Abstract
Listeria monocytogenes can cause serious illness in humans, and subsequent epidemiological investigation requires molecular characterization to allow the identification of specific isolates. L. monocytogenes is usually characterized by serotyping and is subtyped by using pulsed-field gel electrophoresis (PFGE) or ribotyping. DNA microarrays provide an alternative means to resolve genetic differences among isolates, and unlike PFGE and ribotyping, microarrays can be used to identify specific genes associated with strains of interest. Twenty strains of L. monocytogenes representing six serovars were used to generate a shotgun library, and subsequently a 629-probe microarray was constructed by using features that included only potentially polymorphic gene probe sequences. Fifty-two strains of L. monocytogenes were genotyped by using the condensed array, including strains associated with five major listeriosis epidemics. Cluster analysis of the microarray data grouped strains according to phylogenetic lineage and serotype. Most epidemiologically linked strains were grouped together, and subtyping resolution was the same as that with PFGE (using AscI and ApaI) and better than that with multilocus sequence typing (using six housekeeping genes) and ribotyping. Additionally, a majority of epidemic strains were grouped together within phylogenetic Division I. This epidemic cluster was clearly distinct from the two other Division I clusters, which encompassed primarily sporadic and environmental strains. Discriminant function analysis allowed identification of 22 probes from the mixed-genome array that distinguish serotypes and subtypes, including several potential markers that were distinct for the epidemic cluster. Many of the subtype-specific genes encode proteins that likely confer survival advantages in the environment and/or host.
Listeria monocytogenes is a gram-positive bacterial pathogen that is capable of causing significant morbidity and mortality in humans. Listeriosis is primarily a food-borne disease that has a significant impact on specific risk groups, including pregnant women and their fetuses, neonates, and people who are immunosuppressed (11). L. monocytogenes is capable of surviving and replicating under a wide range of environmental conditions, and this, as well as its widespread distribution, makes it particularly hard to eradicate from food-processing plants (11). Due to the severity of listeriosis, the United States maintains a zero-tolerance policy regarding contamination of ready-to-eat food products.
Although 13 serotypes of L. monocytogenes have been described (25), only three serotypes (1/2a, 1/2b, and 4b) cause the vast majority of clinical cases (26). Interestingly, although serotype 1/2a is most frequently isolated from food, serotype 4b causes the majority of human epidemics (12). Thus, many have suggested that there may be a link between serotype and virulence potential.
Numerous molecular subtyping techniques have identified two major phylogenetic divisions within the species. Division I consists of serotypes 1/2b, 3b, 4b, 4d, and 4e, and Division II consists of serotypes 1/2a, 1/2c, 3a, and 3c (1-3, 5, 15, 21). A third division, consisting of serotypes 4a and 4c and a subset of 4b strains, has also been described (8, 22, 27).
Epidemiological investigation of epidemic and sporadic cases of listeriosis requires molecular characterization to allow the identification of specific subtypes. L. monocytogenes subtypes are usually characterized by serotyping and then further subtyped by using the current “gold standard,” pulsed-field gel electrophoresis (PFGE) (16) or ribotyping. Multilocus sequence typing (MLST) has been described as a novel, reproducible, and potentially discriminatory subtyping method (10, 18, 23, 24), and Revazishvili et al. (23) recently demonstrated that MLST was able to differentiate most of the L. monocytogenes strains examined better than PFGE with AscI restriction endonuclease digestion.
DNA subtyping with DNA microarrays may provide an improved alternative to resolve genetic differences that exist among isolates (4, 9, 28). This technique has the added advantage that, unlike PFGE, ribotyping, and MLST, it can identify specific or unique genes associated with strains of interest. For example, Call and colleagues (9) demonstrated that certain strains of L. monocytogenes contained genes responsible for repairing UV-damaged DNA, salt tolerance, and biofilm formation, which would confer an advantage in certain ecological niches such as food production environments.
In the present study, a 629-probe “condensed” microarray was constructed using exclusively polymorphic probes. Fifty-two strains of L. monocytogenes were genotyped using the condensed array to compare the resolution of microarray subtyping to that of PFGE, MLST, and ribotyping and to identify genetic regions that characterize subtypes.
MATERIALS AND METHODS
Bacterial strains and subtyping.
Bacterial strains and sources are listed in Table 1. Listeria innocua strain ATCC 51742 was used as an outgroup for phylogenetic analysis. L. monocytogenes isolates were subtyped by using serotyping and PFGE (with AscI and ApaI restriction endonucleases) as previously described (16).
TABLE 1.
