Abstract
Thirty-three strains of Brevibacillus laterosporus, including three novel strains isolated from Brazilian soil samples, were examined for genetic variability by the use of different PCR-based methods. Molecular markers that could characterize bacterial strains with regards to their pathogenic potential were investigated. In addition, toxicity was assessed by the use of insects belonging to the orders Lepidoptera and Coleoptera and the mollusk Biomphalaria glabrata. Among the targets tested, Biomphalaria glabrata demonstrated the highest degree of sensitivity to B. laterosporus, with some strains inducing 90 to 100% mortality in snails aged 3 and 12 days posteclosion. Larvae of the coleopteron Anthonomus grandis were also susceptible, presenting mortality levels of between 33 and 63%. Toxicity was also noted towards the lepidopteron Anticarsia gemmatalis. In contrast, no mortality was recorded among test populations of Tenebrio molitor or Spodoptera frugiperda. The application of intergenic transcribed spacer PCR and BOX-PCR generated 15 and 17 different genotypes, respectively. None of the molecular techniques allowed the identification of a convenient marker that was associated with any entomopathogenic phenotype. However, a 1,078-bp amplicon was detected for all strains of B. laterosporus when a primer for amplification of the BOXA1R region was used. Similarly, a 900-bp amplicon was generated from all isolates by use of the primer OPA-11 for randomly amplified polymorphic DNA analysis. These amplicons were not detected for other phenotypically related Brevibacillus species, indicating that they represent markers that are specific for B. laterosporus, which may prove useful for the isolation and identification of new strains of this species.
Brevibacillus laterosporus comb. nov. (28), previously classified as Bacillus laterosporus (Laubach 1916), is an aerobic spore-forming bacterium that is characterized by its ability to produce a canoe-shaped lamellar parasporal inclusion adjacent to the spore. Some strains produce crystalline inclusions of various shapes and sizes, which are released separately from spores during lysis of the sporangium. B. laterosporus has the potential to be used as a biological control agent which, in comparison with strains of Bacillus thuringiensis and Bacillus sphaericus, demonstrates a very wide spectrum of biological activities. In this context, toxicity has been observed towards the beetle Lasioderma serricone, the nematode fitoparasite Heterodera glycines, the nematode zooparasite Trichostrongylus colubriformis, and adult forms of the mollusk Dreissena polymorpha (31). In addition, the 50% lethal concentration (LC50) values recorded for the Coleoptera Leptinotarsa serricone and Leptinotarsa decemlineata (Colorado potato beetle) were comparable to that exhibited for Bacillus thuringiensis serovar Tenebrionis (LC50 = 0.1 μg/cm2) (31, 32). Finally, toxicity towards larvae of the mosquitoes Culex quinquefasciatus and Aedes aegypti and the blackfly Simulium vittatum has also been reported (6, 25). In addition, some strains of B. laterosporus produce the medically important substances espergualin (19, 37) and bacithrocins A, B, and C (13).
Despite showing such wide-ranging biological activities, B. laterosporus has not been seriously considered for use in biological control, most probably because the observed mosquitocidal activity is generally much weaker than that of Bacillus thuringiensis serovar israelensis. Yet Orlova et al. (21) demonstrated that crystalliferous strains of B. laterosporus presented LC50 values similar to those attained with Bacillus thuringiensis serovar israelensis in bioassays employing larvae of three species of mosquitoes, with the larvicidal activity of B. laterosporus being associated with spores and crystalline inclusions. Although Bacillus thuringiensis and Bacillus sphaericus have been used as biological control agents in many countries (4, 18, 23, 24), some strains still present problems, including low environmental persistence and a restricted range of targets. Thus, the availability of alternative agents would be highly desirable.
In a previous study (41), the genetic variability of B. laterosporus was demonstrated by the use of randomly amplified polymorphic DNA PCR (RAPD-PCR), multilocus enzyme electrophoresis, and pulsed-field gel electrophoresis. However, no molecular markers associated with specific pathogenicity profiles were identified. The present study was undertaken with the primary objective of further evaluating genetic variability among B. laterosporus isolates through the application of an extended range of molecular techniques. In addition, we sought to examine strains of B. laterosporus for biocidal activities towards insects of the orders Diptera, Lepidoptera, and Coleoptera as well as towards the mollusk Biomphalaria glabrata, which acts as an intermediate vector for the transmission of Schistosoma mansoni to humans (7).
MATERIALS AND METHODS
Strain maintenance and isolation.
The strains used for this work are listed in Table 1. All of these strains are maintained in the culture collection of the Laboratory of Systematic Biochemistry, but they were originally supplied by A. A. Yousten (Department of Biology, Virginia Polytechnic Institute and State University, Blacksburg, Va.), with the exceptions of strain BL 16-92 (kindly donated by Rouldolf R. Azizbekyan, Laboratory of Genetics of Biopesticides, State Institute of Genetics, Moscow, Russia) and the three novel isolates reported in this study. The strains were maintained at −20°C as spore suspensions in 20% glycerol and were activated in nutrient yeast salt medium (NYSM) (6) for 3 to 5 days.
TABLE 1.
