Abstract
We characterized microbial biofilm communities developed over two very closely located but distinct benthic habitats in the Pensacola Bay estuary using two complementary cultivation-independent molecular techniques. Biofilms were grown for 7 days on glass slides held in racks 10 to 15 cm over an oyster reef and an adjacent muddy sand bottom. Total biomass and optical densities of dried biofilms showed dramatic differences for oyster reef versus non-oyster reef biofilms. This study assessed whether the observed spatial variation was reflected in the heterotrophic prokaryotic species composition. Genomic biofilm DNA from both locations was isolated and served as a template to amplify 16S rRNA genes with universal eubacterial primers. Fluorescently labeled PCR products were analyzed by terminal restriction fragment length polymorphism, creating a genetic fingerprint of the composition of the microbial communities. Unlabeled PCR products were cloned in order to construct a clone library of 16S rRNA genes. Amplified ribosomal DNA restriction analysis was used to screen and define ribotypes. Partial sequences from unique ribotypes were compared with existing database entries to identify species and to construct phylogenetic trees representative of community structures. A pronounced difference in species richness and evenness was observed at the two sites. The biofilm community structure from the oyster reef setting had greater evenness and species richness than the one from the muddy sand bottom. The vast majority of the bacteria in the oyster reef biofilm were related to members of the γ- and δ-subdivisions of Proteobacteria, the Cytophaga-Flavobacterium -Bacteroides cluster, and the phyla Planctomyces and Holophaga-Acidobacterium. The same groups were also present in the biofilm harvested at the muddy sand bottom, with the difference that nearly half of the community consisted of representatives of the Planctomyces phylum. Total species richness was estimated to be 417 for the oyster reef and 60 for the muddy sand bottom, with 10.5% of the total unique species identified being shared between habitats. The results suggest dramatic differences in habitat-specific microbial diversity that have implications for overall microbial diversity within estuaries.
Estuarine environments are among the most productive on earth, creating and processing more organic matter each year than comparably sized areas of reefs, forest, grassland, or agricultural land. This productivity supports the majority of the world's commercial and recreational fishery catch. In addition, these ecosystems are surrounded by high densities of human development that threaten the various estuarine habitats (i.e., salt marsh, sea grass, oyster reef) that contribute to this high productivity. Within each habitat, there are species that are characteristic to each and others that are less habitat specific. Thus, within-habitat diversity, or α-diversity, is determined by both species endemic to that habitat and the number of more widely distributed species supported within each habitat. The degree of habitat fidelity or endemism of species will largely determine the β-, or across-habitat, diversity within the estuarine ecosystem.
Investigations of microbial diversity have been limited by lack of appropriate technology. As the cultivability of marine microbes is estimated to be less then 0.1% (25), molecular marine ecology has been instrumental in the assessment of microbial diversity by the application of cultivation-independent diagnostic tools. A great deal of this effort has focused on unique and extreme environments (the open sea, hydrothermal vents, hot springs, etc.), while more mundane habitats close to human habitation are often overlooked. Despite the information being collected about microbial life, we still know little about microbial community structures, their correlations to biogeochemical processes, and their physiological capabilities. Some reports indicate that microhabitat differences as a result of macroscopic organism activity create unique microbial spatial patterns in terms of activity and diversity (2, 16, 17, 47).
Many microorganisms in estuarine environments are particle attached or associated with surfaces forming biofilms (7, 8, 24). These biofilms are complex communities of bacteria, protozoa, microalgae, and micrometazoa in a polymer matrix on submerged surfaces and suspended particles. The tremendous surface area available within intertidal marshes, sea grasses, and oyster reefs in addition to shell, sand, and mud benthos support microbial biofilms and significant aerobic and anaerobic microbial activities. Many of the biogeochemical reactions important to estuarine system functioning are associated with the benthos and particle-attached bacteria, which have been reported to show 10 to 100 times higher metabolic activities than free-living bacteria (7, 8, 24).
In this study, we investigated microbial α-diversity in biofilm communities influenced by an oyster reef and by the adjacent muddy sand bottom. It was hypothesized that the microbial biofilms grown in these habitats would integrate the resources and predation effects localized within each particular habitat and, thus, reflect localized productivity and diversity of available substrates. 16S rRNA libraries were constructed to classify operational taxonomic units (OTUs). Unique OTUs were subsequently sequenced to identify phylogenetic relations to known organisms. This approach was complemented by terminal restriction fragment length polymorphism (T-RFLP) analysis, which has turned out to be a highly reproducible and robust technique yielding high-quality fingerprints of bacterial assemblages (6, 37). Despite potential biases (51), PCR-based approaches like T-RFLP analysis have been shown to provide an accurate reflection of the ratios of 16S rRNA templates in model communities, with defined amounts of 16S rRNA gene copies from selected organisms (30). In the present case, we provide evidence that microbial biofilms analyzed by these techniques reflect habitat-specific responses of microbial communities and, thus, imply a high degree of microbial β-diversity within estuaries. The information content of these microbial biofilms may provide the basis for their use as indicators of estuarine habitat conditions.
MATERIALS AND METHODS
Study area and sample collection.
Polyvinyl chloride frames were constructed to hold slides (2.5 by 7.5 cm) and acrylic plates (19.5 by 9.5 cm) as artificial substrates for biofilm growth 10 to 15 cm off the bottom. Concrete was molded onto the polyvinyl chloride frame for anchors so that samplers would hold their position on the bottom. A 7-day incubation period was found to optimize microbial growth stimulated by allochthonous conditions and minimize colonization by calcareous invertebrates and macroalgae (data not shown). Oyster reef and adjacent muddy sand bottom samples were obtained from the open waters of Escambia Bay, Fla., in a region of scattered oyster reefs. The reef sample was obtained from 30°29′48.3"N, 87°06′53.8"W. The muddy sand bottom sample was obtained from 30°29′44.4"N, 87°06′36.7"W. The incubation covered the 1-week period from 22 to 29 August 2002. Both sites were 2.1 m in depth. The incubation period started with the spring part of the tidal cycle with a 1.94-ft tidal amplitude and extended through the neap tide, with a 0.17-ft amplitude. Temperature ranged from 30.3 to 32.8°C. Salinity ranged from 13.89 to 26.04 ppt.
Biofilm biomass determination by optical density.
Plates were scanned using an Epson 636 flat bed scanner with an Epson EU-14 transparency unit connected to a Power Macintosh G3 with Adobe Photoshop to digitize images. Image analysis was performed with NIH Image (National Institutes of Health) to provide means and standard deviations of pixel density (0 to 256 in gray scale) as relative measures of biomass accumulation and distribution over plate surfaces.
Genomic DNA extraction.
DNA was extracted from at least four independent slides and pooled before PCR amplification to incorporate any between-slide variability. The biofilm pellet was washed off filters with 2% NaCl and harvested by centrifugation at 7,000 rpm for 10 min in a microcentrifuge. Cells were resuspended in 500 μl of extraction buffer (0.05 M NaCl, 50 mM Tris-HCl [pH 7.5], 50 mM EDTA, 0.3 M sucrose, 1.5% hexadecyltrimethyl ammonium bromide) and sonicated with two 15-s bursts (power level 3; Ultrasonics micro tip 415 attached to a model W225R sonicator; Heat Systems Inc., Farmingdale, N.Y.) with cooling on ice between bursts. The mixture was then incubated in the presence of 5 mg of lysozyme/ml, 500 μg of pronase E/ml, 10 U of mutanolysin/ml, RNase A, and 0.4% sodium dodecyl sulfate (SDS) with occasional agitation. After 1 h at 37°C, proteinase K was added to a final concentration of 500 μg/ml, and the mixture was incubated at 37°C for 30 min and then another 30 min at 55°C. SDS concentration was increased to 2%, and NaCl was increased to 0.9 M. N-Laurylsarcosine was added to 1% final concentration, followed by an incubation at 65°C for 30 min. Samples were then cycled three times through freezing at −70°C and thawing at 65°C (5 min each). The cell debris was pelleted for 5 min at 13,000 rpm with a Heraeus Instruments Scientific Products Biofuge 13R centrifuge. For purification of the mixture, an equal volume of phenol (saturated with 10 mM Tris-HCl [pH 8.2]) was added to the supernatant, followed by two extractions with phenol-chloroform-isoamyl alcohol (25:24:1). Traces of phenol were removed by extraction with chloroform-isoamyl alcohol (24:1). Nucleic acids were recovered by addition of 0.7 volumes of 2-propanol and centrifugation for 30 min at 13,000 rpm at room temperature. The DNA pellets were rinsed with 70% ethanol, air dried, and resuspended in 50 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA).
