Abstract
A panel of 23 real-time PCR assays based on TaqMan technology has been developed for the detection and monitoring of 16 different viruses and virus families including human polyomaviruses BK virus and JC virus, human herpesviruses 6, 7, and 8, human adenoviruses, herpes simplex viruses 1 and 2, varicella-zoster virus, cytomegalovirus, Epstein-Barr virus, parvovirus B19, influenza A and B viruses, parainfluenza viruses 1 to 3, enteroviruses, and respiratory syncytial virus. The test systems presented have a broad dynamic range and display high sensitivity, reproducibility, and specificity. Moreover, the assays allow precise quantification of viral load in a variety of clinical specimens. The ability to use uniform PCR conditions for all assays permits simultaneous processing and detection of many different viruses, thus economizing the diagnostic work. Our observations based on more than 50,000 assays reveal the potential of the real-time PCR tests to facilitate early diagnosis of infection and to monitor the kinetics of viral proliferation and the response to treatment. We demonstrate that, in immunosuppressed patients with invasive virus infections, surveillance by the assays described may permit detection of increasing viral load several days to weeks prior to the onset of clinical symptoms. In virus infections for which specific treatment is available, the quantitative PCR assays presented provide reliable diagnostic tools for timely initiation of appropriate therapy and for rapid assessment of the efficacy of antiviral treatment strategies.
The employment of PCR techniques for virus detection and quantification offers the advantages of high sensitivity and reproducibility combined with an extremely broad dynamicrange. A plethora of qualitative and quantitative PCR virus assays have been described, and commercial PCR kits are available for quantitative analysis of a number of clinically important viruses such as human immunodeficiency virus (14, 21), hepatitis B and C viruses (13, 22), and cytomegalovirus (CMV) (6). In addition to permitting the assessment of viral load at a given time point, quantitative PCR tests offer the possibility of determining the dynamics of virus proliferation, monitoring of the response to treatment, and in viruses displaying persistence in defined cell types, distinction between latent and active infection. Moreover, from a technical point of view, the employment of sequential quantitative PCR assays in virus monitoring helps identify false-positive results caused by inadvertent contamination of samples with traces of viral nucleic acids or PCR products (20).
We have established quantitative virus detection assays based on the real-time PCR (RQ-PCR) technology for 16 different viruses or virus families which play an important role in the clinical surveillance of immunosuppressed children. All assays were designed to run under identical PCR conditions to render the diagnostic work as economical as possible.
The RQ-PCR assays are presented in a ready-to-use format, and clinical applications of quantitative virus analysis in immunosuppressed patients are discussed.
MATERIALS AND METHODS
Sample preparation. (i) Nucleic acid extraction.
For the isolation of DNA and RNA, commercially available kits were used, essentially as recommended by the manufacturer. DNA extraction from largely cell-free liquids, except urine, and from peripheral blood leukocytes for the detection of intracellular virus particles was performed by using the QIAamp DNA mini kit (Qiagen GmbH, Hilden, Germany). Isolation of virus DNA from urine was done by using the QIAamp viral RNA mini kit, and isolation of virus DNA from stool was done by using the QIAamp DNA stool mini kit (Qiagen). For the isolation of RNA from each of these sources, the QIAamp viral RNA mini kit was used. The only modifications performed included the adjustment of the input and the elution volumes. For nucleic acid extraction, the input volume for all samples was 200 μl for DNA and 140 μl for RNA and the elution volume was 240 μl for DNA and 120 μl for RNA.
(ii) Reverse transcription.
For reverse transcription of purified viral RNA, a total of 30 μl of viral RNA eluate and 5 μl of nuclease-free water were mixed with 1 mM concentrations of each of the deoxynucleoside triphosphates and 25 μM pd(N)6, and this mixture was incubated at 72°C for 5 min. The denatured RNA was placed on ice for 1 min before the addition of 12 μl of reaction buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 5 mM MgCl2), 10 mM dithiothreitol, 1.5 μl of RNasin (40 U/μl; Promega, Mannheim, Germany), and 1.5 μl of Moloney murine leukemia virus reverse transcriptase (200 U/μl; Invitrogen, Carlsbad, Calif.). The reaction mixture was incubated at 37°C for 45 min, and finally, the enzymes were inactivated by heating at 98°C for 3 min.
Target sequence selection and primer and probe design.
Specific primers and probes were selected and designed by using the Primer Express, version 2.0, software (Applied Biosystems [AB], Foster City, Calif). The oligonucleotide sequences, locations, amplicon lengths, and GenBank accession numbers of the corresponding target genes are displayed in Table 1. As indicated in this table, some of the primers reveal a degenerated code. This was a prerequisite for the detection of viral subspecies differing from each other by single nucleotides.
TABLE 1.