Strains used for this work and toxicity levels towards four insect targets
| Strainb | Toxicitya for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
Aedes aegypti
|
Culex quinque- fasciatus
|
Anticarsia gemmatalis
|
Athonomus grandis
|
|||||
| V | S | V | S | V | S | V | S | |
| Shi 1 | − | − | + | + | + | + | ++ | ++ |
| Shi 2 | − | − | + | + | − | ++ | ++ | ++ |
| Shi 3 | − | − | + | + | + | ++ | ++ | ++ |
| Shi 4 | − | − | + | + | + | ++ | ++ | +++ |
| Shi 5 | ++ | ++ | ++ | ++ | + | ++ | ++ | ++ |
| ATCC 64 | ++ | ++ | + | + | ++ | + | ++ | ++ |
| NRS 1644 | + | + | ++ | ++ | + | + | ++ | ++ |
| NRS 340 | + | + | + | + | ++ | ++ | ++ | ++ |
| NRS 342 | + | + | + | + | ++ | ++ | ++ | ++ |
| CCEB 342 | ++ | ++ | ++ | ++ | + | + | ++ | ++ |
| NRS 590 | ++ | ++ | ++ | ++ | ++ | +++ | ++ | ++ |
| NRS 661 | + | ++ | ++ | ++ | + | + | +++ | +++ |
| Bon 706 | − | − | + | + | + | + | ++ | ++ |
| Bon 707 | − | − | ++ | ++ | + | + | ++ | ++ |
| Bon 708 | − | − | + | + | ND | ++ | ND | ++ |
| Bon 712 | − | − | ++ | ++ | + | + | ++ | +++ |
| BL 16-92 | + | ++ | ++ | ++ | + | + | ++ | ++ |
| NRS 1111 | + | + | + | + | ++ | ++ | ++ | ++ |
| NRS 1264 | ++ | ++ | − | − | ND | ++ | ND | ++ |
| NRS 1267 | + | + | − | − | + | + | + | ++ |
| NRS 1338 | + | + | − | − | ND | + | ND | +++ |
| NRS 1642 | + | + | − | − | + | ++ | ++ | ++ |
| NRS 1643 | + | + | − | − | ++ | + | ++ | ++ |
| NRS 1644 | + | + | − | − | ++ | ++ | ++ | +++ |
| NRS 1645 | + | + | − | − | + | + | ++ | ++ |
| NRS 1646 | + | ++ | − | − | ++ | ++ | ++ | +++ |
| NRS 1647 | ++ | ++ | − | − | ND | + | ND | ++ |
| NRS 1648 | ++ | ++ | − | − | ND | ++ | ND | ++ |
| Montaldi | − | − | − | − | ++ | +++ | ++ | +++ |
| ATCC 6457 | + | + | − | − | + | +++ | ++ | ++ |
| ATCC 9141 | + | + | ++ | ++ | ++ | + | ++ | ++ |
| NI 1c | + | + | − | − | + | + | ++ | ++ |
| NI 2c | − | − | − | − | + | + | +++ | ++ |
| NI 3c | − | − | + | + | + | + | ++ | +++ |
The new isolates, designated NI 1, NI 2, and NI 3, were recovered from soil samples collected in the state of Rio de Janeiro, Brazil, according to protocols recommended by the World Health Organization (40), using NYSM with incubation at 31°C (6). The identity of each isolate was confirmed by performing a sequence of morphological and physiological tests as described previously (8, 9, 28). Bacillus cereus (LFB-Fiocruz 406/NCTC 2599), Bacillus thuringiensis (LFB-Fiocruz 584/IPS-82), and Brevibacillus brevis (LFB-Fiocruz 417/NCTC 2611) from the Culture Collection of Genus Bacillus and Correlated Genera (Laboratory of Bacterial Physiology, Instituto Oswaldo Cruz) were used as controls in some bioassays and for molecular techniques.
Preparation of material for bioassays.
All of the strains that were used for the fermentation process were initially grown in NYSM, with incubation at 31°C for 48 h. A loopful of each culture was then transferred to individual 10- by 12-mm tubes containing 2 ml of distilled water and homogenized. One milliliter was transferred to a 250-ml Erlenmeyer flask containing 100 ml of NYSM and incubated at 31°C for 12 h at 120 rpm. Thereafter, duplicate 5-ml volumes of culture were transferred to two Erlenmeyer flasks with a 2-liter capacity, with each containing 450 ml of a fermentation medium developed at the Laboratory of Bacterial Physiology/IOC/Fiocruz (patent no. BR PI 9501166, 1998, INPI-Fiocruz) (22) supplemented with fructose (33). The cultures were subjected to agitation (200 rpm) in a model 25D shaker (New Brunswick Scientific) at a temperature of 31°C. Each strain was inoculated into two flasks to allow for the production of both vegetative (48 h) and sporulation-phase (72 h) cultures. Direct cell observations (light microscopy) and viability counts were performed at both time points to determine the cell numbers and to confirm the growth phase of each culture. Vegetative cells and spores were recovered by centrifugation with refrigeration at 25,000 × g for 40 min (model HR-1; International High Speed refrigerated centrifuge). The pH of each biomass was adjusted to 4.5 to 5.0 with propionic acid. This material was then stored at 4°C until it was used for bioassays. An exception to this process was the preparation of dried biomass for use in the Biomphalaria glabrata bioassay. In this case, the biomass (sporulated cultures) was dried in a vacuum oven (model 815; Lab Line Instruments) at 45°C under negative pressure (620 mm Hg) for 7 days, and the dry weight of each biomass was recorded.