L. monocytogenes strains subtyped by microarray analysis
| Strain | Other designation(s) | Serotype | Sourcea | Epidemic | Donorb |
|---|---|---|---|---|---|
| A050M | 36046A | 1/2a | Bulk milk | USDA | |
| A061M | 36582B | 1/2a | Bulk milk | USDA | |
| A070M | 37952A | 1/2a | Bulk milk | USDA | |
| A259M | 32490E | 1/2a | Bulk milk | USDA | |
| A437N | FDA 15c03 | 1/2a | Food | FDA | |
| A501N | ILSI 33, TS4, F6854 | 1/2a | Food-Sp | United States, hot dog | ILSI |
| A502S | ILSI 34, TS14, F6900 | 1/2a | Human-Sp | United States, hot dog | ILSI |
| A503E | ILSI 35, J0161 | 1/2a | Human-Epi | United States, deli, 2000 | ILSI |
| A582N | H7788 | 1/2a | Food | United States, hot dog | CDC |
| B339S | DOH 9900104 | 1/2b | Human-Sp | DOH | |
| B345S | DOH 1159 | 1/2b | Human-Sp | DOH | |
| B404U | NADC 2053 | 1/2b | Unknown | USDA | |
| B412N | NADC 9916B | 1/2b | Environmental | USDA | |
| B430N | FDA 2492 | 1/2b | Food | FDA | |
| B439N | FDA 3280 | 1/2b | Food | FDA | |
| B443N | FDA 2475 | 1/2b | Food | FDA | |
| B445N | FDA 2450 | 1/2b | Food | FDA | |
| B507E | ILSI 39, G6003 | 1/2b | Food-Epi | Illinois, 1994 | ILSI |
| B508E | ILSI 40, G6054 | 1/2b | Human-Epi | Illinois, 1994 | ILSI |
| B588E | HPB1983 | 1/2b | Human-Epi | United Kingdom, 1989 | HC |
| C366S | H9333 | 1/2c | Human | CDC | |
| C368S | H9067 | 1/2c | Food | CDC | |
| C622N | RM3000 | 1/2c | Soil | USDA | |
| T590Sc | HPB1031, TS74 | 3b (1/2b) | Human-Sp | HC | |
| F113V | 01-3368A | 4b | Bovine | USDA | |
| F268S | 33027A | 4b | Bulk milk | USDA | |
| F336S | DOH 9900094 | 4b | Human-Sp | DOH | |
| F347S | DOH 1161 | 4b | Human-Sp | DOH | |
| F357S | DOH 2150 | 4b | Human-Sp | DOH | |
| F358S | DOH 2172 | 4b | Human-Sp | DOH | |
| F405N | NADC 575 | 4b | Food | USDA | |
| F428N | FDA 3365 | 4b | Food | FDA | |
| F429N | FDA 3276 | 4b | Food | FDA | |
| F446N | FDA 3515 | 4b | Food | FDA | |
| F447N | FDA 3655 | 4b | Food | FDA | |
| F469E | ILSI 1, ScottA | 4b | Human-Epi | United States, Massachusetts, 1983 | ILSI |
| F470E | ILSI 2, H7550 | 4b | Human-Epi | United States, hot dog, 1998 | ILSI |
| F494E | ILSI 26, TS43, F4565 | 4b | Human-Epi | California, 1985 | ILSI |
| F495E | ILSI 27, TS50, L4760 | 4b | Food-Epi | Halifax, Canada, 1981 | ILSI |
| F496E | ILSI 28, TS27, L4738 | 4b | Human-Epi | Halifax, Canada, 1981 | ILSI |
| F497E | ILSI 29, TS45, L3350 | 4b | Food-Epi | United Kingdom, 1988-1990 | ILSI |
| F498E | ILSI 30, TS38, L3306 | 4b | Human-Epi | United Kingdom, 1988-1990 | ILSI |
| F499E | ILSI 31, TS21, L4486j | 4b | Food-Epi | Switzerland, 1987 | ILSI |
| F505E | ILSI 37, J0144 | 4b | Food-Epi | North Carolina, 2000 | ILSI |
| F581E | H7738 | 4b | Food-Epi | United States, hot dog, 1998 | CDC |
| F583E | HPB850 | 4b | Human-Epi | Switzerland, 1996 | HC |
| F584E | HPB2142 | 4b | Human-Epi | United States, hot dog, 1998 | HC |
| F586E | HPB774 | 4b | Human-Epi | United Kingdom, 1991 | HC |
| F589E | HPB1808 | 4b | Human-Epi | California, 1985 | HC |
| F591E | HPB2262 | 4b | Human-Epi | Italy, 1998 | HC |
| F592E | HPB2182 | 4b | Human-Epi | Canada, 1999 | HC |
| F593E | HPB1026 | 4b | Human-Epi | California, 1985 | HC |
Sp, sporadic; Epi, epidemic.