Amplification of 16S rRNA genes.
Bacterial small subunit rRNA genes were selectively amplified from purified genomic DNA with the bacteria-specific forward primer 27F-TOPO (5′-CACCAGAGTTTGATC[A/C]TGGCTCAG-3′, corresponding to Escherichia coli positions 8 to 25) and the universal reverse primer 1492R (5′-GG[C/T]TACCTTGTTACGACTT-3′, corresponding to E. coli positions 1510 to 1492) (27). The 4-bp sequence CACC at the 5′ end of the forward primer provides the topoisomerase I recognition site, whereas the 3′ end stays blunt due to the lack of such an extension. This allows directional cloning into pENTR/TOPO vectors (Invitrogen Corp., Carlsbad, Calif.). For T-RFLP analysis, the forward primer 27F fam (5′-AGAGTTTGATC[A/C]TGGCTCAG-3′) was labeled at the 5′ end with 6-carboxyfluorescein. PCR amplification was performed by using 50-μl reaction mixtures containing 5 ng of sample DNA, 1.25 U of ExTaq polymerase (Takara.Mirus.Bio, Madison, Wis.), 1× ExTaq buffer (as supplied, containing 2 mM MgCl), a 2.5 mM concentration of each deoxynucleoside triphosphate, 50 μg of bovine serum albumin (Roche Diagnostics, Indianapolis, Ind.), and each primer at a concentration of 0.2 μM. Other reaction mixtures, identical except that they contained no target DNA, were used as negative controls and did not yield products. The PCR conditions used were 1.5 min of initial denaturation followed by 30 cycles at 94°C for 1 min, 45°C for 1 min, and 72°C for 90 s in a Techne thermal cycler with a final extension step of 7 min at 72°C. Product lengths were in the expected range of 1,500 to 1,600 bp. PCR products used for subsequent cloning were obtained using TGO polymerase (Roche Diagnostics). This polymerase with proofreading activity produces blunt ends required for cloning into pENTR/TOPO vectors. Reaction conditions were the same except for the use of the supplied TGO buffer (1× final concentration).
Cloning of 16S ribosomal DNA (rDNA) PCR products.
PCR products obtained from the amplification of total community DNA were separated on 0.8% agarose gels to excise a gel slice comprising products in the range from 1,500 to 1,600 bp in length. The DNA was purified using the QIAquick gel extraction kit (QIAGEN, Valencia, Calif.) following the manufacturer's instructions. DNA products were cloned with a pENTR directional TOPO cloning kit (Invitrogen Corp.) in accordance with the manufacturer's instructions. Ligation mixtures were used to transform competent E. coli TOP10 cells (supplied with the cloning kit). Recombinants were selected by using Luria-Bertani agarose plates containing 50 μg of kanamycin/ml.
Amplified rRNA gene restriction analysis (ARDRA).
Crude cell lysates were prepared from clones by suspending cells from colonies in 30 μl of TE buffer with subsequent lysis at 95°C for 10 min in a heat block. Preparations were centrifuged for 30 s at 10,000 × g, and 2 μl of the supernatant served as a template for PCR reamplification of inserts with primers M13F-20 (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) flanking the insertion sites. The thermal cycling program proceeded as follows: initial denaturation at 94°C for 90 s; 34 cycles consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s; and a final extension step of 72°C for 7 min. Amplicons were restriction digested at 37°C for 4 h in separate reactions using HhaI (CfoI; Roche Diagnostics). The restriction fragments were separated by electrophoresis on 3% (wt/vol) agarose gels (Fisher Scientific) in 1× TAE (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA), stained with ethidium bromide, and visualized with UV illumination. Band patterns were digitized and grouped by similarity using the GelCompare II software package (Applied Maths, Kortrijk, Belgium). PCR products yielding similar patterns were further restricted with MspI and RsaI (both from Roche Diagnostics) to define OTUs. The OTUs were designated ORB1 to ORB188 (for Oyster Reef biofilm) and SBB1 to -153 (for sand bottom biofilm). The estimated percentages of coverage for the two sites studied were calculated as follows: [1 − (n/N)] × 100, where n is the number of unique clones detected in a library of size N. Total species richness for the two habitats (α-diversity) was estimated by fitting the equation y = x/(ax + b), where y is the cumulative number of distinct OTUs, x is number of clones, and a and b are coefficients (44). The cumulative number of unique clones identified was plotted as a function of the cumulative number of clones processed. Inversion of the equation results in 1/y = a + b/x, meaning that for x→infinity, y approaches 1/a. Coefficients a and b were calculated by plotting 1/y against 1/x for the two communities, allowing the extrapolation of the saturation curve and the estimation of species richness.
16S rDNA sequencing and sequence analysis.
E. coli transformants carrying plasmids with 16S rDNA inserts representing unique OTUs identified by ARDRA were grown overnight in Luria-Bertani broth with 50 μg of kanamycin/ml. Plasmid DNA was prepared from the clones with a QIAprep spin miniprep kit (QIAGEN) according to the manufacturer's instructions. High-throughput sequencing was carried out at the Genomics Technology Support Facility (Michigan State University, Lansing) with the M13F-20 primer (5′-GTAAAACGACGGCCAG-3′). rDNA sequences with a range of about 600 to 800 bases were obtained for most clones. All sequences were checked for chimeric artifacts by using the CECK_CHIMERA program in the Ribosomal Database Project II (RDP-II [31]). For sequences that were identified as potentially chimeric, inconsistent secondary structures were checked for potential abnormalities. Sequences with good continuous homology in the critical fusion region with other aligned sequences were regarded as nonchimeric.
Phylogenetic analysis of cloned 16S rDNA sequences.
Unaligned sequences were entered into the BLAST search program (Blastcl3) of the National Center for Biotechnology Information in order to obtain closely related phylogenetic sequences. Phylogenetic trees were constructed with the ClustalW software (available from the server at http://www.ebi.ac.uk/clustalw/) (48) and were displayed using TREEVIEW software (39).
T-RFLP analysis.
16S rRNA genes from community DNA were amplified with primers 27F fam and 1492R. PCR products were visualized on an ethidium bromide-stained 1% agarose gel. Roughly 250 to 300 ng of product (as estimated with a size standard of known concentration) was digested using the same restriction enzymes and conditions as for ARDRA. The digested DNA was precipitated with a 0.1 volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of 99% ethanol, followed by pelleting at 13,000 rpm at 4°C for 30 min with a Heraeus Instruments Scientific Products Biofuge 13R centrifuge. The DNA pellet was washed with 70% ethanol, dried, and resuspended in 10 μl of water. Ten microliters of formamide and 0.5 μl of internal DNA standard (Mapmarker 1000; Bioventures Inc., Murfreesboro, Tenn.) were added prior to denaturation at 95°C for 5 min. The fluorescent label of the terminal fragments was detected on an ABI Prism 310 genetic analyzer (Applied Biosystems) in GeneScan mode (15 kV; 5-s injection; 60°C for 40 min). T-RFs with a peak height less than 50 fluorescence units were excluded from the analysis. Fragment sizes were estimated by using the local Southern method in the GeneScan 3.1 software (Applied Biosystems). The ABI peaks were transformed into abundance peaks based on relative peak areas of the individual signals. Abundance patterns were graphically presented using SigmaPlot software (SPSS).
RESULTS
Growth of biofilms and optical analysis.