Bioassays.
For all bioassays using Lepidoptera and Coleoptera, larvae were fed an artificial diet based on black beans, brewer's yeast, soy protein, wheat germ, and casein, as recommended by Shmidt et al. (29). Biomphalaria glabrata snails were fed lettuce leaves that had previously been washed in 10% acetic acid. The criteria used to define the association between mortality and toxicity were those established by Oliveira et al. (20): low toxicity (+), 1 to 20% mortality; medium toxicity (++), 21 to 50% mortality; and high toxicity (+++), >50% mortality. Standardized quantities of biomass were used in each bioassay, including that for mosquito larvae. Unless otherwise stated, the inocula comprised the resuspended biomass (viable vegetative or sporulated bacterial cells, as determined by plate count [data not shown]), and the material was dissolved in 0.85% NaCl to achieve an optical density of 0.1 (at 600 nm).
Mosquito bioassays.
Bioassays employing mosquito larvae (C. quinquefasciatus and Aedes aegypti) were conducted as previously described by Oliveira et al. (20) and as recommended by the World Health Organization (40).
Anthonomus grandis bioassay.
For the Anthonomus grandis bioassay, the treatment was prepared by mixing 200 μl of bacterial suspension with the insect diet. Treatment and control diets were then spread over the surfaces of 5-cm-diameter petri dishes and allowed to solidify. A total of 60 holes were made in the solidified diets, and an individual second-instar larva was inserted into each hole. The plates were incubated at 27°C, and mortality was monitored for 7 days.
Anticarsia gemmatalis bioassay.
For the Anticarsia gemmatalis bioassay, 150 μl of bacterial suspension was added to the diet, which was held in 50-ml cups, with each cup containing 10 second-instar larvae. A total of 20 cups (10 treatment and 10 control) were used for each test strain of B. laterosporus. After 48 h, dead larvae were removed and the remaining larvae were transferred to new cups containing the control diet. Mortality levels were recorded for 6 days.
Spodoptera frugiperda bioassay.
The Spodoptera frugiperda assay was performed in 24-well tissue culture plates, with one half of each plate containing the treatment diet (supplemented with 30 μl of bacterial suspension) and the other half serving as a control. Each well contained a single second-instar larva which was held there for 48 h. This procedure was used in order to prevent cannibalism among the larvae. Thereafter, the larvae were transferred to individual 50-ml cups containing the control diet, and mortality levels were monitored for 6 days.
Tenebrio molitor bioassay.
Ten second-instar larvae were placed in 5-cm-diameter petri dishes containing 0.2 g of wheat meal with or without the addition of 200 μl of bacterial suspension. Each assay was composed of five replicates per test strain and 10 control plates. After 48 h, live larvae were transferred to 50-ml cups containing the control diet, and the level of mortality was determined over 5 days.
Biomphalaria glabrata bioassay.
The Biomphalaria glabrata assay used freshwater planorbidae snails that were recovered 0 to 3 days and 12 to 15 days posteclosion. Treatment involved the addition of dried biomass (final concentration, 50 mg/liter) to plastic cups, with each cup containing 100 mollusks in 200 ml of dechlorinated tap water. The mollusks were fed lettuce leaves. The room incubation temperature was 27 ± 1°C and the bioassay was conducted for 7 days, with mortality levels being recorded daily.
Preparation of genomic DNAs and PCR-based methods.
Total DNA was extracted by the use of GenomicPrep cell and tissue DNA isolation kits (Amersham Pharmacia Biotech) according to the manufacturer's instructions. DNA concentrations were determined spectrophotometrically in a Gene Quant apparatus (Pharmacia). All PCR-based techniques employed a model 9600 thermocycler (Applied Biosystems). Amplification products were analyzed by separating 12 μl of each reaction mixture in 2.0% (wt/vol) agarose gels at 3.2 V cm−1 in TBE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]), followed by staining with ethidium bromide and examination under UV light. Negative controls (without DNA) were included in all amplifications. The protocol for BOX-PCR utilized the primers and reaction conditions reported by Versalovic et al. (38), with the application of a hot start (7 min at 95°C) to avoid initial mispriming and to enhance the specificity. Amplification of the variable spacer region between the 16S and 23S gene sequences was performed by established methods for intergenic transcribed spacer PCR (ITS-PCR) (10), utilizing 25 ng of genomic DNA as a template. RAPD-PCR was used in order to compare the profiles of the new Brazilian strains and strain BL 16-92 with those of isolates that were previously examined by this method (41). The reaction conditions were the same as those described by Zahner et al. (41) and used the primers OPA-11 (5′ AGG GGT CTT G 3′) and OPA-01 (5′ AAG AGC CCG T 3′) (Operon Technologies).
Detection of 16S ribosomal DNA genes by Southern hybridization.
One microgram of genomic DNA was digested with HindIII. The resulting fragments were separated in a 1.2% agarose gel and blotted onto a nitrocellulose membrane (35). The membrane was hybridized with a digoxigenin (DIG)-labeled DNA probe prepared by a PCR with the cloned 16S rrnB rRNA operon of Escherichia coli as a template (1) by use of a PCR-DIG probe synthesis kit (Roche). The hybridization temperature was 65°C. Immunological detection and colorimetric revelation of the hybrids were performed according to the instructions provided in a DIG nucleic acid detection kit (Roche).