USDA, U.S. Department of Agriculture Agricultural Research Service; DOH, Washington State Department of Health, Shoreline; CDC, Centers for Disease Control and Prevention, Atlanta, Ga.; FDA, U.S. Food and Drug Administration, Bothel, Wash.; ILSI, International Life Sciences Institute. L. monocytogenes strain collection (http://www.foodscience.cornell.edu/wiedmann/listeriadbase.htm); HC, Health Canada.
This isolate has serotyped as both 3b and 1/2b (25; M. K. Borucki, unpublished data).
Automated ribotyping.
Ribotyping was performed with the restriction enzyme EcoRI and the RiboPrinter microbial characterization system (Qualicon Inc., Wilmington, Del.), according to the manufacturer's manual and as previously described (6, 7).
MLST.
Loci were identified by searching for housekeeping genes from both L. monocytogenes and L. innocua via GenBank (http://www.ncbi.nlm.nih.gov). These genes were mapped against the L. monocytogenes EGD genome sequence (13) to provide adequate genome coverage, and primers were chosen to amplify coding regions of 500 to 750 bp under common conditions (3 mM MgCl2 and an annealing temperature of 58°C). Nucleotide sequences were obtained by PCR amplification of coding regions from the following genes: ahs, O-acetylhomoserine sulfhydralase homolog; pstI, phosphenolpyruvate-dependent phosphotransferase enzyme I; lisK, histidine kinase homolog; lhkA, histidine kinase; dhk, dihydroxyacetone kinase; and abcZ, ABC transporter homolog Z. PCR products were purified by using QIAquick 96 PCR purification kits (Qiagen, Valencia, Calif.) and were eluted in ∼60 μl of water; 96-well plates were stored at −20°C. Dye terminator cycle sequencing was performed with the CEQ cycle sequencing kit (Beckman Coulter, Fullerton, Calif.) in 10-μl reaction volumes with 10 to 20 ng of DNA. Sequencing reaction products were ethanol precipitated and dried, and samples were resuspended in 20 μl of formamide prior to separation by capillary electrophoresis with a CEQ2000XL DNA sequencer (Beckman Coulter). Sequence alignment and editing were performed with BioNumerics version 2.5 (Applied Maths, Kortrijk, Belgium). Allele sequence types were identified from 450 to 550 bp from each locus. Unweighted pair group method using arithmetic averages (UPGMA) analysis of categorical information based on the six different allele sequence types for each isolate was performed.
Microarray construction.
A genomic library was constructed from 20 strains representing six serotypes (1/2a [n = 5], 1/2b [n = 4], 1/2c [n = 4], 3a [n = 1], 4b [n = 5], and 4c [n = 1]) and obtained from a variety of sources (human sporadic [n = 10], epidemic [n = 2], environmental [n = 7], and veterinary [n = 1]). Genomic DNA was extracted from the 20 strains by using an Easy DNA kit (Invitrogen, Carlsbad, Calif.). DNA was quantified by UV spectrophotometry, and equal amounts of genomic DNA from each strain were mixed. This pooled genomic DNA was used to construct a random shotgun library (Amplicon Express, Pullman, Wash.). Briefly, 10 μg of DNA was cut with the restriction enzyme CviJI (Chimerx, Milwaukee, Wis.) or by sonification, and fragments of approximately 600 bp were gel isolated, extracted, and ligated into pUC18. Ligation products were transformed into Escherichia coli, and 12,000 positive recombinant clones were picked and arrayed into 96-well plates. Clone inserts were amplified by PCR with M13 primers (55 pmol each), 1.5 μl of bacterial culture (template DNA), 4 U of Taq polymerase with 1× reaction buffer (Fisher, Pittsburgh, Pa.), a 0.2 mM concentration of each deoxynucleoside triphosphate (Eppendorf, Westbury, N.Y.), and 2.5 mM MgCl2 in a 100-μl reaction volume. PCR cycle conditions were 95°C for 5 min; followed by 35 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min; followed by 72°C for 10 min after cycling was completed. The insert size was determined by using gel electrophoresis (1% agarose). PCR products of the correct size (500 to 1,000 bp) were purified with a Montage PCR96 Cleanup kit (Millipore Corp., Bedford, Mass.) and stored at −20°C until ready for printing.