Sequence details of all primer-probe combinations used
| Virus type | Target | Amplicon length (bp) | Oligonucleotide sequence (5′-3′)a
|
Concn (nM) | Nucleotide positions | GenBank accession no. | |||
|---|---|---|---|---|---|---|---|---|---|
| HHVs | |||||||||
| CMV | MIE protein | 76 | AAC TCA GCC TTC CCT AAG ACC A | 300 | 2414-2435 | M21295 | |||
| CAA TGG CTG CAG TCA GGC CAT GG | 200 | 2437-2459 | |||||||
| GGG AGC ACT GAG GCA AGT TC | 300 | 2470-2489 | |||||||
| EBV | BNT p143 | 74 | GGA ACC TGG TCA TCC TTT GC | 300 | 4679-4698 | NC_001345 | |||
| CGC AGG CAC TCG TAC TGC TCG CT (AS) | 200 | 4700-4722 | |||||||
| ACG TGC ATG GAC CGG TTA AT | 300 | 4733-4752 | |||||||
| HHV-6 | DNA polymerase gene | 74 | GAA GCA GCA ATC GCA ACA CA | 300 | 57517-57536 | NC_001664 | |||
| AAC CCG TGC GCC GCT CCC | 200 | 57544-57561 | |||||||
| ACA ACA TGT AAC TCG GTG TAC GGT | 900 | 57568-57590 | |||||||
| HHV-7 | Major capsid protein | 124 | CCC AAC TAT TTA CAG TAG GGT TGG TG | 300 | 84230-84255 | U43400 | |||
| CTA TTT TCG GTC TTT CCA ATG CAC GCA (AS) | 200 | 84258-84284 | |||||||
| TTT AGT TCC AGC ACT GCA ATC G | 900 | 84332-84353 | |||||||
| HHV-8 | ORFc 26 | 111 | GTG CTC GAA TCC AAC GGA TT | 300 | 47308-47327 | U75698 | |||
| TGT TCC CCA TGG TCG TGC C | 200 | 47336-47354 | |||||||
| CGA TAT TTT GGA GTA GAT GTG GTA CAC | 300 | 47392-47418 | |||||||
| HSV-1b | US 4 gene | 166 | TTC TCG TTC CTC ACT GCC TCC C | 900 | 137279-301 | NC001806 | |||
| CGT CTG GAC CAA CCG CCA CAC AGG T (AS) | 200 | 137379-404 | |||||||
| GCA GGC ACA CGT AAC GCA CGC T | 50 | 137423-445 | |||||||
| HSV-2 | Glycoprotein D gene | 71 | CGC CAA ATA CGC CTT AGC A | 300 | 99-117 | AF021342 | |||
| CTC GCT TAA GAT GGC CGA TCC CAA TC | 200 | 123-148 | |||||||
| GAA GGT TCT TCC CGC GAA AT | 300 | 150-169 | |||||||
| VZV | ORF 38 | 82 | AAG TTC CCC CCG TTC GC | 300 | 69313-69329 | X04370 | |||
| CCG CAA CAA CTG CAG TAT ATA TCG TCT CA | 200 | 69336-69364 | |||||||
| TGG ACT TGA AGA TGA ACT TAA TGA AGC | 300 | 69368-69394 | |||||||
| Human AdV | |||||||||
| AdV A | Hexon gene | 135 | GGK CTG GTG CAA TTC GCC | 300 | 17818-17835 | X73487 | |||
| CCA CGG ACA CCT ACT TCA CCC TGG G | 200 | 17840-17864 | |||||||
| CAC GGG CAC AAA ACG CA | 300 | 17936-17952 | |||||||
| AdV B | Hexon gene | 138 | CGC CGG ACA GGA TGC TT | 900 | 45-61 | X76549 | |||
| AGT CCG GGT CTG GTG CAG TTC GCC | 200 | 73-96 | |||||||
| CTA CGG TCG GTG GTC AC | 900 | 166-182 | |||||||
| AdV C | Hexon gene | 138 | ACC TGG GCC AAA ACC TTC TC | 300 | 2884-2903 | J01966 | |||
| AAC TCC GCC CAC GCG CTA GA | 200 | 2910-2929 | |||||||
| CGT CCA TGG GAT CCA CCT C | 900 | 2940-2958 | |||||||
| AdV D | VA RNA gene | 143 | AAA AAC GAA AGC GGT TGA GC | 300 | 2-21 | U10675 | |||
| CCA ATA CCA CGT TAG TCG CGG CT | 200 | 104-126 | |||||||
| CGG GTC GAG ACG GGA GT | 50 | 128-144 | |||||||
| AdV E | Hexon gene | 75 | CAA CAC CTA CTC GTA CAA AGT GCG | 900 | 225-248 | X84646 | |||
| CGC CCA CGG CCA GCG TGT | 200 | 251-268 | |||||||
| TAG GTG CTG GCC ATG TCC A | 300 | 281-299 | |||||||
| AdV F | 1.5 IV-2 gene | 113 | CCC GTG TTT GAC AAC GAA GG | 300 | 31277-31296 | L19443 | |||
| ATC GAC AAG GAC AGT CTG CCA ACA CTA ACG | 200 | 31326-31355 | |||||||
| TTA GAG CTA GGC ATA AAT TCT ACA GCA | 300 | 31363-31389 | |||||||
| Human polyoma- viruses: | |||||||||
| BKV | VP3 gene | 116 | TGT ACG GGA CTG TAA CAC CTG C | 300 | 1525-1546 | V01108 | |||
| TGA AGC ATA TGA AGA TGG CCC CAA C | 200 | 1550-1574 | |||||||
| TTT GGM ACT TGC ACG GG | 300 | 1624-1640 | |||||||
| JCV | Late mRNA gene | 123 | TGA ACC AAA AGC TAC ATA GGT AAG TAA TG | 900 | 474-502 | NC001699 | |||
| TTC ATG GGT GCC GCA CTT GCA | 200 | 523-543 | |||||||
| AAT CCT GTG GCA GCA G | 900 | 581-596 | |||||||
| PVB 19 | VP2 gene | 75 | TGG CCC ATT TTC AAG GAA GT | 300 | 3017-3036 | Z68146 | |||
| CCG GAA GTT CCC GCT TAC AAC | 200 | 3040-3062 | |||||||
| CTG AAG TCA TGC TTG GGT ATT TTT C | 300 | 3067-3091 | |||||||
| Enterovirusesd | |||||||||
| 5′ UTRe gene | 148 | CCC TGA ATG CGG CTA ATC C | 900 | 455-473 | D00820 | ||||
| CGG AAC CGA CTA CTT TGG GTG TCC GTG TTT C | 200 | 535-565 | |||||||
| ARA TTG TCA CCA TAA GCA GCC A | 900 | 581-602 | |||||||
| Respiratory syncytal virus | N gene | 149 | GGC AGT AGA GTT GAA GG | 900 | 1801-1817 | M11486 | |||
| ACT TGC CCT GCA CCA TAG GCA TTC ATA AAC AAT | 200 | 1830-1862 | |||||||
| ACA ACT TGT TCC ATT TCT GC | 300 | 1930-1949 | |||||||
| Influenza viruses | |||||||||
| Influenza A | Mf gene | 132 | CAT GGA ATG GCT AAA GAC AAG ACC | 900 | 126-149 | U49116 | |||
| TTT GTG TTY ACG CTC ACC GTG CCC A | 200 | 184-208 | |||||||
| CCA TTT AGG GCA TTT TGG ACA | 900 | 237-257 | |||||||
| Influenza B | HAg gene | 137 | AGA CCA GAG GGA AAC TAT GCC C | 300 | 134-155 | AB036449 | |||
| ACC TTC GGC AAA AGC TTC AAT ACT CCA | 200 | 219-245 | |||||||
| TCC GGA TGT AAC AGG TCT GAC TT | 900 | 248-270 | |||||||
| Parainfluenza viruses | |||||||||
| PIV-1h | HNi gene | 109 | GTT GTC AAT GTC TTA ATT CGT ATC AAT AAT T | 900 | 1191-1220 | U70948 | |||
| TAG GCC AAA GAT TGT TGT CGA GAC TAT TCC AA | 200 | 1232-1263 | |||||||
| GTA GCC TMC CTT CGG CAC CTA A | 900 | 1278-1299 | |||||||
| PIV-2 | HN gene | 90 | GCA TTT CCA ATC TTC AGG ACT ATG A | 900 | 767-791 | D00865 | |||
| CCA TTT ACC TAA GTG ATG GAA TCA ATC GCA AA | 200 | 795-826 | |||||||
| ACC TCC TGG TAT AGC AGT GAC TGA AC | 900 | 831-856 | |||||||
| PIV-3 | HN gene | 136 | AGT CAT GTT CTC TAG CAC TCC TAA ATA CA | 900 | 779-807 | L25350 | |||
| AAC TCC CAA AGT TGA TGA AAG ATC AGA TTA TGC A | 200 | 828-861 | |||||||
| ATT GAG CCA TCA TAA TTG ACA ATA TCA A | 900 | 887-914 | |||||||
| Positive controls | |||||||||
| SHV | gBj gene | 89 | GGG CGA ATC ACA GAT TGA ATC | 900 | 267-287 | Z68147 | |||
| TTT TTA TGT GTC CGC CAC CAT CTG GAT C | 200 | 305-332 | |||||||
| GCG GTT CCA AAC GTA CCA A | 900 | 337-355 | |||||||
| B2-MG | DNA | 105 | TGA GTA TGC CTG CCG TGT GA (ex 2) | 300 | 343-362 | M17987 | |||
| CCA TGT GAC TTT GTC ACA GCC CAA GAT AGT T (ex 2) | 200 | 364-394 | |||||||
| ACT CAT ACA CAA CTT TCA GCA GCT TAC (intr 2) | 300 | 421-447 | |||||||
| B2-MG | RNA | 82 | TGA GTA TGC CTG CCG TGT GA (ex 2) | 300 | 343-362 | M17987 | |||
| CCA TGT GAC TTT GTC ACA GCC CAA GAT AGT T (ex 2) | 200 | 364-394 | |||||||
| TGA TGC TGC TTA CAT GTC TCG AT (ex 3) | 300 | 1018-1040 |
Sequences are for the forward primer, probe, and reverse primer (top, middle, and bottom, respectively).
