Abstract
Sphingomonas paucimobilis B90A is able to degrade the α-, β-, γ-, and δ-isomers of hexachlorocyclohexane (HCH). It contains the genes linA, linB, linC, linD, linE, and linR, which have been implicated in HCH degradation. In this study, dynamic expression of the lin genes was measured in chemostat-grown S. paucimobilis B90A by RNA dot blot hybridization and real-time reverse transcriptase PCR upon exposure to a pulse of different HCH isomers. Irrespective of the addition of HCH, linA, linB, and linC were all expressed constitutively. In contrast, linD and linE were induced with α-HCH (2 mg/liter) and γ-HCH (7 mg/liter). A sharp increase in mRNA levels for linD and linE was observed from 10 to 45 min after the addition of α- or γ-HCH. Induction of linD and linE was not detectable upon the addition of 0.7 mg of γ-HCH per liter, although the compound was degraded by the cells. The addition of β-HCH (5 mg/liter) or δ-HCH (20 mg/liter) did not lead to linE and linD induction, despite the fact that 50% of the compounds were degraded. This suggests that degradation of β- and δ-HCH proceeds by a different pathway than that of α- and γ-HCH.
Hexachlorocyclohexane (HCH) has been extensively used for the control of insect pests on agriculturally important crops, seeds, and vegetables, in forestry, and in vector control (34). Mainly two forms of HCH, lindane (γ-HCH) and a technical mixture of all isomers, have been applied. Technical grade HCH (33) largely consists of α-HCH (60 to 70%), with β-HCH (5 to 12%), γ-HCH (10 to 15%), and δ-HCH (6 to 10%) (16). As a result of the extensive use of lindane dust and technical HCH over the years and despite the recent ban on the use of HCH, several countries are currently faced with two very serious problems: (i) soil contamination with small amounts of HCH and (ii) highly contaminated sites where lindane was produced and purified or disposed of (5, 25).
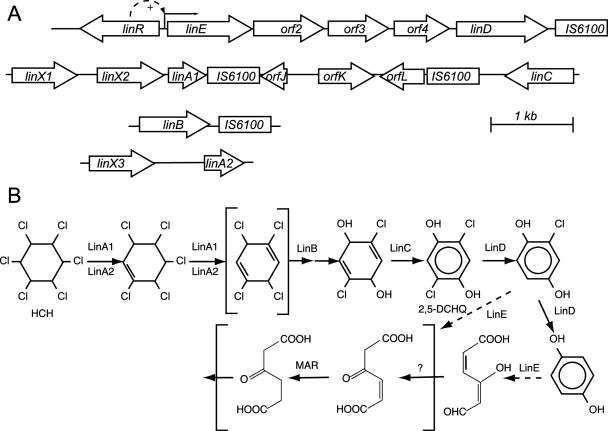
Although HCH is persistent and difficult to biodegrade, a few microorganisms have been isolated which can degrade one or more HCH isomers under aerobic conditions (27, 29, 31). Most of the strains, such as Sphingomonas paucimobilis UT26, degrade α- and γ-HCH but do not degrade β-HCH. Only one strain has been described, S. paucimobilis B90A (9), the parent strain of strain B90 (14, 15), that is able to degrade the four α-, β-, γ-, and δ-HCH isomers, although with different rates and not to completion for the β- and δ-HCH isomers. The degradation pathway for γ-HCH is well established from work on S. paucimobilis strain UT26 (20-24). Degradation of γ-HCH is mediated by the products of the so-called lin genes (23). It is assumed that α-HCH is degraded through the same pathway, but this has not been proven for β- and δ-HCH. S. paucimobilis strains UT26 and B90A have very similar lin gene sequences and organizations (9, 15). In contrast to typical degradation pathways in, for example, Pseudomonas or Ralstonia, where very long polycistronic operons are common (10, 17, 36), the genes for lindane degradation in S. paucimobilis are not organized within one or a few operons (Fig. 1). At least five different transcriptional units encode the lindane degradation pathway in strain UT26, e.g., linXA, linB, linC, and linDE and linR (21, 22). The last three genes form part of a small inducible regulon with linR as its main transcriptional activator (21). As far as is known, one of the main differences between strains UT26 and B90A is that strain B90A contains two copies of the linA gene and three copies of a linX-like gene (9). Since the two linA gene copies in B90A are not part of the same organizational unit, this increases the number of possible transcriptional units for the lindane degradation pathway in strain B90A to six (Fig. 1A). This makes it an interesting system to determine if and how S. paucimobilis B90A is capable of coordinating lin gene expression in response to HCH isomers.