Numerical analysis.
The procedures for numerical analysis followed those provided by Zahner et al. (41), and the value of the cophenetic correlation coefficient (CCC) was obtained with the COPH program of the NTSYS software package (version 2.02; Exeter Software, Setauket, N.Y.).
RESULTS AND DISCUSSION
Our group has attempted to isolate B. laterosporus from a variety of environments on several occasions, but in general we have been unsuccessful. For the present study, we employed a range of growth media (data not shown) to examine 50 different soil samples and recovered a total of three strains of B. laterosporus. Thus, it appears that B. laterosporus shows a more limited distribution than either Bacillus thuringiensis or Bacillus sphaericus. None of the new isolates presented parasporal bodies, although they all demonstrated a degree of biological activity in at least one of the bioassays. The notion that the presence of parasporal bodies is a prerequisite for insect pathogenicity is controversial. In this context, strains of B. laterosporus with and without parasporal bodies have been reported to show biocidal activity towards mosquito larvae. In particular, the strains 16-92 (921) and LAT 006 (615) showed significant activity against larvae of Aedes aegypti and Anopheles stephensis (21, 34). It was demonstrated that both mosquitocidal strains produced parasporal crystals with different sizes and shapes which were believed to be responsible for the observed toxic activity, although no information was provided as to the composition of these structures. On the other hand, mosquitocidal activities have also been observed for strains that do not present crystals (6).
No toxicity was detected in the bioassays examining Tenebrio molitor (yellow mealworm) and Spodoptera frugiperda (velvetbean caterpillar) for any of the test strains. The data produced from all other insect bioassays are presented in Table 1. It should be emphasized that the quantities of biomass resulting from each fermentation were sufficient to allow the use of a standardized product in every bioassay.
Toxicity levels towards the two species of mosquito were similar to those reported previously (6, 16), with all B. laterosporus strains, including the new Brazilian isolates, showing limited toxicities (Table 1). It is worth noting that with the exception of the findings of a single study (21), there are limited data to encourage the utilization of B. laterosporus as an agent for mosquito control. Our data represent a comprehensive evaluation of the mosquitocidal capacity of this bacterium, in terms of both the number of strains examined and a comparison of vegetative and sporulated cells, and they support the view that B. laterosporus has limited potential as an agent for mosquito control.
Anticarsia gemmatalis is an agricultural pest which can be controlled by the use of bioinsecticides based on either bacterial, fungal, or viral agents (15). Our study revealed that all strains of B. laterosporus showed some degree of toxicity towards this insect, with the highest level of toxicity (90% mortality) being shown by the NRS 590 strain. Interestingly, toxic activity (50% mortality) was present in material prepared from vegetative cells and rose to the 90% level for cells that were collected and processed during the sporangium phase. A similar phenomenon was observed with strains Montaldi and ATCC 9141, for which the use of sporulated material was shown to enhance the toxicity. In contrast, vegetative cells of strains ATCC 64, NRS 1643, and ATCC 9141 induced mortality levels of 46, 43, and 48%, respectively, which declined to values of 20, 18, and 13% when the biomass comprised sporulated material. In these cases, the toxin was apparently synthesized during the exponential phase of bacterial growth and underwent a subsequent degradation. However, for the majority of the cases (16 of 29) in which the two forms of biomass were compared, there was no difference in relative toxicity (Table 1). Finally, a single isolate, strain Shi 2, presented a low toxicity (24%) when applied as sporulated material but was not toxic when offered to the insect as vegetative cells.
Toxicity was also demonstrated by all test strains towards Anthonomus grandis (also known as the Mexican cotton boll weevil), an insect that is associated with damage to stored grains (17). Like the data recorded for Anticarsia gemmatalis, there was no clear correlation between the bacterial growth phase and toxicity. Thus, a higher toxicity level was recorded when the diet contained bacteria collected during the sporulation rather than the vegetative phase in the cases of strains Shi 4, Bon 712, NRS 1267, NRS 1644, NRS 1646, Montaldi, and NI 3. Yet the basis for the toxicity shown by the other strains was initiated during the vegetative phase and remained at the same level when these strains were applied as sporulated material (Table 1). An exception to this was strain NI 2, which showed a reduced toxicity when given as sporulated cells. Based on the data presented in Table 1, we suggest that for the majority of the test strains, the insecticidal toxin (or toxins) responsible for the biocidal activity of B. laterosporus is produced during the vegetative phase of the bacterial growth cycle and appears to be maintained in the cell during sporulation. In Bacillus sphaericus, the MTX toxin is also produced during the vegetative phase, but it is subsequently degraded by a sfericase (36).