HSV-1, herpes simplex virus type 1.
ORF, open reading frame.
Includes polioviruses, coxsackie A and B viruses, echoviruses, and enterovirus types 68 to 71.
UTR, untranslated region.
M, matrix protein.
HA, hemagglutinin.
PIV-1, parainfluenza, virus type 1.
HN, hemagglutinin/neuraminidase mRNA.
gB, glycoprotein B.
For the experiments described below, hydrolysis probes labeled with 6-carboxyfluorescein reporter molecules at the 5′ end and 6-carboxy-tetramethylrhodamine quencher molecules at the 3′ end (AB) were used. The optimal concentration of primers was assessed by performing serial PCRs across a concentration range from 50 to 900 nM.
Real-time PCR.
All reactions were set up as singleplex PCRs in a total volume of 25 μl containing 12.5 μl of Universal Master mix (2× concentration, including ROX reference dye and uracil N′-glycosylase [UNG]; AB), 50 to 900 nM concentrations of primers, 200 nM TaqMan probe (Table 1), and 6 μl of genomic DNA or cDNA template. The mixtures were prepared in 96-well optical microtiter plates (AB), centrifuged for 1 min at 272 × g and amplified on the ABI 7700 or 7900 sequence detection system by using the following uniform cycling parameters: 2 min at 50°C (degradation of potentially present contaminating dUTP-containing amplicons by UNG), 10 min at 95°C (inactivation of UNG and activation of AmpliTaq Gold DNA polymerase), and 50 cycles of 15 s at 95°C and 60 s at 60°C (amplification of the specific target sequence).
Specificity.
All primer and probe combinations were tested for potential cross-reactivity with unrelated viral and other microbial sequences based on the available data by the BLAST alignment software. None of the selected primer and probe combinations displayed significant homologies to any other sequences. Moreover, the theoretically conceivable cross-reactivity with human DNA and RNA sequences has been excluded by testing the primer and probe combinations against preparations of human nucleic acids.
Standardization.
For standardization of quantitative virus detection assays, commercially available quantified DNA control panels (Advanced Biotechnologies, Inc., Columbia, Md.), in-house cloned plasmid standards, or high-titer virus preparations derived from culture supernatants were used. The calculation of virus particle numbers was based on spectrophotometric or fluorometric measurement of purified viral DNA or RNA. For the establishment of standard curves, serial logarithmic dilutions covering a range of ≥4 logs were employed, as described in more detail in Results.
Controls. (i) Negative controls.
A number of precautions were undertaken to prevent and control the occurrence of false-positive virus tests. Every clinical RQ-PCR test performed included control reactions lacking template (no-template controls) and reactions including nonhomologous template (no-amplification controls) to test for the presence of contamination or the generation of nonspecific amplification products under the assay conditions used. Moreover, to further reduce the risk of false-positive tests resulting from contamination with PCR products, all PCRs were performed by replacing the nucleotide dTTP with dUTP. Prior to amplification, a digestion step with UNG was carried out to eliminate any contaminating PCR product, if present.
(ii) Positive controls.
In addition to the DNA and cDNA of the respective control virus strain, the following controls were used in each assay to document efficient nucleic acid extraction and absence of enzyme inhibitors in the template preparation. (a) In largely cell-free clinical samples, such as plasma, serum, cerebrospinal fluid, urine, sputum, bronchoalveolar lavage fluid, or stool, a defined quantity of a nonhuman control virus (seal herpes virus [SHV], kindly provided by H. G. M. Niesters, University of Rotterdam, The Netherlands) was spiked into each sample prior to DNA and RNA extraction. Since constant DNA quantities of the control virus are coextracted even when RNA isolation kits are used (H. G. M. Niesters, personal communication; our own unpublished observations), the virus can also serve as a control in RNA virus detection assays. Under the standardized assay conditions used, constant levels of the seal virus were detected, provided that the nucleic acid extraction was efficient and no inhibitors of reverse transcription or PCR amplification were present (see Results). (b) In clinical samples containing cells, such as peripheral blood, buccal swabs, or biopsy material, a human single-copy housekeeping gene (β2-microglobulin [B2-MG]) (Table 1) (19) was coamplified in parallel with the virus sequence of interest.
In instances in which the cycle threshold (Ct) values of the above controls were off scale (below the expected reading), an appropriate correction factor was applied to the calculation of virus copy number in the corresponding clinical samples to compensate for impaired nucleic acid extraction or amplification efficiencies. Negative virus test results in the presence of low-positive (>1 log below normal) or negative SHV or B2-MG controls were regarded as not interpretable.
RESULTS
The sequence information of primers and probes for 23 real-time PCR virus detection assays is displayed in Table 1, together with an indication of their precise positions within the targeted genes. Moreover, the optimal primer and probe concentrations are indicated for each detection assay. All virus PCR tests presented were designed to be conducted under identical cycling conditions, as outlined in Materials and Methods, to facilitate the molecular diagnostic work.