FIG. 1.
Organization of the lin genes in S. paucimobilis strain B90A. (A) Organization of the relevant regions analyzed by DNA sequencing with the lin genes indicated (5). The only known regulatable promoter among the lin genes is in front of linE and is activated by LinR (21). The function of the linX gene product is not known, but based on sequence homology, it was predicted to be a dehydrogenase perhaps similar in function to LinC (24). (B) Simplified version of the degradation pathway for γ-HCH (http://umbbd.ahc.umn.edu/ghch/ghch_image_map.html). Reactions catalyzed by Lin enzymes are indicated as such. When more than one arrow is drawn, reactions are supposed to have intermediate steps involving unstable intermediates. The part shown within brackets remains speculative and is based on analogies to maleylacetate degradation. 2,5-DCHQ, 2,5-dichlorohydroquinone; MAR, maleylacetate reductase.
Here we have studied the expression dynamics of the lin genes in S. paucimobilis B90A. Our first interest was to determine if lin gene expression from the different operons in strain B90A is regulated in a coordinated way in response to γ-HCH. Second, we wanted to know whether low γ-HCH concentrations would still lead to detectable lin gene expression. As a third aspect of this work, we studied lin gene expression in response to the other HCH isomers (α-, β-, and δ-HCH). Gene expression studies were all carried out on chemostat-grown cultures of S. paucimobilis B90A cells, which could be sampled in a rapid time sequence after the addition of the HCH isomers for lin mRNA synthesis and HCH concentration without disturbing the culture growth, as described previously (12, 19). Amounts of lin mRNAs were determined by quantitative dot blot hybridization, and linD and linB levels were determined by quantitative real-time reverse transcriptase PCR (qRT-PCR).
MATERIALS AND METHODS
Bacterial strains and culture medium.
S. paucimobilis B90A (9) was grown at 28°C in mineral salt medium containing the following (per liter): 0.5 g of (NH4)2 HPO4, 0.2 g of MgSO4 · 7H2O, 0.1 g of K2 HPO4, 0.01 g of Ca (NO3)2, and 0.01 g of FeSO4 · 7H2O (pH 7.0), supplemented with 1 g of glucose and 1 g of peptone. Escherichia coli DH5α was used as general host for plasmid cloning. E. coli strains were grown routinely at 37°C on Luria broth Luria agar (28), supplemented with 100 μg of ampicillin per ml when necessary.
Chemostat cultivation of S. paucimobilis B90A.
In order to study lin gene expression, S. paucimobilis B90A was grown in continuous culture in a 0.5-liter reactor under carbon-limited conditions. The reactor had a working volume of 200 ml at a dilution rate of 0.05 h−1 and was maintained at 30°C with a dissolved oxygen concentration at 90% of saturation. Growth medium consisted of mineral salt medium containing 0.1% glucose. S. paucimobilis B90A was grown for at least 7 volume changes before being exposed to HCH isomers. The turbidity of the culture at 600 nm under steady-state conditions was 1.2. HCH was added to the cultures as one single pulse of 500 μl of a solution of the respective HCH isomer in dimethyl sulfoxide, which was introduced directly into the chemostat vessel while continuing the supply of normal growth medium. The final concentrations of the HCH isomers were 2 mg/liter for α-HCH, 5 mg/liter for β-HCH, and 20 mg/liter for δ-HCH; these concentrations approach the aqueous solubilities of the isomers as reported in the literature (35). In induction experiments with γ-HCH, two final concentrations of 7 (the maximal aqueous solubility) and 0.7 mg per liter were established. Experiments with γ-HCH were performed twice independently, and induction experiments with α-, β-, and δ-HCH were performed only once.