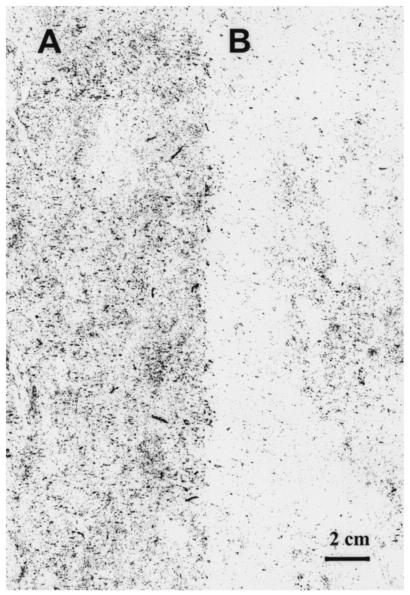
Benthic biofilms from an oyster reef and a muddy sand bottom were harvested after a 7-day incubation. Analysis of digitized images of representative dried substrates indicated the oyster reef biofilms had nearly twice as much biomass by density than the muddy sand bottom biofilms (Fig. 1).
FIG. 1.
Seven-day-old biofilms grown on acrylic plates (19.5 by 9.5 cm) as artificial substrates placed on an oyster reef (A) and on the adjacent sandy mud bottom (B) at an equivalent depth. Average pixel densities ± standard deviations of these biofilms are 43.69 ± 34.90 (oyster reef) and 22.30 ± 17.31 (sandy mud), making the oyster reef sample more dense yet also more variable in biomass coverage. Tube-dwelling amphipods were apparent in the oyster reef sample (dark short lines) but missing from the adjacent sandy mud bottom sample.
Construction and analysis of 16S rDNA libraries.
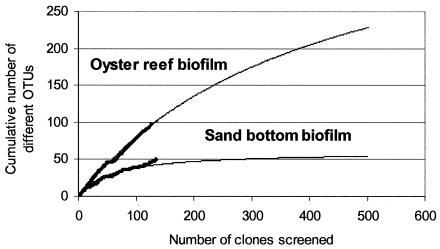
The extracted genomic DNA was amenable to direct amplification of 16S rRNA genes (16S rDNA). The nearly full-length (approximately 1,550-bp) PCR products were cloned in a directional manner and reamplified. Two clone libraries were constructed, representing the bacterial biofilm communities at the two locations. The diversity of the recovered 16S rDNA PCR fragments in the libraries was examined by ARDRA using HhaI as the primary enzyme to distinguish between different OTUs. PCR products with similar patterns were further analyzed by MspI and RsaI restriction to unequivocally distinguish between different OTUs. The restriction patterns were used as a measure of diversity of the two clone libraries. A total of 130 clones were screened for the oyster reef, and 136 clones were screened for the muddy sand bottom, leading to the identification of 97 and 50 different OTUs, respectively. Fourteen OTUs from each biofilm, or 10.53% of the total unique OTUs recovered, occurred in both biofilms. The two libraries were estimated by curve fitting to have recovered 23 and 63% of the total species from oyster reef and muddy sand bottom, respectively. The differences in microbial diversities estimated by plotting the cumulative number of unique OTUs against the number of clones analyzed for each site are shown in Fig. 2. According to this calculation, the possible total number of OTUs was estimated to approach 417 different OTUs for the oyster reef biofilm but only 60 different OTUs for the sandy mud bottom biofilm.
FIG. 2.
Estimation of microbial diversities in biofilms grown over 7 days in an oyster reef setting and a sandy mud bottom setting. The numbers of the cumulative different or unique OTUs were plotted against the number of clones screened. The bold curves were calculated based on the experimental data. The thin lines indicate an extrapolation leading to an estimated diversity of 417 OTUs for the oyster reef biofilm and 60 OTUs for the muddy sand bottom biofilm.
16S rDNA sequence analysis.
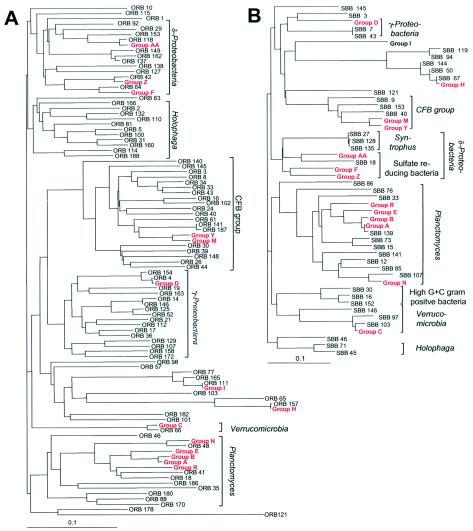
All clones representing distinguishable OTUs were subjected to partial sequence analysis. Four chimeric sequences (three from the oyster reef biofilm and one from the sand bottom biofilm) were identified and were excluded from subsequent analysis. The remaining sequences were aligned, and their phylogenetic positions relative to known sequences were determined. Sequences showed similarities to database entries in the range between 83 and 99%. None was completely identical to any 16S rDNA sequence of a cultured organism or to an environmental clone available from GenBank, indicating that the clone sequences were derived from unknown taxa. This is not surprising, given the immense number of bacteria in the environment, of which only a relatively small have been sequenced and are included in public databases. A total of 122 of the 133 identified unique OTUs were less than 98% similar to database entries. Sequence similarities are presented in phylogenetic trees (Fig. 3).
FIG. 3.
Phylogenetic relationships among 16S rDNA sequences of bacterial clones from biofilms from an oyster reef (A) and a sandy mud bottom (B). OTUs which were found more than once in either of the two communities were classified as a group (A to Z and AA). Groups with members in both communities are indicated in red. Putative divisions are listed outside the brackets. Scale bars represent 0.1 nucleotide substitutions per nucleotide position.
Oyster reef biofilm diversity.
The results of the phylogenetic comparison are summarized in Table 1. Sequenced clones fell into five major lineages of the domain Bacteria: the γ- and δ-subdivisions of Proteobacteria (22.2 and 12.6%), the Cytophaga-Flavobacterium-Bacteroides (CFB) group (22.2%), the Planctomyces phylum (12.7%), and the Holophaga-Acidobacterium phylum (9.5). Two sequences were affiliated with the phylum Verrucomicrobia (2.4%). All other sequences which could not be assigned to any of these groups were reported as “others” (19%).
TABLE 1.