Efficiency and sensitivity of RQ-PCR virus assays.
These parameters were assessed by repeated testing of serial logarithmic dilutions of the standard reference virus strains covering a range of ≥4 logs. The number of virus copies used to prepare the serial dilutions had been determined by spectrophotometric or fluorometric measurement of the genomic DNA and cDNA concentrations of individual virus strain preparations.
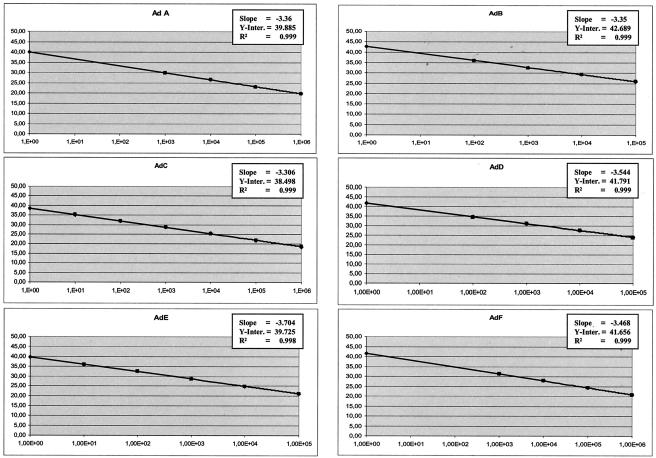
After PCR amplification, the Ct values (crossing point of the amplification curve with the preset threshold of fluorescence detection) of individual dilution steps were plotted against the initial virus copy number, leading to typical standard curves. The standard curves provided information on the amplification efficiency, the consistency of replicate reactions, and the theoretical and actual detection limits of the assay. The amplification efficiencies, defined by the standard curve slopes, were generally at or around 3.5. The consistency of replicates was measured by the correlation coefficient (R2), which indicates the linearity of the Ct values plotted in the standard curves. The R2 indices were higher than 0.990 in all measurements (Fig. 1).
FIG. 1.
Standard curves of the RQ-PCR virus assays described. Serial logarithmic dilutions were analyzed by using standard amplification conditions. The Ct (x axis) of each of the dilutions is plotted against the cycle number (y axis). The slope, y-axis intercept (Y-Inter.), and correlation coefficient are displayed in each graph. HSV 1, herpes simplex virus type 1; B19, PVB19; JCV, JC virus; EV, enterovirus; RSV, respiratory syncytial virus; Inf A, influenza A virus; PIV 1, parainfluenza virus type 1.
The actual sensitivities of the assays were determined by the lowest standard dilution consistently detectable in replicate reactions. In all assays presented, 1E + 02 virus genomes were reproducibly detected.
Reproducibility of RQ-PCR virus assays.
To evaluate the intra-assay variation of the virus tests, control samples across a wide range of virus copy numbers were analyzed concomitantly in triplicate reactions. The coefficients of variation (CVs) were in the range of 0.5% for most of the virus samples analyzed; only samples containing very low virus genome equivalents (<1.00E + 01) showed consistently higher CVs (around 1%). The interassay variation was assessed by investigating a minimum of three different DNA or cDNA aliquots of individual virus samples in independent assays. Comparison of triplicate tests within different runs revealed CVs in the order of 1.6% (Table 2).
TABLE 2.
CVs of virus assays describeda
| PCR target | Quantity | Intra-assay variation
|
Interassay variation (CV2) | |
|---|---|---|---|---|
| CV1a | CV1b | |||
| CMV | 1.00E+05 | 0.22 | 0.21 | 2.41 |
| 1.00E+04 | 0.94 | 1.09 | 2.12 | |
| 1.00E+03 | 0.57 | 0.47 | 1.90 | |
| 1.00E+02 | 0.80 | 0.26 | 1.56 | |
| 1.00E+01 | 0.38 | 2.12 | 1.99 | |
| EBV | 1.00E+06 | 0.30 | 0.63 | 2.96 |
| 1.00E+05 | 1.15 | 0.80 | 2.80 | |
| 1.00E+04 | 0.32 | 0.28 | 1.90 | |
| 1.00E+03 | 0.24 | 0.17 | 1.83 | |
| HHV-6 | 1.00E+06 | 0.27 | 0.39 | 1.42 |
| 1.00E+05 | 0.26 | 0.52 | 1.63 | |
| 1.00E+04 | 0.51 | 0.50 | 1.00 | |
| 1.00E+03 | 0.08 | 0.63 | 0.84 | |
| 1.00E+02 | 0.25 | 1.56 | 1.60 | |
| HHV-7 | 1.00E+05 | 0.85 | 0.41 | 2.45 |
| 1.00E+04 | 0.11 | 0.26 | 1.89 | |
| 1.00E+03 | 0.45 | 0.21 | 1.69 | |
| 1.00E+02 | 0.50 | 0.26 | 1.73 | |
| 1.00E+01 | 0.20 | 0.28 | 1.24 | |
| HHV-8 | 1.00E+06 | 0.52 | 0.46 | 1.07 |
| 1.00E+05 | 0.29 | 0.44 | 0.77 | |
| 1.00E+04 | 0.53 | 0.11 | 0.82 | |
| 1.00E+03 | 0.68 | 0.58 | 0.81 | |
| 1.00E+02 | 0.23 | 0.20 | 0.67 | |
| HSV-1b | 1.00E+05 | 0.38 | 0.10 | 2.60 |
| 1.00E+04 | 0.30 | 0.20 | 1.99 | |
| 1.00E+03 | 0.20 | 0.19 | 1.79 | |
| 1.00E+02 | 0.51 | 0.22 | 1.61 | |
| HSV-2 | 1.00E+05 | 2.42 | 0.62 | 2.05 |
| 1.00E+04 | 0.37 | 0.12 | 1.80 | |
| 1.00E+03 | 0.87 | 0.36 | 1.73 | |
| 1.00E+02 | 0.33 | 0.36 | 1.43 | |
| VZV | 1.00E+05 | 0.64 | 0.46 | 3.67 |
| 1.00E+04 | 0.17 | 0.20 | 3.09 | |
| 1.00E+03 | 0.39 | 0.38 | 3.17 | |
| 1.00E+02 | 0.29 | 0.13 | 2.69 | |
| 1.00E+01 | 2.94 | 1.15 | 3.91 | |
| AdV A | 1.00E+06 | 0.32 | 0.62 | 1.16 |
| 1.00E+05 | 0.18 | 0.24 | 1.08 | |
| 1.00E+04 | 0.18 | 0.37 | 0.85 | |
| 1.00E+03 | 0.40 | 0.17 | 0.84 | |
| AdV B | 1.00E+05 | 0.32 | 0.07 | 1.92 |