Total RNA extraction.
Total RNA from chemostat-grown S. paucimobilis B90A was isolated 50 and 100 min before and 5, 10, 15, 20, 30, 45, 60, 120, 180, and 240 min after the addition of HCH to the culture. Samples (3 ml) were taken directly from the chemostat, immediately pelleted by centrifugation (10,000 × g, 30 s) at 4°C, and resuspended in 50 μl of RNAlater (Ambion, Inc., Austin, Tex.). When all the samples were collected, the cells in RNAlater were again pelleted by centrifugation, resuspended in buffer, and extracted by acid phenol at 60°C as described by Aiba et al. (1). Total nucleic acids were further purified by phenol-chloroform extraction. After that, the RNA was precipitated with ethanol, treated with DNase I to remove traces of contaminating DNA (RNase free; Roche Biochemicals, Rotkreuz, Switzerland), and once more precipitated with ethanol. Aqueous RNA concentrations were measured from their UV absorption at 260 nm in a regular UV-visible light spectrophotometer (Kontron Instruments AG, Zürich, Switzerland).
In vitro synthesis of sense and antisense lin probes.
The linA1, linB, linC, linX, linD, linE, and linR open reading frames (ORFs) were cloned from plasmids pLINA1, pLINB, pLINC, pLINX, pLIND, pLINE, and pLINR (9, 15) in pGEM-7Zf(+) (Promega Corp., Madison, Wis.) which were maintained in E. coli DH5α (28). All the clones were verified by restriction analysis, and the orientation of lin ORFs in pGEM7Zf(+) was analyzed by DNA sequencing. One microgram of each pGEM-derived plasmid was linearized by restriction digestion to obtain an antisense transcript from the respective lin ORF. Antisense-labeled RNAs were then synthesized by in vitro transcription with biotin-16-UTP and either T7 or SP6 RNA polymerase (Roche Biochemicals). After synthesis, the template DNAs were degraded by incubation with RNase-free DNase I (Roche Biochemicals), and the mRNAs were aliquoted, precipitated in absolute ethanol, and stored at −20°C. This procedure was followed for probes of all lin genes, except linA2. This gene is too similar to linA1 and, therefore, cannot be distinguished by hybridization from linA1 (15).
Dot blot hybridization of lin mRNAs.
Total RNA from all the extractions was diluted in diethylpyrocarbonate-treated double-distilled water to final concentrations of 4 and 0.4 μg in 25 μl and blotted onto positively charged nylon membranes (QIAGEN, Basel, Switzerland) in a 96-well dot blot manifold (Gibco Life Technologies, Gaithersburg, Md.). Dilutions of denatured total genomic DNA from S. paucimobilis B90A and of pGEM-T-easy plasmid DNA containing the respective lin inserts were included on the same blot. The membrane was prehybridized for 1 h at 65°C in a solution of 7% sodium dodecyl sulfate, 10 g of bovine serum albumin (Fraction V; Sigma Chemical Co., St. Louis, Mo.) per liter, 0.5 M sodium phosphate buffer (pH 7.2), and 1 mM EDTA (pH 8.0) and then hybridized for 15 h at 65°C in the same solution with one aliquot of in vitro synthesized biotin-labeled single-stranded antisense RNA probes (approximately 100 ng) as described previously (4). Sense RNAs from the same plasmids were used in separate hybridizations as negative controls. After hybridization, the membranes were washed twice at 65°C in 0.1% sodium dodecyl sulfate and 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 30 min and then incubated for detection as described for the Southern-Light chemiluminescence system (Tropix, Bedford, Mass.). Membranes were exposed at room temperature to Hyperfilm (Amersham Life Sciences, Little Chalfont, Buckinghamshire, United Kingdom). Exposed films were scanned and transferred to 8-bit TIFF files, and the densities of the dots on the exposures were quantified with Metaview version 4.5 (Universal Imaging Corporation, West Chester, Pa.). In all cases, the sense RNA hybridizations did not show any signals above the background.