PCR products were purified by sodium acetate precipitation, resuspended in 100 μl of H2O, quantified by UV spectrophotometry, and air dried. Probe DNA was then suspended in print buffer (200 mM Na2HPO4 plus 0.4 M NaCl [pH 11.5]) at a final concentration of 100 ng/μl, using a BIO-ROBOT 8000 instrument (Qiagen). Probes were then printed onto epoxy-coated slides (TeleChem International, Inc., Sunnyvale, Calif.) by using an Omnigrid spotter (GeneMachines, San Carlos, Calif.). PCR products from cloned fragments of L. monocytogenes ribosomal and listeriolysin genes were used as positive controls, and PCR products from a mouse cDNA library were used as negative controls. After printing, the slides were UV cross-linked (120,000 μJ) and stored at room temperature in the dark.
Target preparation and hybridization.
Genomic DNA was extracted from target strains by using a DNeasy tissue kit (Qiagen) and quantified by using UV spectrophotometry. Target DNA (1.5 μg) was nick translated in the presence of biotin-dATP (BioNick labeling system; Invitrogen). The labeled DNA was then ethanol precipitated, resuspended in 150 μl of hybridization buffer consisting of 4× SSC (60 mM NaCl, 0.6 mM Na citrate [pH 7.0]) and 5× Denhardt's solution (0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin), and added to the slide for overnight hybridizations at 55°C. Hybridizations and subsequent amplification steps were done in a GeneTAC hybridization station (Genomic Solutions, Ann Arbor, Mich.). Following target hybridization, the signal was amplified with a Tyramide signal amplification kit (Perkin-Elmer, Boston, Mass.). The slides were washed twice at 23°C for 30 s with TNT buffer (100 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20). Subsequent wash steps used two washes (30 s each) in TNT buffer, and all subsequent manipulations occurred at approximately 23°C. Streptavidin conjugated to horseradish peroxidase (1:100 in hybridization buffer) was incubated on the slide for 30 min, followed by washing and incubation with 10% equine serum (Sigma-Aldrich) in 2× SSC for 30 min. Biotinyl tyramide (1:50 in amplification buffer [tyramide signal amplification biotin system]) was then incubated on each slide for 10 min, followed by washing and a 30-min incubation with 2 μg of streptavidin per ml conjugated to Alexa Fluor 546 (Molecular Probes, Eugene, Oreg.) in 1× SSC-5× Denhardt's solution. The slides were given a final wash, followed by drying and imaging with a ScanArray 4000XL laser scanner (Packard BioChip Technologies, Downers Grove, Ill.).
Signal analysis.
Quantarray software (Packard Biochip Technologies) was used to quantify signal intensity. The final output included median intensity values, and data were normalized by dividing the median signal intensity by the median signal intensity of the ribosomal positive control. Data were managed by using MS Excel (Microsoft Corp., Redmond, Wash.) spreadsheets.
Data analysis.
Our analysis was limited to only those probes that were bimodally distributed such that both positive hybridizations (high signal) and negative hybridizations (low signal) were clearly identified. The selection process was based on a previously published algorithm (14). Briefly, for each probe, intensity values were assigned to either a “low” or a “high” cluster. After intensity values for all hybridization experiments were assigned to these two clusters, cluster averages and standard deviations were calculated. If cluster averages were different by greater than three standard deviations, the probe was considered bimodal. Ninety bimodal probes were selected for analysis by this technique. An additional 30 bimodal probes were selected for analysis as described previously (4).
For dendrogram construction, probes with normalized intensity readings of less than 0.2 were assigned a score of 1, probes with normalized intensity readings of greater than 0.2 but less than 0.4 were scored as 2 (and treated as ambiguous data in the phylogenetic analysis), and probes with normalized intensity readings of greater than 0.4 were scored as 3. A matrix was constructed and processed with PAUP (version 4.0b8a; Sinauer Associates, Inc., Sunderland, Mass.). UPGMA and Treeview (20) were used to construct a dendrogram that summarized genetic relationships between samples. Stepwise discriminant function analysis (DFA) (NCSS 2001 statistical software; NCSS, Kaysville, Utah) was used to identify probes characteristic of divisions and subtypes. Data were also examined by using a spreadsheet (Microsoft Excel) to identify probes that consistently discriminated between various dendrogram clusters.
Sequence analysis.
