitis). In addition, clone analysis identified 14 species not previously reported in CF lung infections (Abiotrophia adiacens, Abiotrophia paraadiacens, Bacteroides gracilis, Eubacterium brachy, Mycobacterium mucilaginosus, Mycoplasma salivarium, Porphyromonas salivae, Prevotella melaninogenica, Prevotella sp. oral clone, Ralstonia taiwanensis, Staphylococcus hominis, Streptococcus anginosus, Treponema vincentii, and Veillonella atypica) (Table 2). This group included five facultative anaerobic species (A. adiacens, A. paraadiacens, R. taiwanensis, S. hominis, and S. anginosus) and eight obligate anaerobic species (B. gracilis, E. brachy, M. salivarium, Porphyromonas sp., P. salivae, P. melaninogenica, Prevotella sp. oral clone, and V. atypica). The presence of these species is consistent with previous studies that found that the CF lung has anaerobic regions (41). This is potentially clinically relevant, as anaerobic species have only recently been implicated in CF lung infections and are not targeted by many current antibacterial treatments.
Comparison of the species detected in this sample set with those previously detected in 103 clones obtained from five adult CF patients (34) reveals both similarities and differences between the two sets of patients. Although only one of the 29 species detected in the two studies was found in patients within both cohorts (P. aeruginosa), in both cases, relatively large proportions of the species detected were anaerobes or species not previously reported in CF lung infections (Table 2). It is important to note that bias can be introduced at the cloning stage of this process and can have a significant impact on the data derived (40). In addition, analysis of clone libraries is a relatively inefficient and expensive process that typically requires the examination of large numbers of clones to obtain detailed ecological information from a sample. Moreover, it is highly labor intensive and, as such, is unsuitable for the analysis of a set of samples taken from a large patient group.
T-RFLP profiling, however, is well suited to the analysis of large groups of samples. In this study, attempts were made to link T-RF band lengths with particular bacterial species on the basis of predictions made by using published sequence data. This process, described previously (34), encompassed more than 160 bacterial species associated with a wide range of environments. In the process of assigning T-RFs to particular bacterial species, it should be noted that the quality of the sequence data used influences the accuracy of the linkage made. As more sequence data become available, however, such linkages will become more robust. Through this analysis, around 20% of the T-RF band lengths resolved in the sample set were assigned to at least one bacterial species. As this process affords no direct verification of species assignation, these data were compared to the 16S rDNA clone analysis data. This is an iterative process, and the proportion of T-RF bands that have corresponding species predictions will increase as further clone analysis of CF sputum samples is performed.
The mean species richness for the samples group was 13.3 (±7.9). To put this in context, soil communities typically have a minimum of 4,000 to 7,000 different bacterial genomes per gram (39). Also at the high end of reported species richness levels are certain areas of the human body. For example, the gut is estimated to contain between 400 and 500 different bacterial species (26). In the gastrointestinal tract, the presence of this diverse array of species is not typically associated with deleterious health effects to the host. Chronic sinusitis is an example of an infectious disease that results from the presence of multiple bacterial species. Culture-based studies have shown that such infections typically involve around three separate bacterial species (5). At the other end of the spectrum of species richness, many types of bacterial infection involve a single species (4, 14). Therefore, the levels of species richness reported here in CF sputa were between these two extremes. However, the data indicate that there is diverse and complex polymicrobial infection in an area of the body that has been considered to be sterile under normal circumstances in healthy individuals (11, 35). In a very-small-scale pilot study of samples obtained from healthy individuals through induced expectoration, we have obtained data that challenge this concept (data not shown). However, interestingly, little or no overlap was found between the bacterial community banding profiles obtained for healthy individuals and those obtained from CF patients.
The mean species richness found in individual patients was almost identical to that for the sample set as a whole (13.4 ± 6.7), suggesting that species richness is relatively consistent between patients. This relative lack of variation suggests that the level of species richness found in the CF lung may reflect certain facets of this environment. However, despite consistency in levels of species richness between patients, the variation found between samples taken from particular patients was relatively high (a mean variation of 9.9 ± 8.0). This fluctuation suggests that the bacterial community present in the CF lung is dynamic and may show marked changes over relatively short time intervals. These changes may reflect such factors as the onset of pulmonary infective exacerbation or antibacterial therapy. In the longer term, they may provide a useful clinical tool for the prediction and quantification of pulmonary infective exacerbations.