Signal intensities of hybridization spots were measured across the exact spot size (i.e., total gray value divided by spot area) but were subsequently standardized for one spot size throughout the membrane. The chromosomal and plasmid DNA standards were used to prepare a standard curve for the amount of DNA versus average gray value per pixel, according to a similar method described by Leveau et al. (19). Signal intensities of RNA samples were converted from the standard curve to equivalent DNA amounts and divided by the amount of RNA spotted. When possible, values derived from different RNA dilutions were averaged, and standard deviations were calculated.
Analytical method.
The disappearance of individual HCH isomers during the induction of chemostat-grown cells of S. paucimobilis B90A was measured by a gas-liquid chromatograph (GC-5890; Perkin Elmer) equipped with an electron capture detector and DB5 capillary column (15). For gas chromatography analysis, chemostat samples (200 μl) were extracted with 500 μl of hexane, of which 1 μl was injected into the gas chromatograph as described previously (15).
Real-time PCR analysis.
Based on results from dot blot hybridization, only linB (an example of a constitutively expressed gene) and linD mRNA (an inducible gene) were chosen for quantification by qRT-PCR analysis. Total RNA was taken from samples of the cultures induced with 7 mg of γ-HCH per liter, which were also used for dot blot studies. All purified RNA samples were used in two separate reactions, one to synthesize cDNA and the other to test for DNA contamination. The reverse transcription step was performed with TaqMan reverse transcription reagents (Applied Biosystems) with (for the cDNA synthesis reaction) and without (for the control reaction) reverse transcriptase with the help of gene-specific primers (Table 1). Mixtures for cDNA synthesis and the control reactions were heated in a standard thermocycler (Applied Biosystems) for 10 min at 25°C and then for 30 min at 48°C (transcription step), followed by 5 min at 95°C (inactivation). Following cDNA synthesis, real-time PCR was performed (ABI Prism 7000; Applied Biosystems) for primer optimization for the linB and linD genes with gene-specific primers (Table 1) in new tubes to which cDNA and control preparation were added, as described above. Once the correct primer concentration was identified for each of the genes, real-time PCR was performed with an initial hold temperature of 95°C for 10 min, followed by 40 amplification cycles of 15 s at 95°C and 1 min at 60°C.
TABLE 1.
PCR primers used for real-time RT-PCR analysis
| Primer sequence (5′ to 3′) | Designation | GenBank accession no. |
|---|---|---|
| GTC CGA ATC GCC CAT GCC GA | RTlinB | AY150581 |
| GTC GAA CCC TTC GGA ATC TT | RTlinD | AY150583 |
| TGG CGA GAA GAA ATT CAT TGA GA | linBf | AY150581 |
| TCG CCG GTC CCT TCA TC | linBr | AY150581 |
| GAA CTG TTC CAC TTC GTG TTC TCA | linDf | AY150583 |
| GGT CAC GCC CTT CTC CAT TA | linDr | AY150583 |
RESULTS AND DISCUSSION
Constitutive expression of the linA, linB, linC, and linX genes.