Probes of interest were retrieved from the clone library and sequenced by using two-pass automated sequencing, and data were analyzed by using DNASTAR (DNASTAR, Madison, Wis.). Nucleotide sequences were compared to existing nucleotide and protein sequences present in the GenBank database by using BLASTn and BLASTx searches. Seven of these probe sequences were selected to identify how sequence divergence was reflected by signal intensity on the microarray. PCR primers were designed to amplify a 500- to 600-bp region of the corresponding sequences from 15 L. monocytogenes isolates representing the two primary phylogenetic divisions. The resulting PCR products were sequenced, and percent sequence similarity was calculated.
Nucleotide sequence accession numbers.
The DNA sequences of the MLST loci have been deposited in GenBank under accession numbers AY622010 through AY622039 (abcZ), AY622040 through AY622069 (ahs), AY622070 through AY622099 (dhk), AY622100 through AY622129 (lhkA), AY622130 through AY622159 (lisK), and AY622160 through AY622189 (ptsI).
RESULTS
A shotgun library was constructed by mixing equal molar amounts of genomic DNAs from 20 strains (six serotypes) of L. monocytogenes (4, 9). A 2,000-probe screening microarray was constructed from the clone library and screened for polymorphic probes by hybridizing the genomic DNAs from 80 strains of L. monocytogenes to the array. These strains were obtained from diverse sources (human epidemic, human sporadic, environmental, and veterinary) and included seven serotypes (1/2a, 1/2b, 1/2c, 3a, 4a, 4b, and 4e). The 685 probes identified as polymorphic were sequenced, and nucleotide sequences were compared to identify replicate probes. Six hundred twenty-nine probes were identified as unique, and the closest protein match for each probe was identified by using BLASTx searches against GenBank. The probes were then used to construct a condensed array consisting entirely of polymorphic and characterized probes.
Fifty-two L. monocytogenes strains were hybridized to the condensed array, and subsequent data analysis identified 130 bimodally distributed probes. Data analyses were limited to these probes to maximize the likelihood of identifying subtype- or division-specific DNA sequences.
Phylogenetic divisions and subgroups.
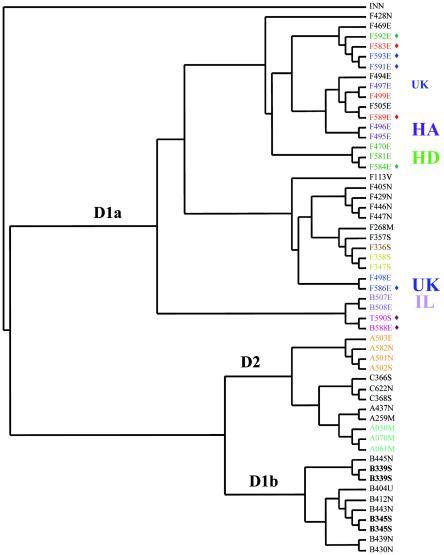
Comparative microarray analysis grouped strains according to previously described phylogenetic divisions, and all strains were grouped by serotype (Fig. 1). Division I (D1) consisted of two main subgroups (D1a and D1b). Interestingly, the D1b subgroup, consisting of human sporadic and environmental serotype 1/2b strains, clustered more closely to Division II (D2) strains than to D1a strains. However, DFA and subsequent sequence analysis were unable to identify probes with sequences unique to D2 and D1b. Indeed, sequence analysis of 13 of the 14 probes revealed that the majority of sequence differences occurred between the major divisions (D1 and D2). Three probes that differentiated between serovars 1 and 4 (probes 55 and 205) or between serotypes (probe 1083) were identified, and it is likely that serovar-specific probes may have influenced the topological position of the D1b subcluster.
FIG. 1.
UPGMA representation of genetic relationships between 52 L. monocytogenes isolates and one L. innocua isolate (INN) based on hybridization data derived from 130 bimodally distributed microarray probes. Phylogenetic divisions are indicated as D1a and D1b (Division I) and D2 (Division II). Isolates B339S and B345S (in boldface) were tested for processing and analysis reproducibility in two separate experiments. Isolates with the same AscI and ApaI PFGE restriction patterns are shown in the same color. Nine isolates were subtyped by PFGE, MLST, and ribotyping and are labeled with a diamond symbol. Ten isolates from the following four different epidemics were tested for subtype grouping: the 1981 Halifax epidemic (HA) (isolates F495E and F496E), the 1994 Illinois epidemic (IL) (isolates B507E and B508B), the 1998 multistate frankfurter-associated epidemic (HD) (isolates F470E, F581E, and F584E), and the 1988 to 1990 United Kingdom epidemic (UK) (isolates F497E, F498E, and F586E).