| 1.00E+04 | 0.34 | 0.06 | 1.79 | |
| 1.00E+03 | 0.39 | 0.26 | 1.73 | |
| 1.00E+02 | 0.23 | 0.17 | 1.27 | |
| AdV C | 1.00E+05 | 0.50 | 0.51 | 0.56 |
| 1.00E+04 | 0.40 | 0.31 | 0.75 | |
| 1.00E+03 | 0.07 | 0.15 | 0.45 | |
| 1.00E+02 | 0.21 | 0.22 | 0.43 | |
| 1.00E+01 | 0.75 | 0.81 | 0.73 | |
| AdV D | 1.00E+05 | 0.53 | 0.68 | 1.79 |
| 1.00E+04 | 0.09 | 0.45 | 1.92 | |
| 1.00E+03 | 0.37 | 0.15 | 1.49 | |
| 1.00E+02 | 0.42 | 0.11 | 1.08 | |
| AdV E | 1.00E+05 | 0.18 | 0.57 | 1.17 |
| 1.00E+04 | 0.17 | 0.15 | 0.78 | |
| 1.00E+03 | 0.63 | 0.43 | 0.74 | |
| 1.00E+02 | 0.32 | 0.33 | 0.50 | |
| 1.00E+01 | 0.45 | 0.58 | 1.34 | |
| AdV F | 1.00E+06 | 0.13 | 0.30 | 0.87 |
| 1.00E+05 | 0.19 | 0.32 | 1.17 | |
| 1.00E+04 | 0.22 | 0.15 | 1.02 | |
| 1.00E+03 | 0.13 | 0.27 | 0.82 | |
| BKV | 3.00E+06 | 0.31 | 0.21 | 1.02 |
| 3.00E+05 | 0.36 | 0.46 | 1.08 | |
| 3.00E+04 | 0.22 | 0.27 | 0.86 | |
| 3.00E+03 | 0.33 | 0.39 | 0.96 | |
| 3.00E+02 | 0.25 | 0.16 | 0.79 | |
| JCV | 1.60E+05 | 0.21 | 0.18 | 0.53 |
| 1.60E+04 | 0.53 | 0.30 | 0.56 | |
| 1.60E+03 | 0.33 | 0.27 | 0.52 | |
| 1.60E+02 | 0.30 | 0.24 | 0.35 | |
| 1.60E+01 | 0.61 | 1.15 | 0.83 | |
| PVB19 | 1.00E+06 | 0.56 | 0.57 | 2.33 |
| 1.00E+05 | 0.35 | 0.37 | 2.49 | |
| 1.00E+04 | 0.38 | 0.25 | 2.41 | |
| 1.00E+03 | 0.43 | 0.37 | 2.07 | |
| Enterovirus | 1.00E+07 | 0.24 | 0.45 | 1.58 |
| 1.00E+06 | 0.43 | 0.44 | 1.31 | |
| 1.00E+05 | 0.63 | 0.40 | 1.18 | |
| 1.00E+04 | 0.62 | 0.42 | 1.08 | |
| 1.00E+03 | 1.75 | 2.57 | 2.39 | |
| RSVc | 1.00E+05 | 0.67 | 0.39 | 3.20 |
| 1.00E+04 | 0.55 | 0.21 | 2.06 | |
| 1.00E+03 | 0.39 | 0.40 | 1.66 | |
| 1.00E+02 | 0.58 | 0.59 | 2.43 | |
| Influenza A virus | 1.00E+06 | 0.34 | 0.25 | 2.52 |
| 1.00E+05 | 0.34 | 0.20 | 1.99 | |
| 1.00E+04 | 0.83 | 0.50 | 1.78 | |
| 1.00E+03 | 0.60 | 0.22 | 1.31 | |
| Influenza B virus | 1.00E+04 | 0.70 | 0.73 | 2.78 |
| 1.00E+03 | 0.30 | 0.31 | 2.36 | |
| 1.00E+02 | 0.39 | 0.40 | 2.10 | |
| 1.00E+01 | 1.12 | 1.16 | 2.13 | |
| PIV-1d | 1.00E+05 | 0.88 | 0.26 | 1.50 |
| 1.00E+04 | 0.18 | 0.19 | 1.46 | |
| 1.00E+03 | 0.43 | 0.09 | 1.30 | |
| 1.00E+02 | 0.34 | 0.21 | 1.20 | |
| 1.00E+01 | 0.52 | 0.32 | 1.03 | |
| PIV-2 | 1.00E+05 | 0.06 | 0.24 | 1.84 |
| 1.00E+04 | 0.41 | 0.19 | 1.22 | |
| 1.00E+03 | 0.17 | 0.29 | 1.33 | |
| 1.00E+02 | 0.18 | 0.19 | 1.20 | |
| 1.00E+01 | 0.73 | 0.94 | 1.25 | |
| PIV-3 | 1.00E+04 | 0.32 | 0.26 | 0.57 |
| 1.00E+03 | 0.82 | 0.57 | 0.72 | |
| 1.00E+02 | 0.55 | 0.50 | 0.59 | |
| 1.00E+01 | 0.44 | 1.56 | 1.29 | |
CVs observed in RQ-PCR analyses of different virus quantities are presented. The intra-assay variation was calculated by comparing the Ct values of two simultaneously amplified triplicate reactions (CV1a versus CV1b). The interassay variation was assessed by comparing triplicate reactions of different RQ PCR runs (CV2). The mean values for intra-and interassay variation across all virus assays are as follows: CV1a, 0.46; CV1b, 0.43; CV2, 1.55.
HSV-1, herpes simplex virus type 1.
RSV, respiratory syncytial virus.
PIV-1, parainfluenza virus type 1.
The results obtained document the high precision and low variation within and between individual virus tests.
Quantification of virus copy numbers in clinical samples.
The virus loads in individual patient specimens were investigated by testing the most recent and, if available, a previously quantified patient sample in duplicate reactions, together with the appropriate external virus standard preparations. The efficiency of virus DNA and RNA isolation from clinical samples and the possible presence of reverse transcriptase or polymerase inhibitors were monitored by using internal controls, as described in Materials and Methods. These controls permitted appropriate correction in the calculation of virus copy numbers in the specimen investigated. For the calculation of virus particles in the sample tested, the slope (s) and the y-axis intercept (Y) (the y-axis intercept is the point at which the standard curve intersects with the ordinate; it indicates the theoretical detection limit of the reaction by revealing the Ct expected in the presence of a single target molecule in the sample) of the corresponding standard curve and the Ct of the target virus amplification were used according to the following equation: P0 = Inverse log(Ct − Y/s), where P0 is the number of virus copy equivalents in the PCR prior to amplification.