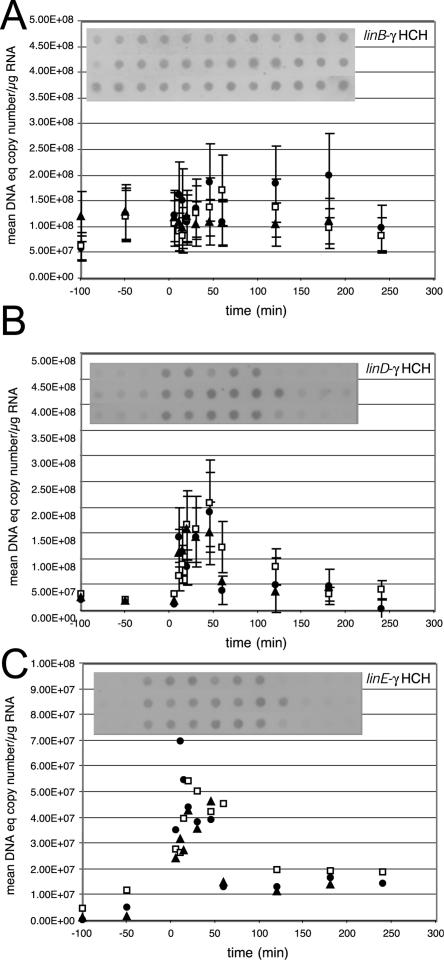
In order to find out whether the different lin operons in strain B90A were coordinately regulated in response to γ-HCH induction, chemostat-grown cells of S. paucimobilis B90A were treated with a single dose of γ-HCH at a concentration of 7 mg/liter. This concentration (25 μM) allows its maximum solubility in water but is still relatively low in comparison to the inducer concentrations usually applied in gene expression studies, which are normally tested in concentrations above 100 μM (8). The formation of individual lin gene transcripts before and up to 4 h after induction was analyzed by quantitative dot blotting with antisense labeled lin probes. Quantitative dot blotting is a simple but relatively accurate technique to monitor gene induction (12, 19). In contrast to the tfd or hbp regulons which were previously studied by this method (12, 19), most of the lin genes of S. paucimobilis B90A were expressed to the same level, irrespective of the addition of γ-HCH (Fig. 2A). Hence, we conclude that linA (here taken as the combination of both linA1 and linA2), linB, linC, and linX are constitutively expressed and are not under the control of lindane-inducible promoters. This situation is similar to the expression of the lin genes in strain UT26, although the lin genetic organizations in strains UT26 and B90A are not identical (9). Approximations based on standard DNA curves showed that the amounts of linA, linB, and linC transcripts were in the range of 0.5 × 108 to 2.0 × 108 mean DNA equivalent copy numbers per microgram of RNA (Fig. 2A). The range measured for linX was slightly lower (1 × 107 to 5 × 107) (data not shown). Despite standard deviations of, in some cases, 20% for individual spot measurements (calculated from hybridization intensities of different RNA dilutions on the same blot), independently repeated induction experiments resulted in the same relative transcript levels.
FIG. 2.
Amounts of lin-specific mRNAs before and after induction with γ-HCH (7 mg/liter). At time zero a single HCH dose was added to the chemostat culture of S. paucimobilis B90A. Panels show mRNAs probed with different antisense mRNAs, as indicated in the right top of each panel. The corresponding autoradiogram used for scanning is included within the graphs. The mRNA amounts are represented as mean DNA equivalent copy number (by comparing to a DNA standard, as explained in Materials and Methods) per microgram of RNA loaded in each spot. A different symbol is used for data from each of three independent induction experiments.
Inducible expression of linD and linE.