Distribution of OTUs in the oyster reef biofilm 16S rDNA clone library and phylogenetic relationships
| Group and OTUa | 16S rDNA identification of closest neighbor (accession no.) | Sequence similarity (%) | Abundanceb in community (%) | Length of 5′ T-RFd (HhaI, in bp) |
|---|---|---|---|---|
| γ-Proteobacteria | ||||
| Group Dc (ORB 49) | Uncultured bacterium Br-z23 (AF506993) | 99 | 3.97 | 211.5 |
| Group P (ORB 19) | Beta proteobacterium A1020 (AF236013) | 93 | 3.17 | 145.0 |
| Group S (ORB 14) | Uncultured proteobacterium clone Bol37 (AY193142) | 97 | 1.59 | 96.6 |
| Group U (ORB 154) | Gamma proteobacterium PII_GH4.2.G5 (AY162068) | 99 | 1.59 | 211.8 |
| ORB 4 | Uncultured bacterium Br-z23 (AF506993) | 99 | 0.79 | ND |
| ORB 17 | Unidentified gamma proteobacterium (AB015254) | 92 | 0.79 | ND |
| ORB 21 | Dechloromarinus chlorophilus (AF170359) | 95 | 0.79 | ND |
| ORB 36 | Unidentified gamma proteobacterium (AB015254) | 92 | 0.79 | ND |
| ORB 52 | Uncultured gamma proteobacterium Sva0091 (AJ240987) | 96 | 0.79 | ND |
| ORB 101 | Uncultured bacterium clone Hw124 (AF497583) | 92 | 0.79 | ND |
| ORB 107 | Pseudoalteromonas sp. AS-43 (AJ391204) | 99 | 0.79 | ND |
| ORB 112 | Uncultured gamma proteobacterium partial (AJ535246) | 95 | 0.79 | ND |
| ORB 125 | Uncultured gamma proteobacterium Sva0091 (AJ240987) | 95 | 0.79 | ND |
| ORB 129 | Thalassomonas viridans (AJ294747) | 95 | 0.79 | ND |
| ORB 146 | Uncultured proteobacterium clone Bol37 (AY193142) | 98 | 0.79 | ND |
| ORB 158 | Gamma proteobacterium MBIC3957 (AB021682) | 86 | 0.79 | ND |
| ORB 163 | Uncultured beta proteobacterium partial (AJ422174) | 89 | 0.79 | ND |
| ORB 172 | Marine gamma proteobacterium MKT112 gene (AB076562) | 86 | 0.79 | ND |
| ORB 182 | H. obtusa (X58198) | 88 | 0.79 | ND |
| Total for group | 22.17 | |||
| δ-Proteobacteria | ||||
| Group Fc (ORB 23) | Desulfobacterium catecholicum (AJ237602) | 95 | 2.38 | 91.2 |
| Group Zc (ORB 143) | Uncultured delta proteobacterium Sva1041 (AJ240984) | 94 | 0.79 | 90.8 |
| Group AAc (SBB 68) | Olavius algarvensis sulfate-reducing endosymbiont (AF328857) | 94 | 0.79 | 92.0 |
| ORB 29 | Uncultured delta proteobacterium partial (AJ535245) | 96 | 0.79 | ND |
| ORB 42 | Uncultured delta proteobacterium Sva1041 (AJ240984) | 93 | 0.79 | ND |
| ORB 64 | Unidentified eubacterium (AJ007373) | 92 | 0.79 | ND |
| ORB 92 | Uncultured delta proteobacterium Sva0485 (AJ241001) | 95 | 0.79 | ND |
| ORB 118 | Olavius algarvensis sulfate-reducing endosymbiont (AF328857) | 93 | 0.79 | ND |
| ORB 127 | Geobacter sp. Ala-6 (AF019929) | 87 | 0.79 | ND |
| ORB 137 | Uncultured delta proteobacterium Sva1033 (AJ240983) | 95 | 0.79 | ND |
| ORB 138 | Bdellovibrio sp. JS10 (AF084863) | 91 | 0.79 | ND |
| ORB 149 | Uncultured delta proteobacterium Sva0566 (AJ241000) | 98 | 0.79 | ND |
| ORB 153 | Delta proteobacterium S2552 (AF468969) | 89 | 0.79 | ND |
| ORB 162 | Uncultured delta proteobacterium Sva0566 (AJ241000) | 94 | 0.79 | ND |
| Total for group | 12.65 | |||
| CFB group | ||||
| Group K (ORB 30) | Uncultured CFB group bacterium FL13A07 (AF446287) | 91 | 2.38 | 91.6 |
| Group X (ORB 34) | CFB group bacterium RW262 (AF493694) | 93 | 2.38 | >1,000 |
| Group W (ORB 39) | Uncultured bacterium clone s22 (AY171331) | 96 | 1.59 | 93.0 |
| Group V (ORB 43) | CFB group bacterium RW262 (AF493694) | 90 | 1.59 | 95.6 |
| Group L (ORB 141) | Flavobacteriaceae strain SW084 (AF493687) | 93 | 1.59 | 93.9 |
| Group Yc (ORB 119) | Salt marsh clone LCP-72 (AF286039) | 97 | 0.79 | 96.3 |
| Group Mc (ORB 120) | Salt marsh clone LCP-72 (AF286039) | 96 | 0.79 | 96.0 |
| ORB 3 | Uncultured CFB group bacterium (AJ441241) | 90 | 0.79 | ND |
| ORB 8 | Uncultured Cytophagales (AF361205) | 92 | 0.79 | ND |
| ORB 16 | Uncultured CFB group bacterium partial (AJ441238) | 93 | 0.79 | ND |
| ORB 24 | Uncultured bacterium clone ARKIA-105 (AF468278) | 89 | 0.79 | ND |
| ORB 26 | Uncult. CFB group bacterium clone SM2D04 (AF445721) | 91 | 0.79 | ND |
| ORB 33 | CFB group bacterium RW262 (AF493694) | 90 | 0.79 | ND |
| ORB 40 | Uncultured Cytophagaceae bacterium clone 1-13 (AY094494) | 90 | 0.79 | ND |
| ORB 44 | Uncultured CFB group bacterium clone SM2D04 (AF445721) | 91 | 0.79 | ND |
| ORB 61 | Flavobacteriaceae bacterium BIA (AY177722) | 92 | 0.79 | ND |
| ORB 102 | Uncultured bacterium clone CARB_ESS_9 (AY239540) | 90 | 0.79 | ND |
| ORB 140 | Uncultured Cytophagales bacterium clone CD4G5 (AY038505) | 90 | 0.79 | ND |
| ORB 145 | Bacteroidetes bacterium GMD13F04 (AY162116) | 89 | 0.79 | ND |
| ORB 148 | Unidentified Cytophagales partial (AJ007871) | 88 | 0.79 | ND |
| ORB 187 | Flavobacteriaceae strain SW084 (AF493687) | 93 | 0.79 | ND |
| Total for group | 22.17 | |||
| Planctomyces phylum | ||||
| Group Bc (ORB 9) | Uncultured planctomycete EC-149 (AF287048) | 97 | 1.59 | 561.5 |
| Group Ac (SBB 58) | Uncultured bacterium clone 33-FL67B99 (AF469406) | 95 | 1.59 | 560.8 |
| Group Nc (ORB 6) | Uncultured planctomycete EC-149 (AF287048) | 96 | 0.79 | >1,000 |
| Group Rc (ORB 97) | Uncultured bacterium gene (AB100005) | 95 | 0.79 | 842.6 |
| Group Ec (ORB 156) | Uncultured bacterium clone 33-FL67B99 (AF469406) | 92 | 0.79 | 560.5 |
| ORB 18 | Uncultured planctomycete partial (BX294700) | 92 | 0.79 | ND |
| ORB 35 | Uncultured bacterium #0319-7F4 (AF234144) | 89 | 0.79 | ND |
| ORB 41 | Uncultured Pirellula clone 6N14 (AF029078) | 95 | 0.79 | ND |
| ORB 48 | Planctomyces maris (strain DSM 8797T) (AJ231184) | 91 | 0.79 | ND |
| ORB 88 | Uncultured Crater Lake bacterium CL500-15 (AF316773) | 92 | 0.79 | ND |
| ORB 103 | Uncultured planctomycete partial (BX294885) | 95 | 0.79 | ND |
| ORB 170 | Uncultured planctomycete partial (BX294710) | 89 | 0.79 | ND |
| ORB 180 | Uncultured planctomycete partial (BX294710) | 88 | 0.79 | ND |
| ORB 186 | Uncultured soil bacterium clone S165 (AF507705) | 88 | 0.79 | ND |
| Total for group | 12.66 | |||
| Holophaga-Acidobacterium phylum | ||||
| Group O (ORB 2) | Uncultured bacterium clone NMS8.79WL (AF432679) | 93 | 1.59 | 355.9 |
| ORB 5 | Uncultured Holophaga-Acidobacterium Sva0725 (AJ241003) | 93 | 0.79 | ND |
| ORB 31 | Sulfate-reducing bacteria gene for 16S rDNA (X80922) | 86 | 0.79 | ND |
| ORB 81 | Holophaga sp. oral clone CA002 (AF385537) | 87 | 0.79 | ND |
| ORB 100 | Uncultured Holophaga-Acidobacterium Sva0725 (AJ241003) | 96 | 0.79 | ND |
| ORB 110 | Uncultured Holophaga sp. (AJ535239) | 95 | 0.79 | ND |
| ORB 114 | Uncultured Holophaga-Acidobacterium Sva0515 (AJ241004) | 92 | 0.79 | ND |
| ORB 132 | Hydrothermal vent eubacterium PVB_OTU_9A (U15118) | 95 | 0.79 | ND |
| ORB 160 | Uncultured Holophaga-Acidobacterium Sva0725 (AJ241003) | 89 | 0.79 | ND |
| ORB 166 | Uncultured delta proteobacterium clone WCB41 (AY217485) | 93 | 0.79 | ND |
| ORB 188 | Uncultured Holophaga-Acidobacterium Sva0515 (AJ241004) | 89 | 0.79 | ND |
| Total for group | 9.49 | |||
| Verrucomicrobia | ||||
| Group Cc (ORB 15) | Unidentified eubacterium LD29 (AF009975) | 96 | 1.59 | 218.6 |
| ORB 66 | Unidentified eubacterium LD29 (AF009975) | 95 | 0.79 | ND |
| Total for group | 2.38 | |||
| Others | ||||
| Group J (ORB 77) | Odontella sinensis complete chloroplast genome (Z67753) | 96 | 3.97 | 843.1 |
| Group Ic (ORB 123) | Uncultured marine eubacterium HstpL35 (AF159636) | 97 | 2.38 | >1,000 |
| Group Q (ORB 57) | Uncultured delta proteobacterium CtaxPhil-2 (AF259630) | 96 | 1.59 | 25.7 |
| Group Hc (SBB 138) | Bacterial sp. (X89322) | 97 | 0.79 | 80.9 |
| ORB 1 | Uncultured myxobacterium Sva1009 (AJ297456) | 95 | 0.79 | ND |
| ORB 10 | Salt marsh clone LCP-68 16S (AF286033) | 96 | 0.79 | ND |
| ORB 46 | Uncultured soil bacterium PBS-II-37 partial (AJ390447) | 90 | 0.79 | ND |
| ORB 63 | Uncultured proteobacterium clone ccs202 (AY133065) | 90 | 0.79 | ND |
| ORB 65 | Bacterial sp. gene for 16S rRNA (isolate 4911) (X89322) | 93 | 0.79 | ND |
| ORB 98 | Uncultured high-G + C gram-positive bacterium Sva0389 (AJ240976) | 93 | 0.79 | ND |
| ORB 111 | Uncultured marine eubacterium HstpL35 (AF159636) | 97 | 0.79 | ND |
| ORB 115 | Uncultured bacterium clone Bol11 (AY193132) | 92 | 0.79 | ND |
| ORB 121 | Uncult. candidate division WS3 bacterium clone LD1-PA13 (AY114311) | 91 | 0.79 | ND |
| ORB 157 | Bacterial sp. gene for 16S rRNA (isolate 4911) (X89322) | 97 | 0.79 | ND |
| ORB 165 | Uncultured marine eubacterium HstpL35 (AF159636) | 98 | 0.79 | ND |
| ORB 178 | Uncultured Acidobacterium-Holophaga (AF423363) | 96 | 0.79 | ND |
| Total for group | 19.00 |
OTUs were named with the prefix ORB for the oyster reef biofilm and SBB for the muddy sand bottom habitat. Groups comprise more than one member.