Examples of clinical application in immunosuppressed patients.
The panel of virus tests presented covers viral pathogens of well established or supposed importance in pediatric patients with severe immunosuppression. The following examples were derived from virus monitoring of immunocompromised children after allogeneic stem cell transplantation (SCT) and illustrate the clinical utility of the RQ-PCR virus detection assays in this particular clinical setting.
(i) Rapid diagnosis of viral cause of disease symptoms.
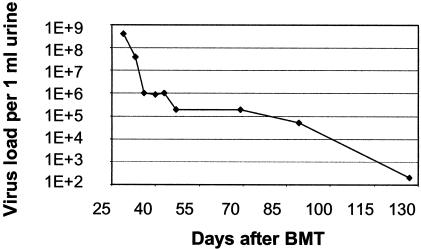
Severe inflammation of the urinary bladder in immunocompromised patients may occur as a result of infection with a variety of bacterial and viral pathogens. The polyomavirus BK virus (BKV) is a relatively common cause of hemorrhagic cystitis in children undergoing allogeneic SCT. Although no specific antiviral treatment is currently available for this type of infection, detection and monitoring of the virus during the course of disease is of clinical relevance with regard to identification of the cause of the symptoms observed and with regard to differential diagnosis from other pathogens requiring specific treatment. Figure 2 illustrates the detection and surveillance of BKV load in serial urine samples of a patient displaying hemorrhagic cystitis after allogeneic SCT. All tests for other pathogens in the urine were negative. The causative agent and the course of infection under symptomatic therapy could be documented by RQ-PCR monitoring.
FIG. 2.
Kinetics of BKV load during hemorrhagic cystitis. Documentation of BKV infection of the urinary bladder and clearance of the virus by serial RQ-PCR analysis of urine samples during the posttransplant period. The virus load (y axis) is plotted against the time after bone marrow transplantation (BMT) (x axis).
(ii) Latent virus infection and reactivation.
Following primary exposure, a number of viruses, including members of the herpesvirus family and adenoviruses, may persist as latent infections in peripheral blood (PB) leukocytes.
In view of the high prevalence of infections with these viruses, low viral copy numbers documenting persistence are detectable in B cells of the PB in a large proportion of healthy individuals. In immunosuppressed patients, reactivation of latent virus infection, associated with serious clinical symptoms, represents a frequent finding (33). In these instances, the viruses proliferate within the affected cells and are released into the extracellular compartment. Since viral reactivation may lead to life-threatening complications in patients with impaired immune responses, early diagnosis is of major clinical importance to permit timely initiation of appropriate antiviral treatment.
Epstein-Barr virus (EBV) is commonly detectable at low copy numbers in PB lymphocytes when present in the latent state. Reactivation of the virus in immunocompromised patients can lead to a lymphoproliferative disease, which may result in the occurrence of fatal malignant lymphoma (15, 32). In these instances, increasing copy numbers of the virus are detectable within PB lymphocytes and rising levels of free virus are detectable in plasma. Early detection of EBV proliferation kinetics provides a basis for timely initiation of preemptive treatment (30, 31). In the example presented, a retrospective analysis of a child who died from EBV-associated lymphoma is shown (Fig. 3). Increasing levels of the virus in PB were documented by RQ-PCR over a period of several weeks before the lymphoma was diagnosed clinically.
FIG. 3.
Kinetics of EBV load in PB in a posttransplant lymphoproliferative disease. Serial RQ-PCR analysis documents the reactivation of a latent EBV infection by revealing constantly increasing virus copy numbers. This retrospective analysis of virus proliferation kinetics, which heralded the development of EBV-associated malignant lymphoma, underlines the potential of molecular detection and monitoring of EBV load to provide a basis for early initiation of preemptive antiviral treatment. BMT, bone marrow transplantation.
(iii) Early detection of invasive viral infection.
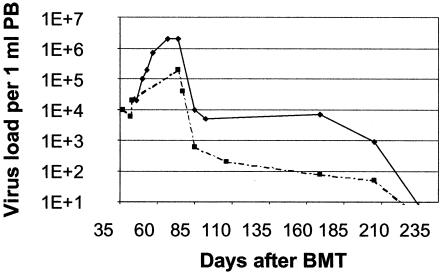
Adenoviral (AdV) infections are frequently observed in immunosuppressed children during the posttransplant period (2) and are associated with high morbidity and mortality. Earlier observations made in our laboratory indicate that invasive AdV infections require a very early onset of antiviral treatment (20). In a number of instances, an example of which is shown in Fig. 4, invasive AdV infections with rapid proliferation kinetics can be detected in serial PB samples by RQ-PCR. Detection of the virus and the observation of increasing viral load precede the onset of clinical symptoms by a median of 3 weeks (20).
FIG. 4.
Kinetics of AdV load in PB. The curve shows the appearance and expansion of AdV in PB. Observation of the first 10-fold increase in virus load (arrow) preceded the onset of clinical symptoms (star) by more than 3 weeks. The monitoring of AdV in PB may therefore serve as a basis for early initiation of preemptive antiviral treatment. BMT, bone marrow transplantation.
(iv) Documentation of response to antiviral treatment.
In immunocompromised patients suffering from potentially life-threatening virus infections, the surveillance of the success of treatment is of paramount importance for appropriate clinical management, including the choice and duration of appropriate antiviral therapy (4, 23, 27). The example displayed in Fig. 5 shows the documentation of decreasing CMV levels both in plasma and PB leukocytes, followed by elimination of the virus below the limit of detection, in response to antiviral treatment with ganciclovir.
FIG. 5.
Kinetics of CMV load in PB in response to antiviral treatment. Monitoring of CMV by RQ-PCR in serial plasma (solid line) and PB leukocyte (dashed line) samples during the posttransplant period reveals viral reactivation by rising levels of CMV DNAemia and subsequent clearance of the virus following antiviral therapy. BMT, bone marrow transplantation.
DISCUSSION
Inthis paper, we present real-time PCR assays for qualitative and quantitative analysis of 16 viruses or virus families which appear to be of particular importance in the clinical management of immunosuppressed children undergoing high-dose chemotherapy or allogeneic SCT. Qualitative virus detection revealing merely the presence or absence of a viral pathogen is not sufficient in a variety of clinical situations. In many instances, the rate of disease development was shown to be related directly to the viral DNA or RNA levels detected in plasma, serum, or PB lymphocytes. The association between disease progression and viral load is well established for infections with a number of viruses, particularly human immunodeficiency virus and hepatitis B and C viruses (3, 11, 22).