The level of species richness for the sample set as a whole was relatively high, with 248 distinct T-RF band lengths resolved. This is particularly significant, since the number of bacterial species that have previously been reported in CF lung infections is relatively small (8). The identification of the species that these T-RF bands represent could have dramatic implications for the way in which CF is viewed and treated and therefore represents another important area for future research.
The relative signal intensity (band volume) was determined for every T-RF band detected in the sample set. These band volumes were used to provide a measure of relative species prevalence. However, it should be noted that there are other factors apart from the number of organisms of each species present in the sample that can affect relative band volumes. Chief among these is variation in the ribosomal operon copy number between different bacterial species. The range of copy numbers is potentially relatively high. For example, Rickettsia prowazekii and Mycoplasma pneumoniae possess only one ribosomal operon (1, 3), whereas Clostridium paradoxum has 15 ribosomal operon copies per genome (30). However, the number of genomes in the majority of species is likely to cluster within a much narrower range.
In previous studies, it has been shown that, in T-RFLP profiles generated from model DNA mixtures composed of different pure cultures, the intensity of the signal representing each species was proportional to the number of ribosomal operon copies possessed by that species (17). Unfortunately, it was not possible to take ribosomal operon copy number into account when interpreting band volume data for two reasons. As indicated above, the majority of T-RF bands detected in these samples are yet to be identified. Moreover, data relating to the number of ribosomal operons possessed by different species, including relatively well-characterized species, are often scarce. However, the volumes of many of the T-RF bands resolved in this study differed by more than an order of magnitude. Further, in many cases, dominant T-RF bands corresponded to species known to have a relatively low operon copy number. It was, therefore, felt that a cautious interpretation of relative species prevalence could be made.
Although the bacterial communities present in CF sputa were shown here to be relatively diverse, the total band volume detected in a profile was not distributed evenly between its constituent species. Instead, in the majority of cases, a single species was dominant, typically representing 59.2% (±21.3%) of the total lane volume, with the next most dominant species representing 15.3% (±8.3%). The fact that 19 different species occupied this dominant position within the 71 samples analyzed suggests strongly that this is not merely due to their possession of a higher than average ribosomal operon copy number.
Significantly, in 28% of samples, the numerically dominant T-RF band corresponded to a bacterial species not traditionally considered to be clinically significant in CF lung infections. For example, in 4.2% of samples, a 574-base T-RF band had the highest band volume. This band length corresponded with that predicted for two different species, Ralstonia pickettii and Oligella urethralis. Of these, only R. pickettii has previously been reported in the respiratory secretions of CF patients (9). However, regardless of whether such species exhibit pathogenic behavior, their presence in the lung at such high levels is likely to be clinically relevant, for example, because they elicit an immune response. Moreover, their presence in the bacterial community may affect other bacterial species through both positive and negative interaction.
In addition, the presence, at a relatively low prevalence, of a wide range of bacterial species not traditionally associated with CF lung infection may be equally significant. In some instances, such species were only detected in a small number of patients and may represent chance infections by opportunistic pathogens able to establish themselves in large numbers once within the lung. An example is the 92-base T-RF band, corresponding to the size predicted for Bilophila wadsworthia. This is a species associated with appendicitis, abscesses, bacteremia, and biliary tract sepsis (21) which has also been reported to be present in the gastrointestinal tract (2). It is tempting to speculate whether the presence of this species in the CF lung may be related to gastric aspiration induced by strenuous coughing.
In other cases, however, these species were detected in a high proportion of individuals. For example, a 104-base T-RF band, corresponding to the predictions made for a number of different Prevotella species, was resolved in 47.2% of patients on at least one occasion. The presence of such species may be significant for several reasons. First, some may be highly pathogenic and have an impact on patient health that is disproportionate to their numbers. Second, the presence of these species may support the colonization of the lung by other, potentially more virulent, organisms. This emphasizes the requirement of considering the bacteria colonizing the CF lung to be part of a complex community, the dynamics of which are, in part, determined by the interactions of the species within it. Consequently, this has an impact on the way in which specific therapeutic agents should be administered against such a community.