Schistosome infections, caused primarily by Schistosoma mansoni, Schistosoma hematobium, and Schistosoma japonicum, affect over 200 million people, mainly in the developing world, resulting in severe morbidity and mortality (7). The freshwater snail Biomphalaria glabrata is an intermediate host and natural vector for Schistosoma mansoni. The bioassay employed in this study is technically demanding in terms of the number of snails used, and for that reason a limited number of strains of B. laterosporus were examined. The same reason motivated the use of a single form of biomass (spores). Although only 12 strains of B. laterosporus were used for this bioassay, the majority (10 of 12) were found to be highly toxic for larvae of Biomphalaria glabrata (Table 2). These data are extremely interesting since chemotherapy is ineffective at preventing reinfection, and therefore the development of complementary control measures, including the control of snail populations, would be highly beneficial. We observed that the age of the larvae was an important factor in determining their susceptibility to the toxic effect, with older (12 to 15 days posteclosion) animals being markedly more resistant to the presence of the spores than were younger larvae (Table 2). This was exemplified by strains NRS 1264 and Shi 1, which were both able to kill 100% of mollusks aged 0 to 3 days posteclosion but only killed 17 and 30%, respectively, when the older mollusks were employed as a target. Interestingly, strain NRS 1111 was able to kill both age groups of mollusks highly effectively (Table 2).
TABLE 2.
Toxic activities of 12 strains of B. laterosporus against larvae of different ages of the mollusk Biomphalaria glabrata
| Straina | Mortality (%) of snails
|
|
|---|---|---|
| 0-3 days posteclosion | 12-15 days posteclosion | |
| NRS 1111 | 100 | 94 |
| Shi 1 | 100 | 30 |
| NRS 1264 | 100 | 17 |
| ATCC 9141 | 98 | 9 |
| Shi 5 | 94 | 12 |
| BL 16-92 | 94 | 4 |
| ATCC 6457 | 92 | 41 |
| Shi 2 | 70 | 5 |
| Bon 707 | 55 | 6 |
| Shi 3 | 55 | 0 |
| NRS 1267 | 10 | 0 |
| NRS 1642 | 5 | 0 |
Dry biomass was produced from sporulated cultures (see the text for details).
Nucleic acid amplification and detection strategies have been used for the characterization and evaluation of genetic diversity of many bacteria, and such applications improve diagnostic approaches and clinical management and in many cases evolve into standard laboratory and point-of-care testing protocols (39). In the cases of bacteria that are used for biological control and screening programs for new strains, it would be desirable to identify molecular markers that facilitate identification and that correlate with particular pathogenic phenotypes. In order to expand upon the results obtained by Zahner et al. (41), we employed a range of PCR-based techniques to further characterize our collection of B. laterosporus strains at the molecular level.
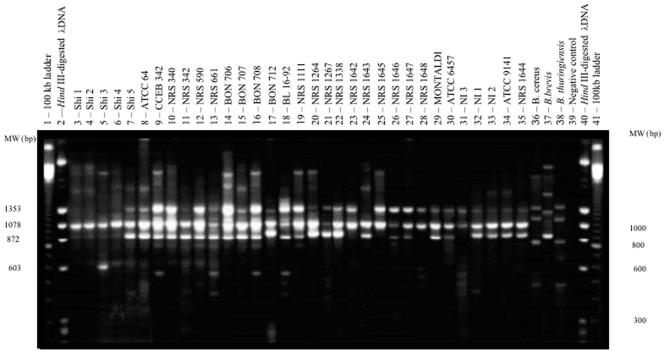
Repetitive element PCR (rep-PCR) profiles based on BOX regions have been used for typing, systematics, and epidemiology studies (12, 26, 27, 30). In the present study, we employed the primer BOXA1R for the examination of B. laterosporus strains and included B. brevis, Bacillus thuringiensis, and Bacillus cereus as controls, as the first is phylogenetically close to B. laterosporus and the others are part of the bacterial genus from which B. laterosporus was previously allocated. A total of 17 genotypes were formed among the B. laterosporus strains, and a common band of 1,078 bp was observed for all genotypes (Fig. 1). This observation could be applied to the characterization of B. laterosporus at the species level since no congruence was observed between the profiles recorded for the B. laterosporus strains and that of the phenotypically related B. brevis. As expected, Bacillus cereus and Bacillus thuringiensis presented the same genetic profile, supporting the findings of comparative 16S rRNA sequence analysis (2) which demonstrated that they are phylogenetically closely related and clearly distinct from the B. laterosporus group. Repetitive sequences have in some cases been considered to contribute to the virulence of a pathogen, as exemplified by the antigenic variation shown by Neisseria gonorrhoeae (14). For this reason, we examined the possibility that this method could identify markers associated with virulence towards the specific targets used in this study. However, our data did not reveal any correlation between pathogenicity and rep-PCR genetic profiles.
FIG. 1.
Banding profiles obtained with B. laterosporus strains after amplification with the BOXA1R primer.
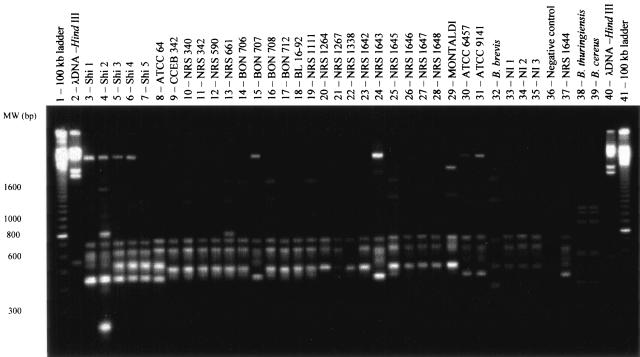
According to Gürtler and Stanisich (10), the intergenic transcribed spacer sequence (ITS region) may show differences between species and even between strains of the same species. Indeed, this region of the bacterial genome has been targeted in many studies and was shown to be valuable for the characterization of individuals within the same species (3, 5, 11). Using this technique, we found a total of 15 genotypes among the 33 strains of B. laterosporus that were analyzed. The majority of the genotypes resulting from the amplification of the ITS region did not exhibit polymorphism (for bands in the range of 750 to 450 bp) at the B. laterosporus intraspecies level. Minor exceptions were observed with some strains for which bands of >700 bp were present (Fig. 2). However, like the case for the rep-PCR approach, no correlation was established between ITS profiles and pathogenicity.