Relative abundance was calculated as follows: relative abundance = (n/N) × 100, where n is the number of clones representing the same OTU and N is the total number of clones in the library. The added relative abundances for each phylogenetic group are shown in bold.
OTU shared between biofilms from the oyster reef habitat and the muddy sand bottom habitat.
Lengths of 5′ T-RFs were experimentally determined.
Sandy mud bottom biofilm diversity.
A summary of ARDRA data from the sandy mud bottom site is listed in Table 2. This library exhibited five highly abundant OTUs, which could be assigned to the groups of the Planctomyces phylum (OTUs B and A, with abundances of around 15%, and OTU E, with 7.4%), the Verrucomicrobia (OTU C, with 13.3%), and the γ-subdivision of Proteobacteria (OTU D, with 9.6%). The numeric dominance of these OTUs rendered the corresponding phylogenetic groups most abundant, with the Planctomyces phylum making up nearly 46% of the community. The Planctomyces group also represented the most diverse subgroup in this habitat, with 14 members. Minor portions were affiliated to the δ-Proteobacteria and the CFB group (both 3.7%) and the Holophaga-Acidobacterium division (1.5%). Other OTUs which could not be affiliated with any of these groups are listed as others (14.1%).
TABLE 2.
Distribution of OTUs in the sand bottom biofilm 16S rDNA clone library and phylogenetic relationships
| Group and OTUa | 16S rDNA identification of closest neighbor (accession no.) | Sequence similarity (%) | Abundanceb in community (%) | Length of 5′ T-RFd (HhaI, in bp) |
|---|---|---|---|---|
| γ-Proteobacteria | ||||
| Group Dc (ORB 49) | Uncultured bacterium Br-z23 (AF506993) | 99 | 9.63 | 211.5 |
| SBB 7 | Pseudoalteromonas sp. AS-43 (AJ391204) | 99 | 0.74 | ND |
| SBB 43 | Pseudoalteromonas sp. AS-43 (AJ391204) | 99 | 0.74 | ND |
| Total for group | 11.11 | |||
| δ-Proteobacteria | ||||
| Group Fc (ORB 23) | Desulfobacterium catecholicum (AJ237602) | 95 | 1.48 | 91.2 |
| Group Zc (ORB 143) | Uncultured delta proteobacterium Sva1041 (AJ240984) | 94 | 0.74 | 90.8 |
| Group AAc (SBB 68) | Olavius algarvensis sulfate-reducing endosymbiont (AF328857) | 94 | 0.74 | 92.0 |
| SBB 18 | Uncultured Desulfuromonadales bacterium (AY177804) | 89 | 0.74 | ND |
| Total for group | 3.7 | |||
| CFB group | ||||
| Group Mc (ORB 120) | Salt marsh clone LCP-72 (AF286039) | 96 | 0.74 | 96.0 |
| Group Yc (ORB 119) | Salt marsh clone LCP-72 (AF286039) | 97 | 0.74 | 96.3 |
| SBB 9 | Uncultured bacterium partial clone Hyd89-65 (AJ535255) | 96 | 0.74 | ND |
| SBB 40 | Flexibacter aggregans subsp. catalaticus gene (AB078042) | 94 | 0.74 | ND |
| SBB 153 | CFB group bacterium RW262 (AF493694) | 88 | 0.74 | ND |
| Total for group | 3.7 | |||
| Planctomyces phylum | ||||
| Group Bc (ORB 9) | Uncultured planctomycete EC-149 (AF287048) | 97 | 15.56 | 561.5 |
| Group Ac (SBB 58) | Uncultured bacterium clone 33-FL67B99 (AF469406) | 96 | 14.81 | 560.8 |
| Group Ec (ORB 156) | Uncultured bacterium clone 33-FL67B99 (AF469406) | 92 | 7.41 | 560.5 |
| Group Nc (ORB 6) | Uncultured planctomycete EC-149 (AF287048) | 96 | 0.74 | >1,000 |
| Group Rc (ORB 97) | Uncultured bacterium gene (AB100005) | 95 | 0.74 | 842.6 |
| SBB 12 | Uncultured planctomycete clone CY0ARA031G01 (BX294851) | 90 | 0.74 | ND |
| SBB 15 | Uncultured Pirellula clone 6N14 (AF029078) | 95 | 0.74 | ND |
| SBB 33 | Marine eubacterial sp. (aggregate agg8) PCR generated (L10942) | 94 | 0.74 | ND |
| SBB 73 | Uncultured Pirellula clone 6N14 (AF029078) | 93 | 0.74 | ND |
| SBB 76 | Uncultured bacterium clone SG2-79 (AY135912) | 93 | 0.74 | ND |
| SBB 85 | Uncultured Planctomyces clone 7F15 (AF029079) | 98 | 0.74 | ND |
| SBB 107 | Uncultured planctomycete EC-149 (AF287048) | 97 | 0.74 | ND |
| SBB 139 | Uncultured bacterium clone 33-FL67B99 (AF469406) | 97 | 0.74 | ND |
| SBB 141 | Planctomyces sp. partial (X81956) | 91 | 0.74 | ND |
| Total for group | 45.92 | |||
| Holophaga-Acidobacterium | ||||
| SBB 45 | Uncultured Holophaga-Acidobacterium Sva0725 (AJ241003) | 96 | 0.74 | ND |
| SBB 71 | Uncultured Holophaga-Acidobacterium Sva0725 (AJ241003) | 93 | 0.74 | ND |
| Total for group | 1.48 | |||
| Verrucomicrobia | ||||
| Group Cc (ORB 15) | Unidentified eubacterium LD29 (AF009975) | 96 | 13.33 | 218.6 |
| SBB 97 | Unidentified eubacterium LD29 (AF009975) | 93 | 0.74 | ND |
| SBB 103 | Unidentified eubacterium LD29 (AF009975) | 95 | 0.74 | ND |
| SBB 146 | Uncultured Verrucomicrobia Arctic 95D-9 (AY028220) | 92 | 0.74 | ND |
| Total for group | 15.55 | |||
| Syntrophus | ||||
| SBB 27 | Unidentified bacterium partial (AJ518378) | 91 | 0.74 | ND |
| SBB 128 | Unidentified bacterium partial, clone Neu4P1-54 (AJ518378) | 91 | 0.74 | ND |
| SBB 135 | Unidentified bacterium partial, clone Neu4P1-54 (AJ518378) | 90 | 0.74 | ND |
| Total for group | 2.22 | |||
| Gram-positive branch | ||||
| SBB 16 | Uncultured high-G + C gram-positive bacterium Sva1007 (AJ241019) | 92 | 0.74 | ND |
| SBB 30 | Uncultured hydrocarbon seep bacterium BPC063 (AF154093) | 96 | 0.74 | ND |
| SBB 152 | Uncultured high-G + C gram-positive bacterium Sva0389 (AJ240976) | 96 | 0.74 | ND |
| Total for group | 2.22 | |||
| Others | ||||
| Group G (SBB 94) | Bacterial sp. (X89322) | <300 bp | 3.7 | 212.6 |
| Group Hc (SBB 138) | Bacterial sp. (X89322) | <200 bp | 2.22 | 80.9 |
| Group T (SBB 144) | Bacterial sp. (X89322) | <200 bp | 1.48 | 81.0 |
| Group Ic (ORB 123) | Uncultured marine eubacterium HstpL35 (AF159636) | 97 | 0.74 | >1,000 |
| SBB 3 | Uncultured bacterium clone BT60DS4BD12 (AF365742) | 98 | 0.74 | ND |
| SBB 46 | Uncultured bacterium partial, clone JG34-KF-153 (AJ532721) | 91 | 0.74 | ND |
| SBB 50 | Bacterial sp. gene (isolate 4911) (X89322) | 96 | 0.74 | ND |
| SBB 67 | Bacterial sp. gene (X89322) | 96 | 0.74 | ND |
| SBB 86 | Uncultured bacterium partial (AJ306763) | 90 | 0.74 | ND |
| SBB 119 | Bacterial sp. gene (X89322) | 93 | 0.74 | ND |
| SBB 121 | Uncultured epsilon proteobacterium clone 33-FL58B00 (AF468777) | 93 | 0.74 | ND |
| SBB 145 | Uncultured bacterium gene clone Rs-H34 (AB089123) | 91 | 0.74 | ND |