To allow serotype or source clusters to be easily visualized on the dendrogram (Fig. 1), strains were coded by serotype (A ([1/2a], B [1/2b], C [1/2c], F [4b], or T [3b]), three-digit lab strain identification number, and source (E [epidemic], S [sporadic], N [environmental or food], M [bulk milk], or V [veterinary]). Interestingly, most strains within serotype 4b grouped according to source as well as serotype, with a majority of serotype 4b epidemic strains forming a monophyletic group within D1a.
Stepwise DFA was used to identify 22 probes that differed among divisions and subclusters. Thirteen of these probes were further investigated by PCR and sequence analysis (Table 2). Sequence data revealed that five of the probes were division specific, four were subcluster specific, and four were serovar or serotype specific.
TABLE 2.
Protein similarity and sequence analysis of L. monocytogenes subtype- specific probes
| Probe | Protein similaritya | Putative function | Probe specificity |
|---|---|---|---|
| 1117 | bvr locus | Virulence gene regulation | Divisionb |
| 951 | Internalin-like (LPXTG motif) | Cell surface | Divisionc |
| 887 | lisRK | Stress tolerance, virulence | Divisionc |
| 875 | ORFA (LPXTG motif), L. innocua | Divisionb | |
| 657 | TagD | Cell wall synthesis | Divisionb |
| 523 | ABC transporter, permease | Molecule transport | Divisionc |
| 203 | Imo0737 | Divisionb | |
| 199 | Imo1434 | Divisionc | |
| 119 | Internalin-like (LPXTG motif) | Cell surface | Divisionb |
| 57 | Multidrug efflux transporter | Molecule transport | Divisionc |
| 1133 | UV damage repair protein | Stress response | Division and subclusterc,d |
| 837 | Exinuclease ABC | Stress response | Division and subclusterc |
| 321 | Internalin-like (LPXTG motif) | Cell surface | Division and subclusterc |
| 141 | Imo1965 | Division and subclusterc | |
| 1263 | RNase PH | Temperature response | Serovarb |
| 1229 | Two-component sensor protein KdpD | Osmoregulation | Serovarb |
| 1083 | Helicase | Replicative DNA helicase, DnaC | Serotype |
| 891 | Autolysin | Cell wall lysis | Serovarc |
| 553 | Glycosyltransferase | Cell wall synthesis | Serovarb |
| 265 | MurZ, Rho | Cell wall synthesis, transcription factor | Serovarb |
| 205 | Rhamnose synthetase | Cell wall synthesis | Serovarc |
| 55 | Amidase 4b protein | Cell adherence | Serovarc |
BLASTx searches showed that all probes had the greatest sequence similarity to L. monocytogenes proteins unless noted otherwise.
Specificity as determined by microarray analysis.
Specificity as determined by microarray and sequence analyses.
Sequence data specific for both division and subclusters.
Reproducibility and resolution.
To verify that assay replicates yielded similar results, genomic DNAs from isolates B339S and B345S were extracted, purified, labeled, and hybridized in two separate experiments. As expected, the replicates for both strains clustered together (Fig. 1).
Resolution of the condensed array was compared to that of the current gold standard, PFGE with AscI and ApaI restriction endonuclease digestion (16), by characterizing a panel of 28 strains by using both techniques. Resolution was similar for the two techniques, with both microarray analysis and PFGE dividing the 28 strains into 10 distinct subtypes (Fig. 1). Additionally, nine epidemiologically unrelated strains were grouped into four subtypes by using ribotyping and MLST with six housekeeping genes (Table 3). These strains separated into five distinct groups when characterized by microarray analysis and PFGE (with AscI and ApaI).
TABLE 3.
L. monocytogenes strains of different origin subtyped by five different methods
| Strain | Previous designationa | Yr | Origin | Serotype | MLST typeb | Ribotype | PFGE
|
Microarray resultc | |
|---|---|---|---|---|---|---|---|---|---|
| AscI | ApaI | ||||||||
| F583E | HPB850 | 1996 | Switzerland | 4b | 222422 | DUP-1038 | A | Ad | + |
| F589E | HPB1808 | 1985 | California | 4b | 222422 | DUP-1038 | A | a | + |
| F586E | HPB774 | 1991 | United Kingdom | 4b(x) | 632222 | DUP-1042 | B | b | + |
| F593E | HPB1026 | 1985 | California | 4b | 632222 | DUP-1042 | B | b | − |
| F591E | HPB2262 | 1998 | Italy | 4b | 632222 | DUP-1042 | C | Be | − |
| T590S | HPB1031 | NAf | United States | 3b (1/2b) | 333223 | DUP-1042 | D | c | − |