In immunosuppressed patients, particularly after allogeneic SCT, the need to quantitatively monitor infections with CMV and EBV has been long appreciated (6, 8, 18). The clinical relevance of a number of other viral infections in this setting is less well established, but there is a growing body of evidence indicating that viruses such as AdV (20, 26), human herpesvirus 6 (HHV-6) (24), varicella-zoster virus (VZV) (12), parvovirus B19 (PVB19) (28), and others (1, 7, 10) merit careful monitoring in these patients. Molecular diagnostic assays, such as the tests presented herein, are therefore of increasing clinical importance.
To render the screening for multiple viruses more practicable, all tests presented were conceived to permit target amplification and quantification under identical PCR conditions. The examples shown represent important paradigms of clinical application of quantitative virus detection assays in immunocompromised patients. As demonstrated, quantitative virus tests are useful when assessing a clinically suspected viral cause of infection because the documentation of rising virus copy numbers, in the absence of other detectable pathogens, provides support for a role of the virus as a causative agent. Moreover, the observation of increasing viral load in sequential assays virtually excludes the possibility of false interpretation of positive PCR results resulting from contamination with amplification products or traces of viral nucleic acids harboring the amplifiable sequence.
Another clinically important application of quantitative virus tests is the possibility of differentiating between latent infection and reactivation. Persisting viruses may occur after primary infection in healthy immunocompetent individuals, as well as in asymptomatic patients (16), and cause universally positive results in qualitative PCR assays. Mere detection of viral pathogens by qualitative PCR may not be relevant to the clinical outcome in these individuals, but consecutive assessment of the virus load seems to play an important role in the diagnosis and prognosis of patients with viral reactivation by providing a basis for timely initiation of appropriate treatment (5, 6, 9, 25, 29). Sequential assessment of viral load by means of RQ-PCR is a helpful parameter for clinical decision making, particularly if molecular detection and documentation of proliferation kinetics of the virus precede the onset of clinical symptoms. This has been demonstrated for a number of virus infections (17, 20, 31).
Finally, the ability of quantitative virus tests to facilitate monitoring of the response to antiviral treatment is an invaluable tool in the clinical care of immunocompromised patients, providing a means of controlling of the appropriate choice and the necessary duration of therapy. Quantitative virus testing has therefore become an indispensable diagnostic instrument in many clinical situations. The real-time PCR tests presented provide a contribution to the rapidly growing field of molecular investigation of viral infections as a basis for improved patient care.
REFERENCES
- 1.Abdallah, A., K. E. Rowland, S. K. Schepetiuk, L. B. To, and P. Bardy. 2003. An outbreak of respiratory syncytial virus infection in a bone marrow transplant unit: effect on engraftment and outcome of pneumonia without specific antiviral treatment. Bone Marrow Transplant. 32:195-203. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, H., T. Kurosu, C. Sakashita, T. Inoue, S. Mori, K. Ohashi, S. Tanikawa, H. Sakamaki, Y. Onozawa, Q. Chen, H. Zheng, and T. Kitamura. 2001. Adenovirus is a key pathogen in hemorrhagic cystitis associated with bone marrow transplantation. Clin. Infect. Dis. 32:1325-1330. [DOI] [PubMed] [Google Scholar]
- 3.Cervia, J., B. Kaplan, S. Schuval, and S. Weiss. 2003. Virologic testing in the management of perinatal HIV exposure. AIDS Read. 13:39-47. [PubMed] [Google Scholar]
- 4.Dini, G., E. Castagnola, P. Comoli, M. J. van Tol, and J. M. Vossen. 2001. Infections after stem cell transplantation in children: state of the art and recommendations. Bone Marrow Transplant. 28(Suppl. 1):S18-S21. [DOI] [PubMed] [Google Scholar]
- 5.Emery, V. C., A. V. Cope, E. F. Bowen, D. Gor, and P. D. Griffiths. 1999. The dynamics of human cytomegalovirus replication in vivo. J. Exp. Med. 190:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emery, V. C., C. A. Sabin, A. V. Cope, D. Gor, A. F. Hassan-Walker, and P. D. Griffiths. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 355:2032-2036. [DOI] [PubMed] [Google Scholar]
- 7.Fischmeister, G., P. Wiesbauer, H. M. Holzmann, C. Peters, M. Eibl, and H. Gadner. 2000. Enteroviral meningoencephalitis in immunocompromised children after matched unrelated donor-bone marrow transplantation. Pediatr. Hematol. Oncol. 17:393-399. [DOI] [PubMed] [Google Scholar]
- 8.Gartner, B. C., H. Schafer, K. Marggraff, G. Eisele, M. Schafer, D. Dilloo, K. Roemer, H. J. Laws, M. Sester, U. Sester, H. Einsele, and N. Mueller-Lantzsch. 2002. Evaluation of use of Epstein-Barr viral load in patients after allogeneic stem cell transplantation to diagnose and monitor posttransplant lymphoproliferative disease. J. Clin. Microbiol. 40:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gustafsson, A., V. Levitsky, J. Z. Zou, T. Frisan, T. Dalianis, P. Ljungman, O. Ringden, J. Winiarski, I. Ernberg, and M. G. Masucci. 2000. Epstein-Barr virus (EBV) load in bone marrow transplant recipients at risk to develop posttransplant lymphoproliferative disease: prophylactic infusion of EBV-specific cytotoxic T cells. Blood 95:807-814. [PubMed] [Google Scholar]
- 10.Hassan, I. A., R. Chopra, R. Swindell, and K. J. Mutton. 2003. Respiratory viral infections after bone marrow/peripheral stem-cell transplantation: the Christie hospital experience. Bone Marrow Transplant. 32:73-77. [DOI] [PubMed] [Google Scholar]
- 11.Hodinka, R. L. 1998. The clinical utility of viral quantitation using molecular methods. Clin. Diagn. Virol. 10:25-47. [DOI] [PubMed] [Google Scholar]