FIG. 2.
Banding profiles obtained with B. laterosporus strains after amplification of the ITS.
The RAPD-PCR technique was used in order to compare the profiles of the new Brazilian strains with those reported previously (41). In agreement with the previously published results, the primer OPA-11 presented a high level of polymorphism and amplified DNAs from all B. laterosporus strains tested. In addition, it generated a single band of approximately 900 bp for all B. laterosporus samples that may serve as a molecular marker for this species, as already suggested (41). Primer OPA-01 was chosen based on the previous observation that it amplified sequences associated with mosquitocidal strains (41). Amplicons were detected for all three novel strains, confirming the observation that they all possessed mosquitocidal activities, albeit at very low levels (Table 1). Strains NI 1 and NI 2 presented identical genetic profiles when the OPA-01 primer was used, but some minor differences were detected between these isolates when the OPA-11 primer was used (data not shown). Interestingly, the three Brazilian strains generated RAPD profiles which were dissimilar to those generated by any of the other strains. Thus, in order to verify the taxonomic position of the Brazilian isolates, we undertook a comparative ribotype analysis. As expected, with the exceptions of Bacillus thuringiensis and Bacillus cereus, which presented the same genotype, each species (B. laterosporus, B. brevis, and Bacillus thuringiensis/Bacillus cereus) produced a single pattern. In addition, the intraspecific patterns obtained among B. laterosporus strains were identical (data not shown).
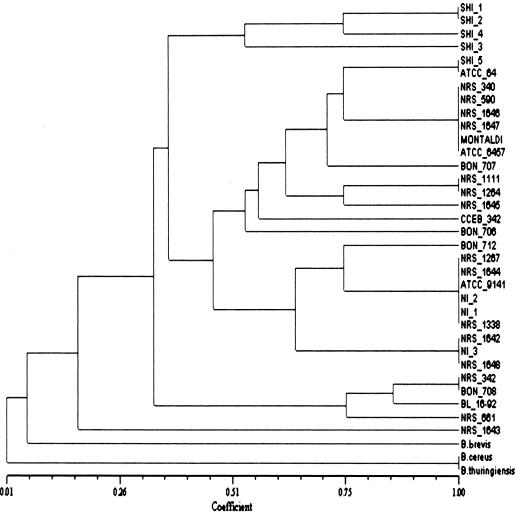
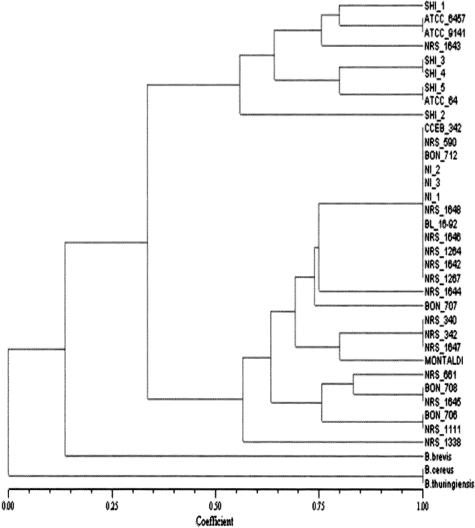
The results of the numerical analysis performed on the BOXA1R amplification profiles are presented in Fig. 3. This dendrogram was based on the results from SSm, since its CCC was 0.97. The dendrogram shown in Fig. 4 was generated by use of the Jaccard coefficient (CCC = 0.95) to analyze the ITS-PCR data. Both dendrograms reflect the low degree of polymorphism among B. laterosporus strains. The dendrogram generated from the RAPD analysis was in agreement with previously reported results (41). The introduction of the novel Brazilian strains and BL16-92 into the database used for the numerical analysis did not interfere with the overall topography of the dendrograms (not shown). An analysis of the dendrograms based on the PCR methods did not reveal any correlation between the pathogenicity of the strains for any given host and any of the clusters formed. Moreover, the results of BOXA1R PCR and ITS-PCR did not generate any molecular marker that was associated with pathogenic activity. Indeed, the amplicons generated by the utilization of the OPA-01 primer remained the only indication of some link between biological activity and the genetic profile, as suggested previously (41). However, the value of this observation is reduced by the fact that mosquito larvae are not considered to represent a promising target for biological control by the use of B. laterosporus, with the notable exception of a single study (21).
FIG. 3.
Dendrogram showing the relationship among B. laterosporus isolates based on the UPGMA method and determined according to Jaccard's similarity coefficients obtained from PCR patterns by using the BOXA1R primer sequence.
FIG. 4.