The enormous complexity of bacterial species within the CF lung that is described here has a significant impact on the way we think about the pathogenesis of CF lung infections. The full significance of these findings has yet to be determined, although these previously unreported species may be important. While many questions remain unanswered, it is likely that these new species affect the host in a number of ways. The direct impacts on the host, such as severe pulmonary infection, are obvious. However, these species may also play important roles in the exuberant, immune-mediated, inflammatory cascade that is elicited by the presence of bacteria in CF patients. Moreover, the interaction of these species with the traditional pathogens may well involve both positive and negative feedback mechanisms, which determine the frequency and severity of CF infective exacerbations. For example, studies with animal models have shown that P. aeruginosa pathogenicity is significantly increased in the presence of avirulent, oropharyngeal flora (12). Further investigation is now required to document temporal changes in bacterial profiles generated from individual patients to correlate these variations with clinical parameters. Such a study will help to improve our understanding of the inadequacies of present antibiotic therapies and allow us to improve our management of infective episodes.
REFERENCES
- 1.Anderson, S. G. E., A. Zomorodipour, H. H. Winkler, and C. G. Kurland. 1995. Unusual organization of the rRNA genes in Rickettsia prowazekii. J. Bacteriol. 177:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, E. J., P. J. Summanen, J. Downes, M. C. Roberts, H. Wexler, and S. M. Finegold. 1989. Bilophila wadsorthia, gen. nov. and sp. nov., a unique Gram-negative anaerobic rod recovered from appendicitis specimens and human faeces. J. Gen. Microbiol. 135:3405-3411. [DOI] [PubMed] [Google Scholar]
- 3.Bercovier, H., O. Kafri, and S. Sela. 1986. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem. Biophys. Res. Commun. 136:1136-1141. [DOI] [PubMed] [Google Scholar]
- 4.Bonsell, S. Isolated knee joint infection with Neisseria meningitidis. Orthopedics 25:537-539. [DOI] [PubMed]
- 5.Brook, I. 1989. Bacteriology of chronic maxillary sinusitis in adults. Ann. Otol. Rhinol. Laryngol. 98:426-428. [DOI] [PubMed] [Google Scholar]
- 6.Bruce, K. D., and M. R. Hughes. 2000. Terminal restriction fragment length polymorphism monitoring of genes amplified directly from bacterial communities in soils and sediments. Mol. Biotechnol. 16:261-269. [DOI] [PubMed] [Google Scholar]
- 7.Clement, B. G., L. E. Kehl, K. L. DeBord, and C. L. Kitts. 1998. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J. Microbiol. Methods 31:135-142. [Google Scholar]
- 8.Coenye, T., J. Goris, T. Spilker, P. Vandamme, and J. J. LiPuma. 2002. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J. Clin. Microbiol. 40:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenye, T., P. Vandamme, and J. J. LiPuma. 2002. Infection by Ralstonia pickettii in cystic fibrosis patients: identification of R. Pickettii and R. mannitolilytica by polymerase chain reaction. Emerg. Infect. Dis. 8:692-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conese, M., and B. M. Assael. 2001. Bacterial infections and inflammation in the lungs of cystic fibrosis patients. Pediatr. Infect. Dis. J. 20:207-213. [DOI] [PubMed] [Google Scholar]
- 11.DeKoster, J. A., and P. S. Thorne. 1995. Bioaerosol concentrations in noncompliant, compliant, and intervention homes in the Midwest. Am. Ind. Hyg. Assoc. J. 56:573-580. [Google Scholar]
- 12.Duan, K., C. Dammel, J. Stein, H. Rabin, M. G. Surette. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50:1477-1491. [DOI] [PubMed] [Google Scholar]
- 13.Dunbar, J., S. White, and L. J. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filler, G., J. H. Ehrich, E. Strauch, and L. Beutin. 2000. Acute renal failure in an infant associated with cytotoxic Aeromonas sobria isolated from patient's stool and from aquarium water as suspected source of infection. J. Clin. Microbiol. 38:469-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilljam, H., A. S. Malmborg, and B. Strandvik. 1986. Conformity of bacterial growth in sputum and contamination free endobronchial samples in patients with cystic fibrosis. Thorax 41:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Health Protection Agency. 2003. Standard operating procedure 8. Investigation of sputum. Issue 5.1. Standards Unit, Evaluations and Standards Laboratory, Specialist and Reference Microbiology Division, Health Protection Agency, London, United Kingdom.