| Total for group | 14.06 |
OTUs were named with the prefix 5 for the oyster reef habitat and the prefix 6 for the muddy sand bottom habitat. Groups comprise more than one member.
Relative abundance was calculated as follows: relative abundance = (n/N) × 100, where n is the number of clones representing the same OTU and N is the total number of clones in the library. The added relative abundances for each phylogenetic group are shown in bold.
OTU shared between biofilms from the oyster reef habitat and the muddy sand bottom habitat.
Lengths of 5′ T-RFs were experimentally determined.
T-RFLP.
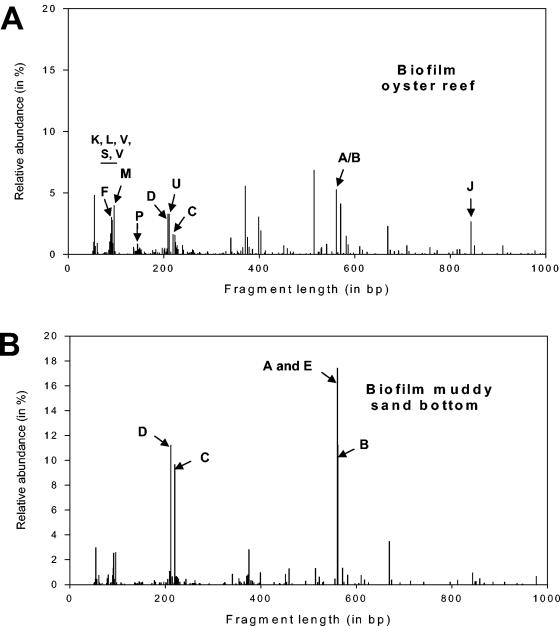
Bacterial community structures were further analyzed by performing T-RFLP analysis, producing molecular fingerprints. The amplified DNA was the same as that used for the ARDRA. Almost identical community profiles were consistently produced from the same sample. Similarly, only little variation was observed as a result of variation between replicate PCR amplifications or restriction digestions. The community structure of the oyster reef biofilm (Fig. 4A) appeared to be much more diverse, with a higher number of peaks and a more even distribution. The biofilm from the muddy sand bottom (Fig. 4B), on the other hand, appeared to be a lot less diverse and was dominated by five terminal fragments (A, B, C, D, and E). These T-RFs could be assigned to the corresponding community members by T-RFLP analysis of the individual clones (data not shown). OTUs A, B, and E all produced fragments of 561 bp, indicating that some of the peaks in the T-RFLP patterns were derived from multiple species.
FIG. 4.
Histograms of 5′ T-RFs generated by HhaI restriction of PCR-amplified 16S rRNA genes from microbial biofilm communities grown in an oyster reef habitat (A) and a muddy sand bottom habitat (B). Fragment length (in base pairs) is shown on the x axis up to 1,000 bp. Abundance (peak area of individual T-RF signal as a percentage of total peak area of all T-RF signals) is shown on the y axis. Arrows indicate T-RFs, which could be assigned to bacterial OTUs. The OTUs K, L, V, S, and V could not be assigned due to signal clustering in the corresponding length ranges. Their approximate location is indicated by the bar in panel A.
DISCUSSION
This study has found substantial differences in the composition of microbial communities, based on biomass accumulation and the two components of ecological diversity, richness and evenness, in biofilms grown on defined surfaces over two very closely located but distinct habitats. The diversity estimation (Fig. 2) suggests that the microbial content of the biofilm in the oyster reef habitat is about seven times as species rich as the non-oyster biofilm, although the curve fit may have slightly underestimated the species richness of the muddy sand biofilm. Given the short incubation time (7 days) of the glass slides, it was assumed that the majority of the community members would by necessity have been actively growing or colonizing the surface in significant numbers from the source habitats. The microbial communities sampled on these standardized substrates were also assumed to reflect integration of the environmental conditions of these habitats but were not intended to be comprehensive samples of the inherent prokaryotic diversity.
The clone library approach with subsequent sequencing and T-RFLP analysis proved to be complementary molecular techniques for the assessment of microbial diversity within these samples. Like other PCR-based techniques, they contain several sources of well-known inherent biases. These include potential variability in rDNA copy number between organisms (ranging from 1 to 14 [15]), different efficacies of cell lysis during the genomic DNA extraction, different amplifiabilities of template molecules, and chimera formation (4, 28, 45, 51). Although one has to bear in mind that the techniques utilizing the amplification of 16S rDNA may not exactly reflect the relative abundances of organisms in the original samples, they still provide a valuable means to assess microbial community structures. Possible biases were minimized by pooling PCR products from several independent reactions and by applying a relatively harsh lysis procedure with a combination of sonication and enzymatic and detergent treatment, as well as freeze-thawing. Extraction efficacy was not maximized at the expense of shearing, compared with other milder methods (data not shown). Fragmentation of template DNA greatly increases chimera formation (26, 38). Although only five chimeric sequences were identified positively, the CHECK_CHIMERA software revealed that chimeras may have been formed for some additional sequences. However, none of the methods for chimera detection is foolproof.
The community composition suggested by ARDRA data correlates nicely with the community composition suggested by T-RFLP analysis. By applying the same scale in the two abundance patterns (Fig. 4), the evenness of the oyster reef biofilm was pronounced relative to that of the sand bottom biofilm, dominated by five highly abundant OTUs. Species richness of communities can theoretically also be estimated from a T-RFLP fingerprint by determining the number of T-RFs. However, this requires that identical DNA amounts be subjected to T-RFLP analysis, as the number of peaks above the noise threshold (we chose 50 relative fluorescence units) is dependent on the amount of digested DNA. We loaded 250 to 300 ng of digested 16S rDNA amplicons on the automated sequencer and compared three independent T-RFLP analyses. The HhaI digests resulted in an average of 162 T-RFs for the oyster reef biofilm, compared to 123 T-RFs for the sand bottom biofilm, indicating a higher degree of complexity in “ribotype diversity” for the former. However, this approach significantly underestimates species richness as determined from ARDRA, a result of rare species producing weak signals below the detection threshold of the T-RFLP analysis and the superimposition of multiple species within some peaks.