| B588E | HPB1983 | 1998 | Canada | 1/2b | 333223 | DUP-1042 | D | Ce | − |
| F584E | HPB2142 | 1998 | United States | 4b | 225226 | DUP-1044 | E | d | + |
| F592E | HPB2182 | 1999 | Canada | 4b | 225226 | DUP-1044 | E | De | + |
F. Pagotto et al., unpublished data.
Gene and number of alleles: abcZ, 6; ahs, 6; dhk, 5; lhkA, 6; lisK, 7; ptsI, 8.
Microarray analysis able (+) or not able (−) to further differentiate isolates.
One band missing.
One extra band.
NA, not available.
A panel of 10 isolates associated with four different epidemics were subtyped by using the condensed array. Most (9 of 10) epidemiologically linked isolates grouped together on the dendrogram (Fig. 1).
DISCUSSION
Microarray analysis grouped strains by phylogenetic divisions and serotype. However, a subcluster of Division I, D1b, grouped more closely to D2 than to D1a. This subcluster consisted of serotype 1/2b strains from human sporadic and environmental sources. This grouping was consistent even when data were analyzed using three different cluster algorithms (UPGMA, neighbor joining, and Ward's minimum variance), using different intensity range scores (i.e., with <0.15 scored as 1), or simply using normalized intensity data to produce the dendrogram (Ward's minimum variance). Two probes (probes 55 and 891) identified by DFA as differentiating D1b from D1a were sequenced and found to be serovar specific (differentiated serovar 1 from serovar 4) (Table 2). Therefore, it likely that a combination of probe differences makes D1b appear more similar to D2 than to D1a.
D1 strains were separated into four main subclusters, with D1a containing three subclusters and D1b consisting of a single 1/2b subcluster. One of the subclusters within D1a included 15 of the 17 serotype 4b strains associated with epidemics (Fig. 1). DFA was used to identify three probes that are most useful in defining this subcluster, and further analysis of these probes is under way.
Strains epidemiologically linked to particular epidemics were included in the microarray analysis to determine whether microarray subtyping did indeed group these strains together. Isolates obtained from patients and implicated foods from the 1981 Halifax epidemic (F495E and F496E), the 1994 Illinois epidemic (B507E and B508B), and the 1998 multistate epidemic (F470E, F581E, and F584E) grouped according to epidemic (Fig. 1). Two of the three strains associated with the 1988 to 1990 United Kingdom epidemic also grouped together. Investigation of the later outbreak identified pÂté as the likely source of an observed upsurge in listeriosis cases; however, no samples of pÂté eaten by patients with listeriosis were available for subtyping (19). Interestingly, the two strains from this outbreak that did cluster together were both obtained from patients, whereas strain F497E, a strain also associated with this epidemic but in a separate cluster, was a food isolate.
Strain A503E, a serotype 1/2a isolate that caused a multistate deli meat-associated epidemic in 2000, clustered with three other 1/2a strains (Fig. 1). Two of these strains are particularly interesting, because one (A501N) was isolated from the same food-processing plant in 1988 as A503E and another (A502S) was from a human sporadic case associated with A501N (17).
The resolutions of four different subtyping methods were compared using a subset of strains (Fig. 1; Table 3). Microarray analysis and PFGE subtyping showed the highest resolution, MLST had moderate subtyping resolution, and ribotyping had the lowest resolution. The microarray analysis subtyping resolution was similar to that of PFGE with two enzymes, the current gold standard for molecular subtyping of L. monocytogenes strains (16). Nevertheless, occasionally the two techniques placed strains in different groups (Fig. 1; Table 3). This is not surprising, because the two techniques sample the genome differently.
Microarray analysis and subsequent DFA processing of data resulted in the identification of 22 subtype-specific probes. Thirteen of these probes were further analyzed by PCR and sequence analysis (Table 2). Sequence analysis indicated that the microarray hybridization was capable of detecting approximately 10% sequence divergence between strains. These data agree with the microarray sensitivity threshold reported previously (9), although microarray sensitivity is obviously dependent on hybridization conditions, sequence content, and signal analysis.
The 22 probes identified as important for division and subtype definition included seven probes with sequence similarity to cell wall-associated proteins (probes 119, 205, 265, 321, 553, 657, 891, and 951). Three of these were serovar or serotype specific (Table 2). Five probes had sequence similarity to proteins important for survival in the environment or host (probes 57, 837, 1133, 1229, and 1263), and four probes were similar in sequence to virulence-associated proteins (probes 55, 875, 887, and 1117).