- 17.Hiraishi, A., M. Iwasaki, and H. Shinjo. 2000. Terminal restriction pattern analysis of 16S rRNA genes for the characterization of bacterial communities of activated sludge. J. Biosci. Bioeng. 90:148-156. [DOI] [PubMed] [Google Scholar]
- 18.http://rdp.cme.msu.edu/html/.
- 19.http://www.ncbi.nlm.nih.gov/blast/.
- 20.Jackson, R., and P. B. Pencharz. 2003. Cystic fibrosis. Best Pract. Res. Clin. Gastroenterol. 17:213-235. [DOI] [PubMed] [Google Scholar]
- 21.Kasten, M. J., J. E. Rosenblatt, and D. R. Gustafson. 1992. Bilophila wadsworthia bacteraemia in two patients with hepatic abscesses. J. Clin. Microbiol. 30:2502-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konstan, M. W., and M. Berger. 1993. Infection and inflammation of the lung in cystic fibrosis, p. 219-276. In P. B. Davis (ed.), Lung biology in health and disease, vol. 64. Cystic fibrosis. Marcel Dekker, New York, N.Y.
- 23.Konstan, M. W., K. A. Hillard, T. M. Norwell, and M. Berger. 1994. Bronchoalveolar lavage findings in cystic fibrosis patients with stable, clinically mild lung disease suggests ongoing infection and inflammation. Am. J. Respir. Crit. Care Med. 150:448-454. [DOI] [PubMed] [Google Scholar]
- 24.Liu, W., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphism of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magurran, A. E. 1988. Ecological diversity and its measurement. Croom Helm, London, United Kingdom.
- 26.Moore, W. E., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhlebach, M. S., P. W. Stewart, M. W. Leigh, and T. L. Noah. 1999. Quantitation of inflammatory responses to bacteria in young cystic fibrosis and control patients. Am. J. Respir. Crit. Care Med. 160:186-191. [DOI] [PubMed] [Google Scholar]
- 28.Peet, R. K. 1974. The measurement of species diversity. Annu. Rev. Ecol. Syst. 5:285-307. [Google Scholar]
- 29.Poulin, R. 1996. Patterns in the evenness of gastrointestinal helminth communities. Int. J. Parasitol. 26:181-186. [DOI] [PubMed] [Google Scholar]
- 30.Rainey, F. A., N. L. Ward-Rainey, P. H. Janssen, and H. Hippe. 1996. Clostridium paradoxum DSM 7308(T) contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology 142:2087-2095. [DOI] [PubMed] [Google Scholar]
- 31.Ramsey, B. W. 1996. Management of pulmonary disease in patients with cystic fibrosis. N. Engl. J. Med. 180:179-188. [DOI] [PubMed] [Google Scholar]
- 32.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rock, T. D. 1987. The study of microorganisms in situ: progress and problems. Symp. Soc. Gen. Microbiol. 41:1-17. [Google Scholar]
- 34.Rogers, G. B., C. A. Hart, J. R. Mason, M. Hughes, M. J. Walshaw, and K. D. Bruce. 2003. Bacterial diversity in cystic fibrosis lung infections 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 41:3548-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, J. J., S. M. Travis, E. P. Greenberg, and M. J. Welsh. 1996. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell 85:229-236. [DOI] [PubMed] [Google Scholar]
- 36.Thomassen, M. J., J. D. Klinger, S. J. Badger, D. W. Heeckeren, and R. C. Stern. 1984. Cultures of thoracotomy specimens confirm usefulness of sputum cultures in cystic fibrosis. J. Pediatr. 104:352-356. [DOI] [PubMed] [Google Scholar]
- 37.Thomson, R. B. 1999. Laboratory diagnosis of respiratory infections. Curr. Opin. Infect. Dis. 12:115-119. [DOI] [PubMed] [Google Scholar]
- 38.Tiquia, S. M., J. Lloyd, D. A. Herms, H. A. J. Hoitink, and F. C. Michel, Jr. 2002. Effects of mulching and fertilization on soil nutrients, microbial activity and rhizosphere bacterial community structure determined by analysis of TRFLPs of PCR-amplified 16S rRNA genes. Appl. Soil Ecol. 21:31-48. [Google Scholar]
- 39.Torsvik, V., Goskyr, J., and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 41.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Döring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]