Analysis of the ARDRA data revealed that bacteria with closest phylogenetic neighbors among the Planctomyces phylum formed by far the most abundant group in the sandy bottom biofilm. The surprisingly strong dominance was mainly due to three OTUs, called groups A, B, and E. Members of the environmentally important order Planctomycetales are ubiquitously distributed in both terrestrial habitats and in fresh- and saltwater environments and may be correlated with slight eutrophication (18, 19, 29, 36, 50, 52). They are often attached to particles (13) and may function as fundamental components in the anaerobic degradation of organic compounds (18).
The other prominent OTUs (groups C and D) identified both by ARDRA and T-RFLP in the sand bottom biofilm are members of Verrucomicrobia and the ubiquitously distributed γ-subdivision of Proteobacteria. Verrucomicrobia has been identified as a numerically abundant component of soil microbial communities in numerous sites around the world (5). In marine environments this phylogenetic group has been found in marine sediments (49) as well as in marine snow (40).
In the oyster reef biofilm, the members of the γ-subdivision of Proteobacteria and the CFB group were identified as the most abundant phylogenetic groups, with abundances around 23 and 22% (Tables 1 and 2). While the γ-Proteobacteria were found to be a significant part of the sand bottom biofilm (around 11%), the CFB group was only a minor group (less than 4%). Due to their ability to produce exopolysaccharide slime and extracellular enzymes that enable them to degrade particulate organic matter (21, 41, 42), their presence in biofilms is not surprising. Cytophaga spp. also exhibit gliding motility and are therefore thought to live primarily on surfaces (20, 32). Members of the CFB group (particularly the genus Cytophaga) were related to mainly particle-attached bacteria from the Columbia River, where they were found to be substantially more abundant in the estuary compared to the river and the adjacent coastal ocean (9). In marine environments, significant numbers of Cytophaga-Flavobacterium members have been found in the water column associated with macroscopic marine aggregates (13) or with diatom assemblages in Antarctic sea ice (3). Organisms of this phylogenetic group have also been found in relatively large numbers in anaerobic environments like Wadden Sea sediments (29). These gram-negative bacteria are thought to be specialized for the degradation of complex macromolecules (22, 41, 42). They were found to be the most prominent organisms in marine sediments in a study using cyanobacterial biomass to enrich anaerobic sediment as a simulation of the input of eutrophic phytoplankton detritus (43). In Escambia Bay, cyanobacterial biomass and production dominate the system in warmer months (34). The presence of CFB group members mainly in the reef-derived biofilm was not surprising, given that oysters are filter feeders producing significant quantities of organic-rich pseudofeces in addition to true feces and soluble nutrients from digestion and respiration of suspended particulate matter (1, 10, 11). Thus, an oyster reef represents a source of complex organic as well as inorganic nutrients within an estuarine system. Our results suggest that even in a region of the estuary with significant oyster reef cover, the effects may be highly localized.
Roughly twice as many community members were affiliated with the δ-Proteobacteria in the oyster reef biofilm than with the sand bottom biofilm. This phylogenetic group is interesting, as δ-Proteobacteria include obligately anaerobic sulfate reducers, which may grow in low-oxygen regions of the biofilms (35). Sulfate is the single most important terminal electron acceptor in anoxic marine environments, due to its ready availability. In many estuarine sediments roughly half of the total respiration is associated with sulfate reduction (12).
Roughly 9 and 1% of the clones in the oyster reef biofilm and the sand bottom biofilm, respectively, appeared to be affiliated with the relatively newly recognized bacterial division Holophaga-Acidobacterium. These organisms are widely distributed in different soils (23), but they are also present in lake sediments (46, 54) and marine habitats (53). Only two OTUs representing 2.4% of the community seemed to belong to the Verrucomicrobia (compared to 15.5% in the sand bottom biofilm [see above]).
The relative abundances of individual OTUs seen in the ARDRA were reflected in the T-RFLP data. Each of the five most prominent OTUs of the sand bottom biofilm represents a signal in the corresponding T-RFLP abundance plot. The T-RFs at around 218 and 211 bp can be assigned to groups C and D, respectively, whereas the T-RFs around position 560 bp represent Planctomyces groups A, B, and E. The assignment was performed by determining the positions of the T-RFs for the reamplified 16S rDNA from the corresponding individual clones. Whereas signals for C and D were clearly distinguishable, the signals representing A, B, and E all clustered around 561 bp. The T-RFs for A and E ran even at exactly identical positions, adding up to a hybrid peak. Sequence data confirmed that all three OTUs produced fragments of this length after in silico HhaI restriction. However, they became distinguishable after RsaI restriction, with signals at 112 and 471 bp, respectively (data not shown).
In the oyster reef biofilm, the assignment of individual peaks to identified OTUs is much more difficult due to the greater number of signals. In accordance with the ARDRA data, none of the identified OTUs seemed to be overwhelmingly prominent compared to those of the sand bottom biofilm. Clustering of peaks can be explained by members of the same phylogenetic group often producing terminal fragments of very similar sizes for restriction sites located in conserved sequences. This complicates the assignment of individual T-RFs and reflects the bias in T-RFLP data discussed earlier. Some of the more abundant T-RFs of the oyster reef fingerprint could not be assigned with the help of the individual 16S rDNA clones representing the major groups based on ARDRA. One explanation could be the presence of false or “pseudo-T-RFs” (14). These are signals which can be identified in T-RFLP profiles, but they do not represent the actual T-RFs. Single-stranded amplicons, which form during PCR, are thought to form transient double-stranded secondary structures which are accessible to restriction enzymes. The occurrence of pseudo-T-RFs has been observed for environmental samples (14). Another potential source for the discrepancy between ARDRA and T-RFLP results could be that the PCR products used for cloning were gel extracted but the ones used for T-RFLP analysis were not. 16S rDNA amplicons with lengths varying substantially from the “normal” length of around 1,550 bp could potentially have been lost, although in excising DNA from gels allowance was made for possible length heterogeneity of 16S rDNA invisible to the eye. Moreover, the proofreading TGO polymerase used to produce blunt-ended products for cloning might have slightly different amplification characteristics than the Taq polymerase used for generating products for T-RFLP. Different Taq polymerases can produce variations in T-RFLP profiles (37). Nonetheless, the fingerprints confirmed the overall community structures suggested by ARDRA.
One possible basis for the difference in community structure we observed that is not accounted for by our data is that we may be examining different succession stages of community development. The greater productivity of the oyster reef community may provide for faster rates of growth and greater biomass accumulation driving community structure to a further endpoint in our 7-day incubations. However, this possibly does not agree with observations of these two habitats at the macro scale, where similar differences in species richness and evenness are also apparent (33). In most cases, microbial ecology mirrors macro ecology rather than following a unique set of rules.
Although 16S analysis does not allow direct conclusions on phenotypic capabilities and metabolic activities, the rapidly accumulating information about bacterial communities in distinct environments may eventually lead to correlations between environmental parameters and community profiles. Due to the pronounced differences in the bacterial populations in the two examined habitats, it is tempting to speculate that the greater richness and evenness in bacterial diversity in the oyster reef setting reflects a greater organic and inorganic nutrient availability from the macrofauna and autochthonous production found on the oyster reef, in part stimulated by enhanced ammonium regeneration and production of pseudofeces (11). The availability of a more complex nutrient spectrum would support higher diversity of metabolic activities. The roles of such foci of production within the context of the highly dynamic and productive water column of most estuaries have not been well defined.
The results of this study suggest that habitat-specific, or α-diversity, differences in microbial flora may be significant and influence the β-diversity of microbial organisms and the processes they mediate within estuarine ecosystems.
Acknowledgments
Laura Pennington, Natasha Rondon, and Linda Martin are acknowledged for field and laboratory assistance. We also thank Hui Wang for her expert help in programming.