In conclusion, these data indicate that microarray analysis has a resolution similar to that of PFGE and better than those of MLST with housekeeping genes and ribotyping. Microarray analysis accurately clustered epidemiologically linked strains. Most epidemic-related strains formed a monophyletic cluster within Division I. Additionally, microarray analysis allowed identification of 22 probes that simultaneously distinguish divisions, serotypes, and subtypes.
Acknowledgments
Funding was provided by the USDA Agricultural Research Service (grant CWU 5348-32000-017-00D) and the Agricultural Animal Health Program (College of Veterinary Medicine, Washington State University).
We gratefully acknowledge the excellent technical assistance provided by James Reynolds, Kevin Tyler, Edith Orozco, Dave Tibbals, and Melissa Krug. L. monocytogenes isolates were kindly provided by Lewis Graves (Centers for Disease Control and Prevention), Jinxin Hu (Washington State Department of Health), Karen Jinneman (U.S. Food and Drug Administration), Lisa Gorski (USDA Agricultural Research Service), and Martin Wiedmann (Cornell University).
REFERENCES
- 1.Aarts, H. J., L. E. Hakemulder, and A. M. Van Hoef. 1999. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int. J. Food Microbiol. 49:95-102. [DOI] [PubMed] [Google Scholar]
- 2.Bibb, W. F., B. Schwartz, B. G. Gellin, B. D. Plikaytis, and R. E. Weaver. 1989. Analysis of Listeria monocytogenes by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Int. J. Food Microbiol. 8:233-239. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, W. F., B. G. Gellin, R. Weaver, B. Schwartz, B. D. Plikaytis, M. W. Reeves, R. W. Pinner, and C. V. Broome. 1990. Analysis of clinical and food-borne isolates of Listeria monocytogenes in the United States by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Appl. Environ. Microbiol. 56:2133-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. [DOI] [PubMed] [Google Scholar]
- 5.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce, J. L., R. J. Hubner, E. M. Cole, C. I. McDowell, and J. A. Webster. 1995. Sets of EcoRI fragments containing ribosomal RNA sequences are conserved among different strains of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 92:5229-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce, J. L. 1996. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 50:77-81. [Google Scholar]
- 8.Cai, S., D. Y. Kabuki, A. Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 11.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilot, P., A. Genicot, and P. Andre. 1996. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 34:1007-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 14.González S., M. J. Krug, M. Nielson, Y. Santos, and D. R. Call. 2004. Simultaneous detection of marine fish pathogens using multiplexed PCR and DNA microarrays. J. Clin. Microbiol. 42:1414-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graves, L. M., B. Swaminathan, M. W. Reeves, S. B. Hunter, R. E. Weaver, B. D. Plikaytis, and A. Schuchat. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J. Clin. Microbiol. 32:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 17.Kathariou, S. 2003. Foodborne outbreaks of Listeriosis and epidemic-associated lineages of Listeria monocytogenes, p. 243-256. In M. E. Torrence and R. E. Isaacson (ed.), Microbial food safety in animal agriculture. Iowa State Press, Ames.
- 18.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLauchlin, J., S. M. Hall, S. K. Velani, and R. J. Gilbert. 1991. Human listeriosis and pate: a possible association. Br. Med. J. 303:773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 21.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 23.Revazishvili, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salcedo, C., L. Arreaza, B. Alcala, L. de la Fuente, and J. A. Vazquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schonberg, A., E. Bannerman, A. L. Courtieu, R. Kiss, J. McLauchlin, S. Shah, and D. Wilhelms. 1996. Serotyping of 80 strains from the W.H.O. multicentre international typing study of Listeria monocytogenes. Int. J. Food Microbiol. 32:279-287. [DOI] [PubMed] [Google Scholar]
- 26.Tappero, J. W., A. Schuchat, K. A. Deaver, L. Mascola, J. D. Wenger, et al. 1995. Reduction in the incidence of human listeriosis in the United States. Effectiveness of prevention efforts? JAMA 273:1118-1122. [DOI] [PubMed] [Google Scholar]
- 27.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, C., M. Zhang, J. Ju, J. Nietfeldt, J. Wise, P. M. Terry, M. Olson, S. D. Kachman, M. Wiedmann, M. Samadpour, and A. K. Benson. 2003. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations. J. Bacteriol. 185:5573-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]