Dendrogram showing the relationship among B. laterosporus isolates based on the UPGMA method and determined according to Jaccard's similarity coefficients obtained from PCR patterns obtained after amplification of the ITS.
In summary, our characterization of B. laterosporus by different molecular methods revealed an elevated level of similarity between isolates, demonstrating a low level of genetic polymorphism within this species and indicating that the isolates are clonally related, as suggested previously (41). The pathogenic potential of B. laterosporus against invertebrates has been known for some time, yet there is a dearth of literature concerning the biological control potential of this species. The broad spectrum of activity and the absence of definitive data with regard to the molecular basis of toxicity make this species a worthy topic of investigation. The bioassay data for Anticarsia gemmatalis, Anthonomus grandis, and Biomphalaria glabrata presented in this study encourage further experimentation to more fully evaluate the potential use of B. laterosporus as a bioinsecticide for the control of these organisms.
Acknowledgments
We acknowledge Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior for fellowship support to E.J.O. and the Instituto Oswaldo Cruz-Fiocruz for providing materials.
REFERENCES
- 1.Aquino de Muro, M., W. J. Mitchell, and F. G. Priest. 1992. Differentiation of mosquito-pathogenic strains of Bacillus sphaericus from non-toxic varieties by ribosomal RNA gene restriction patterns. J. Gen. Microbiol. 138:1159-1166. [DOI] [PubMed] [Google Scholar]
- 2.Ash, C., J. A. E. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 3.Bakshi, C. S., V. P. Sing, M. Malek, B. Sharma, and R. K. Singh. 2002. Polymerase chain reaction of 16S-23S spacer region for rapid identification of Salmonella serovars. Acta Vet. Hung. 50:161-166. [DOI] [PubMed] [Google Scholar]
- 4.Becker, N. 2000. Bacterial control of vector-mosquitoes and black flies, p. 383-398. In J.-F. Charles, A. Delécluse, and C. Nielsen-LeRoux (ed.), Entomopathogenic bacteria: from laboratory to field application. Kluwer Academic Publishers, London, United Kingdom.
- 5.Clementino, M. M., I. Filippis, C. R. Nascimento, R. Branquinho, C. L. Rocha, and O. B. Martins. 2001. PCR analysis of tRNA intergenic spacer, 16S-23S internal transcribed spacer, and randomly amplified relationships of Enterobacter cloacae strains. J. Clin. Microbiol. 39:3865-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favret, M. E., and A. A. Yousten. 1985. Insecticidal activity of Bacillus laterosporus. J. Invertebr. Pathol. 45:195-203. [DOI] [PubMed] [Google Scholar]
- 7.FUNASA. 2000. Status of the prevention and control of transmissible diseases in Brazil (Situação da prevenção e controle das doenças transmissíveis no Brasil) [Online.] http://www.funasa.gov.br.
- 8.Gibson, T., and R. E. Gordon. 1974. Endospore-forming rods and cocci, p. 529-550. In R. E. Buchman and N. E. Gibbons (ed.), Bergey's manual of determinative bacteriology, 8th ed. Williams & Wilkins Co., Baltimore, Md.
- 9.Gordon, R. E., W. C. Haynes, and C. H. N. Pang. 1973. The genus Bacillus. ARS-USDA agriculture handbook no. 427, p. 1-283. U.S. Government Printing Office, Washington, D.C.
- 10.Gürtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 11.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, J. R., C. Clabots, M. Azar, D. J. Boxrud, J. M. Besser, and J. R. Thur. 2001. Molecular analysis of a hospital cafeteria-associated salmonellosis outbreak using modified repetitive element PCR fingerprinting. J. Clin. Microbiol. 39:3452-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiyama, T., T. Umino, Y. Nakamura, Y. Itezono, S. Sawairi, T. Satoh, and K. Yokose. 1994. Bacithrocins A, B and C, novel thrombin inhibitors. J. Antibiot. (Tokyo) 47:959-968. [DOI] [PubMed] [Google Scholar]
- 14.Lupski, J. R., and G. M. Weinstock. 1992. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J. Bacteriol. 174:4525-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melo, I. S., and J. L. Azevedo (ed.). 2000. Controle biológico (biological control). Embrapa Meio Ambiente, Jaguariúna, Brazil.
- 16.Montaldi, F. A., and I. L. Roth. 1990. Parasporal bodies of Bacillus laterosporus sporangia. J. Bacteriol. 172:2168-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nardo, E. A. B., and D. M. F. Capalbo. 1997. Avaliação ecotoxicológica de agentes microbianos de controle de pragas (ecotoxicological evaluation of microbial agents for pest control). O Biológico 59:63-68. [Google Scholar]
- 18.Navon, A. 2000. Bacillus thuringiensis application in agriculture, p. 355-367. In J.-F. Charles, A. Delécluse, and C. Nielsen-LeRoux (ed.), Entomopathogenic bacteria: from laboratory to field application. Kluwer Academic Publishers, London, United Kingdom.