This research has been supported by a grant from the U.S. Environmental Protection Agency's Science to Achieve Results Estuarine and Great Lakes Coastal Initiative through funding to the CEER-GOM project, U.S. EPA agreement EPA/R-82945801.
REFERENCES
- 1.Bartol, I. K., R. Mann, and M. Luckenbach. 1999. Growth and mortality of oysters (Crassostrea virginica) on constructed intertidal reefs: effects of tidal height and substrate level. J. Exp. Mar. Biol. Ecol. 237:157-184. [Google Scholar]
- 2.Berner, E. K., and R. A. Berner. 1996. Global environment: water, air, and geochemical cycles, p. 284-311. Prentice Hall, Englewood Cliffs, N.J.
- 3.Bowman, J. P., S. A. McCammon, M. V. Brown, D. S. Nichols, and T. A McMeekin. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63:3068-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brakenhoff, R. H., J. G. G. Schoenmakers, and N. H. Lubsen. 1991. Chimeric cDNA clones: a novel PCR artifact. Nucleic Acids Res. 19:1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley, D. H., and T. M. Schmidt. 2001. Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soil. FEMS Microbiol. Ecol. 35:105-112. [DOI] [PubMed] [Google Scholar]
- 6.Clement, B. G., L. E. Kehl, K. L. Debord, and C. L. Kitts. 1998. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J. Microbiol. Methods 31:135-142. [Google Scholar]
- 7.Crump, B. C., and J. A. Baross. 1996. Particle-attached bacteria and heterotrophic plankton in the Columbia River estuary. Mar. Ecol. Prog. Ser. 138:265-273. [Google Scholar]
- 8.Crump, B. C., C. A. Simenstad, and J. A. Baross. 1998. Particle-attached bacteria dominate the Columbia River estuary. Aquat. Microb. Ecol. 14:7-18. [Google Scholar]
- 9.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dame, R. F., and S. Libes. 1993. Oyster reefs and nutrient retention in tidal creeks. J. Exp. Mar. Biol. Ecol. 171:251-258. [Google Scholar]
- 11.Dame, R. F., R. G. Zingmark, L. H. Stevenson, and E. Haskin. 1984. Oyster reefs as processors of estuarine materials. J. Exp. Mar. Biol. Ecol. 83:239-247. [Google Scholar]
- 12.Day, J. W., C. A. S. Hall, W. M. Kemp, and A. Yanez-Arancibia. 1989. Estuarine ecology, p. 90-96. John Wiley and Sons, New York, N.Y.
- 13.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924-934. [Google Scholar]
- 14.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Findlay, R. H., M. B. Trexler, J. B. Guckert, and D. C. White. 1990. Laboratory study of disturbance in marine sediments: response of a microbial community. Mar. Ecol. Prog. Ser. 62:121-133. [Google Scholar]
- 17.Findlay, R. H., M. B. Trexler, and D. C. White. 1990. Response of a benthic microbial community to biotic disturbance. Mar. Ecol. Prog. Ser. 62:135-148. [Google Scholar]
- 18.Fuerst, J. A. 1995. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology 141:1493-1506. [DOI] [PubMed] [Google Scholar]
- 19.Fuerst, J. A., H. G. Gwilliam, M. Lindsay, A. Lichanska, C. Belcher, J. E. Vickers, and P. Hugenholtz. 1997. Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon. Appl. Environ. Microbiol. 63:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gherna, R., and C. R. Woese. 1992. A partial phylogenetic analysis of the “Flavobacter-Bacteroides” phylum: basis for taxonomic restructuring. Syst. Appl. Microbiol. 15:513-521. [DOI] [PubMed] [Google Scholar]
- 21.Haack, S. K., and J. A. Breznak. 1993. Cytophaga xylanolytica sp. nov., a xylan-degrading, anaerobic gliding bacterium. Arch. Microbiol. 159:6-15. [Google Scholar]
- 22.Holmes, B. 1991. The genera Flavobacterium, Sphingobacterium, and Weeksella, p. 3620-3630. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer-Verlag, Berlin, Germany.
- 23.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeffrey, W. H., and J. H. Paul. 1986. Activity measurements of planktonic microbial and microfouling communities in a eutrophic estuary. Appl. Environ. Microbiol. 51:157-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kogure, K. U., U. Simidu, and N. Taga. 1980. Distribution of viable marine bacteria in neritic seawater around Japan. Can. J. Microbiol. 26:318-323. [DOI] [PubMed] [Google Scholar]
- 26.Kopczynski, E. D., M. M. Bateson, and D. M. Ward. 1994. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl. Environ. Microbiol. 60:746-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 28.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 29.Llobet-Brossa, E., R. Rossello-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by T-RFLP analysis of SSU rRNA and mcrA genes using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 33.Möbius, K. 1877. Die Auster und die Austernwirtschaft. Berlin, Germany. (English translation by the U.S. Fish Commission, 1880, p. 683-751.)
- 34.Murrell, M. C., and E. M. Lores. 2004. Phytoplankton and zooplankton seasonal dynamics in a subtropical estuary: importance of cyanobacteria. J. Plankton Res. 26:371-382. [Google Scholar]
- 35.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neef, A., R. Amann, H. Schlesner, and K. H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 37.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 38.Paabo, S., D. M. Irwin, and A. C. Wilson. 1990. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 265:4718-4721. [PubMed] [Google Scholar]
- 39.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 40.Rath, J., K. Y. Wu, G. J. Herndl, and E. F. DeLong. 1998. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat. Microb. Ecol. 14:261-269. [Google Scholar]
- 41.Reichenbach, H. 1989. Genus 1. Cytophaga Winogradsky 1929, 577, (AL) emend, p. 2015-2050. In J. T. Staley, M. P. Bryant, N. Pfenning, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 3. Williams & Wilkins, Baltimore, Md.
- 42.Reichenbach, H. 1992. The order Cytophagales, p. 3631-3675. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. M. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 43.Rossello-Mora, R., B. Thamdrup, H. Schäfer, R. Weller, and R. Amann. 1999. The response of the microbial community of marine sediments to organic carbon input under anaerobic conditions. Syst. Appl. Microbiol. 22:237-248. [DOI] [PubMed] [Google Scholar]
- 44.Sekiguchi, Y., Y. Kamagata, K. Syutsubo, A. Ohashi, H. Harada, and K. Nakamura. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655-2665. [DOI] [PubMed] [Google Scholar]
- 45.Shuldiner, A. R., A. Nirula, and J. Roth. 1989. Hybrid DNA artifacts from PCR of closely related target sequences. Nucleic Acids Res. 17:4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spring, S., R. Schulze, J. Overmann, and K. H. Schleifer. 2000. Identification and characterization of ecologically significant prokaryotes in the sediment of freshwater lakes: molecular and cultivation studies. FEMS Microbiol. Rev. 24:573-590. [DOI] [PubMed] [Google Scholar]
- 47.Steward, C. C., S. C. Nold, D. B. Ringelberg, D. C. White, and C. R. Lovell. 1996. Microbial biomass and community structures in the burrows of bromophenol producing and non-producing marine worms and surrounding sediments. Mar. Ecol. Prog. Ser. 133:149-165. [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urakawa, H., K. Kita-Tsukamato, and K. Ohwada. 1999. Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology 145:3305-3315. [DOI] [PubMed] [Google Scholar]
- 50.Vergin, K. L., E. Urbach, J. L. Stein, E. F. DeLong, B. D. Lanoil, and S. J. Giovannoni. 1998. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Von Wintzingerode, F., U. B. Goebel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples-pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 52.Wang, J., C. Jenkins, R. I. Webb, and J. A. Fuerst. 2002. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl. Environ. Microbiol. 68:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weidner, S., W. Arnold, E. Stackebrandt, and A. Puhler. 2000. Phylogenetic analysis of bacterial communities associated with leaves of the seagrass Halophila stipulacea by a culture-independent small-subunit rRNA gene approach. Microb. Ecol. 39:22-31. [DOI] [PubMed] [Google Scholar]
- 54.Zwisler, W., N. Selje, and M. Simon. 2000. Seasonal patterns of the bacterioplankton community composition in a large mesotrophic lake. Aquat. Microb. Ecol. 31:211-222. [Google Scholar]