- 12.Ishizaki, Y., J. Tezuka, S. Ohga, A. Nomura, N. Suga, R. Kuromaru, K. Kusuhara, Y. Mizuno, N. Kasuga, and T. Hara. 2003. Quantification of circulating varicella zoster virus-DNA for the early diagnosis of visceral varicella. J. Infect. 47:133-138. [DOI] [PubMed] [Google Scholar]
- 13.Jungkind, D. 2001. Automation of laboratory testing for infectious diseases using the polymerase chain reaction-our past, our present, our future. J. Clin. Virol. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Kumar, R., N. Vandegraaff, L. Mundy, C. J. Burrell, and P. Li. 2002. Evaluation of PCR-based methods for the quantitation of integrated HIV-1 DNA. J. Virol. Methods 105:233-246. [DOI] [PubMed] [Google Scholar]
- 15.Lankester, A. C., M. J. van Tol, J. M. Vossen, A. C. Kroes, and E. Claas. 2002. Epstein-Barr virus (EBV)-DNA quantification in pediatric allogenic stem cell recipients: prediction of EBV-associated lymphoproliferative disease. Blood 99:2630-2631. [DOI] [PubMed] [Google Scholar]
- 16.Larsson, S., C. Soderberg-Naucler, F. Z. Wang, and E. Moller. 1998. Cytomegalovirus DNA can be detected in peripheral blood mononuclear cells from all seropositive and most seronegative healthy blood donors over time. Transfusion 38:271-278. [DOI] [PubMed] [Google Scholar]
- 17.Leruez-Ville, M., M. Ouachee, R. Delarue, A. S. Sauget, S. Blanche, A. Buzyn, and C. Rouzioux. 2003. Monitoring cytomegalovirus infection in adult and pediatric bone marrow transplant recipients by a real-time PCR assay performed with blood plasma. J. Clin. Microbiol. 41:2040-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Limaye, A. P., M. L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183:377-382. [DOI] [PubMed] [Google Scholar]
- 19.Lion, T. 2001. Current recommendations for positive controls in RT-PCR assays. Leukemia 15:1033-1037. [DOI] [PubMed] [Google Scholar]
- 20.Lion, T., R. Baumgartinger, F. Watzinger, S. Matthes-Martin, M. Suda, S. Preuner, B. Futterknecht, A. Lawitschka, C. Peters, U. Potschger, and H. Gadner. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114-1120. [DOI] [PubMed] [Google Scholar]
- 21.O'Doherty, U., W. J. Swiggard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76:10942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pas, S. D., E. Fries, R. A. De Man, A. D. Osterhaus, and H. G. Niesters. 2000. Development of a quantitative real-time detection assay for hepatitis B virus DNA and comparison with two commercial assays. J. Clin. Microbiol. 38:2897-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reusser, P. 2002. Challenges and options in the management of viral infections after stem cell transplantation. Support. Care Cancer 10:197-203. [DOI] [PubMed] [Google Scholar]
- 24.Sashihara, J., K. Tanaka-Taya, S. Tanaka, K. Amo, H. Miyagawa, G. Hosoi, T. Taniguchi, T. Fukui, N. Kasuga, T. Aono, M. Sako, J. Hara, K. Yamanishi, and S. Okada. 2002. High incidence of human herpesvirus 6 infection with a high viral load in cord blood stem cell transplant recipients. Blood 100:2005-2011. [PubMed] [Google Scholar]
- 25.Savoie, A., C. Perpete, L. Carpentier, J. Joncas, and C. Alfieri. 1994. Direct correlation between the load of Epstein-Barr virus-infected lymphocytes in the peripheral blood of pediatric transplant patients and risk of lymphoproliferative disease. Blood 83:2715-2722. [PubMed] [Google Scholar]
- 26.Schilham, M. W., E. C. Claas, W. van Zaane, B. Heemskerk, J. M. Vossen, A. C. Lankester, R. E. Toes, M. Echavarria, A. C. Kroes, and M. J. van Tol. 2002. High levels of adenovirus DNA in serum correlate with fatal outcome of adenovirus infection in children after allogeneic stem-cell transplantation. Clin. Infect. Dis. 35:526-532. [DOI] [PubMed] [Google Scholar]
- 27.Schutten, M., and H. G. Niesters. 2001. Clinical utility of viral quantification as a tool for disease monitoring. Expert Rev. Mol. Diagn. 1:153-162. [DOI] [PubMed] [Google Scholar]
- 28.Segovia, J. C., G. Guenechea, J. M. Gallego, J. M. Almendral, and J. A. Bueren. 2003. Parvovirus infection suppresses long-term repopulating hematopoietic stem cells. J. Virol. 77:8495-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sirvent-Von Bueltzingsloewen, A., P. Morand, M. Buisson, G. Souillet, H. Chambost, J. L. Bosson, and P. Bordigoni. 2002. A prospective study of Epstein-Barr virus load in 85 hematopoietic stem cell transplants. Bone Marrow Transplant. 29:21-28. [DOI] [PubMed] [Google Scholar]
- 30.van Esser, J. W., H. G. Niesters, S. F. Thijsen, E. Meijer, A. D. Osterhaus, K. C. Wolthers, C. A. Boucher, J. W. Gratama, L. M. Budel, H. B. van der, A. M. van Loon, B. Lowenberg, L. F. Verdonck, and J. J. Cornelissen. 2001. Molecular quantification of viral load in plasma allows for fast and accurate prediction of response to therapy of Epstein-Barr virus-associated lymphoproliferative disease after allogeneic stem cell transplantation. Br. J. Haematol. 113:814-821. [DOI] [PubMed] [Google Scholar]
- 31.van Esser, J. W., H. G. Niesters, H. B. van der, E. Meijer, A. D. Osterhaus, J. W. Gratama, L. F. Verdonck, B. Lowenberg, and J. J. Cornelissen. 2002. Prevention of Epstein-Barr virus-lymphoproliferative disease by molecular monitoring and preemptive rituximab in high-risk patients after allogeneic stem cell transplantation. Blood 99:4364-4369. [DOI] [PubMed] [Google Scholar]
- 32.van Esser, J. W., H. B. van der, E. Meijer, H. G. Niesters, R. Trenschel, S. F. Thijsen, A. M. van Loon, F. Frassoni, A. Bacigalupo, U. W. Schaefer, A. D. Osterhaus, J. W. Gratama, B. Lowenberg, L. F. Verdonck, and J. J. Cornelissen. 2001. Epstein-Barr virus (EBV) reactivation is a frequent event after allogeneic stem cell transplantation (SCT) and quantitatively predicts EBV-lymphoproliferative disease following T-cell-depleted SCT. Blood 98:972-978. [DOI] [PubMed] [Google Scholar]
- 33.Wagner, H. J., M. Wessel, W. Jabs, F. Smets, L. Fischer, G. Offner, and P. Bucsky. 2001. Patients at risk for development of posttransplant lymphoproliferative disorder: plasma versus peripheral blood mononuclear cells as material for quantification of Epstein-Barr viral load by using real-time quantitative polymerase chain reaction. Transplantation 72:1012-1019. [DOI] [PubMed] [Google Scholar]