- 19.Nemoto, K., M. Hayashi, J. Ito, F. Abe, T. Takita, T. Nakamura, T. Takeuchi, and H. Umezawa. 1987. Effect of spergualin in autoimmune disease in mice. J. Antibiot. 40:1448-1451. [DOI] [PubMed] [Google Scholar]
- 20.Oliveira, E. J., S. E. A. Silva, and L. Rabinovitch. 1998. A standardized protocol for the rapid detection of gelatin hydrolysis by Bacillus sphaericus. Isr. J. Entomol. 32:141-146. [Google Scholar]
- 21.Orlova, M. V., T. A. Smimova, L. A. Ganushkina, V. Y. Yacubovich, and R. R. Azizbekyan. 1998. Insecticidal activity of Bacillus laterosporus. Appl. Environ. Microbiol. 64:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabinovitch, L., C. M. B. Silva, R. S. A. Aives, R. A. G. B. Consoli, B. S. Santos, and M. A. Lamounier. 1998. Produção de bioinseticidas à base de Bacillus thuringiensis e Bacillus sphaericus, p. 479-783. Anais do VI SICONBIOL, Rio de Janeiro, Brazil.
- 23.Régis, L., and C. Nielsen-LeRoux. 2000. Management of resistance to bacterial vector control, p. 419-439. In J.-F. Charles, A. Delécluse, and C. Nielsen-LeRoux (ed.), Entomopathogenic bacteria: from laboratory to field application. Kluwer Academic Publishers, London, United Kingdom.
- 24.Régis, L., M. H. N. L. Silva-Filha, C. M. F. Oliveira, E. M. Rios, S. D. Silva, and A. F. Furtado. 1995. Integrated control measures against Culex quinquefasciatus, the vector of filariasis in Recife. Mem. Inst. Oswaldo Cruz 90:115-120. [DOI] [PubMed] [Google Scholar]
- 25.Rivers, D. B., C. N. Vann, H. L. Zimmack, and D. H. Dean. 1991. Mosquitocidal activity of Bacillus laterosporus. J. Invertebr. Pathol. 58:444-447. [DOI] [PubMed] [Google Scholar]
- 26.Serwecinska, L., T. Cieslikowski, M. Pytlos, A. Jaworski, and W. Kaca. 1998. Genomic fingerprinting of Proteus species using repetitive sequence based PCR (rep-PCR). Acta Microbiol. Pol. 47:313-319. [PubMed] [Google Scholar]
- 27.Seurinck, S., W. Verstraete, and S. D. Siciliano. 2003. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl. Environ. Microbiol. 69:4942-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shida, O., B. Takagi, K. Kadowaki, and K. Komagata. 1996. Proposal for two new genera, Brevibacillus gen. nov. and Aneurinibacillus gen. nov. Int. J. Syst. Bacteriol. 46:939-946. [DOI] [PubMed] [Google Scholar]
- 29.Shmidt, F. G. V., R. G. Monnerat, M. Borges, and R. S. Carvalho. 2001. Metodologia de criação de insetos para avaliação de agentes entomopatogênicos. Circular técnica no. 11. Embrapa, Brasília, Brazil.
- 30.Silva, K. R. A., L. Rabinovitch, and L. Seldin. 1999. Phenotypic and genetic diversity among Bacillus sphaericus strains isolated in Brazil, potentially useful as a biological control agent against mosquito larvae. Res. Microbiol. 150:153-160. [DOI] [PubMed] [Google Scholar]
- 31.Singer, S. 1996. The utility of strains of morphological group II Bacillus. Adv. Appl. Microbiol. 42:219-261. [DOI] [PubMed] [Google Scholar]
- 32.Singer, S. 1981. Potential of Bacillus sphaericus and related spore-forming bacteria for pest control, p. 283-292. In M. D. Burges (ed.), microbial control of pests and plant diseases. Academic Press, New York, N.Y.
- 33.Singer, S., T. B. Bair, T. B. Hammill, A. M. Verte, M. M. Correa-Ochoa, and A. D. Stambaugh. 1994. Fermentation and toxin studies of the molluscocidal strains of Bacillus brevis. J. Ind. Microbiol. 13:12-119. [DOI] [PubMed] [Google Scholar]
- 34.Smimova, T. A., I. B. Minenkova, M. V. Orlova, M. M. Lecadet, and R. R. Azizbekyan. 1996. The crystal-forming strains of Bacillus laterosporus. Res. Microbiol. 147:343-350. [DOI] [PubMed] [Google Scholar]
- 35.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 36.Thanabalu, T., and A. G. Porter. 1995. Efficient expression of a 100-kilodalton mosquitocidal toxin in protease-deficient recombinant Bacillus sphaericus. Appl. Environ. Microbiol. 61:4031-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umezawa, K., and T. Takeuchi. 1987. Spergualin: a new antitumour antibiotic. Biomed. Pharmacother. 41:227-232. [PubMed] [Google Scholar]
- 38.Versalovic, J., M. Schneider, F. J. Druijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 39.Versalovic, J., and J. R. Lupski. 2002. Molecular detection and genotyping of pathogens: more accurate and rapid answers. Trends Microbiol. 10:S15-S21. [DOI] [PubMed] [Google Scholar]
- 40.World Health Organization. 1987. Report of an informal consultation on the detection, isolation, identification and ecology of biocontrol agent disease vectors. TDR/BCV/IC-GE/87-3, annex II, p. 12. World Health Organization, Geneva, Switzerland.
- 41.Zahner, V., L. Rabinovitch, P. Suffys, and H. Momen. 1999. Genotypic diversity among Brevibacillus laterosporus strains. Appl. Environ. Microbiol. 65:5182-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]