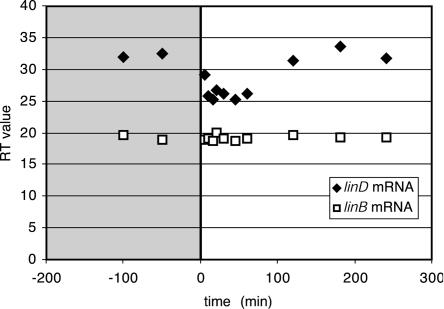
In contrast to the expression of linA, linB, linC, and linX, that of linD and linE appeared to be inducible (Fig. 2B and C). The amount of transcripts while the cells were growing on glucose was only around or below 107 DNA equivalent copy numbers per μg of RNA (Fig. 2B and C), but a sharp increase of linD and linE transcripts became visible 10 and 5 min, respectively, after the addition of γ-HCH. The faster appearance of the linE transcript compared to linD mRNA might be due to the fact that it is the first gene of that polycistronic mRNA and the time required for RNA polymerase to reach linD is longer, which is similar to previous observations of other systems (9, 19, 21). The relative induction level for linD and linE with 7 mg of γ-HCH per liter compared to expression on glucose alone was about 10-fold higher (although the uninduced expression level could not be measured accurately due to the low transcript abundance). To more accurately measure induction of the linD transcript, we used qRT-PCR, which resulted in a value of the threshold cycle number which was six lower at the peak of mRNA abundance (Fig. 3). Theoretically, this would correspond to a 64-fold increase in induction (i.e., 26). Both the dot blot hybridizations and qRT-PCR showed that abundance of the linB mRNA remained the same throughout the experiment. The 64-fold induction increase measured with qRT-PCR is similar to the magnitude of induction observed for the tfdA, tfdCI, and tfdCD mRNAs in Ralstonia eutropha after the addition of 10 μM 2,4-dichlorophenoxyacetate (2,4-D) (19). Probably, since the uninduced level cannot be measured very accurately by dot blotting, the ratios for the linE and linD transcripts were underestimated by that technique. A slight difference in the transcript levels of linE and linD was calculated even though both mRNAs are part of the same original transcript, which might be due to different mRNA stabilities, as observed before for the tfd system in R. eutropha (19).
FIG. 3.
Real-time PCR analysis of linB and linD gene expression in S. paucimobilis B90A. The mRNAs were purified from the induction experiment with γ-HCH (7 mg/liter) as described in the legend of Fig. 2. The time sequence is that described in the legend of Fig. 2. The y axis shows the threshold cycle number during which exponential amplification starts.
The increased level of linD and linE mRNA was only temporary and disappeared after 1 to 2 h, paralleling the degradation of γ-HCH (Fig. 4). This pulse-like transcript appearance is probably a reflection of the time course of the intracellular concentration of the inducing compound, which—by analogy to that shown for strain UT26 (21)—might be 2,5-dichlorohydroquinone, a metabolite formed during γ-HCH degradation. One can envision that upon first entry of γ-HCH in the cells, the metabolite is produced and accumulates due to constitutive LinA, LinB, and LinC activity (Fig. 1). Once formed, 2,5-dichlorohydroquinone will induce LinR-mediated expression from the linE promoter, which in turn will lead to formation of the dichlorohydroquinone metabolizing enzymes and breakdown of the inducer. If we assume that the rate of transcription from the linE promoter is directly related to the intracellular concentration of 2,5-dichlorohydroquinone, the net effect during γ-HCH degradation will be an initial sharp increase of linE and linD mRNAs, followed by a decrease. A comparison, for example, of the amount of the linA transcript and that of linD makes clear that the level of constitutive expression is similar to or only slightly less than the maximum expression level of the inducible lin genes reached with 7 mg of γ-HCH per liter. This might mean that metabolites from HCH are produced faster than they can be degraded by the inducible pathways. No significant fluctuations were observed for the linR transcript, but the general abundance of the linR mRNA was much lower than for the other lin genes (data not shown).
FIG. 4.
Disappearance of HCH isomers after pulsing to a continuous culture of S. paucimobilis B90A (depicted as the percentage of HCH remaining compared to the initial concentration). The washout curve depicts the theoretical dilution of HCH in the chemostat due to medium replenishment. Initial established concentrations were as follows: α-HCH, 2 mg/liter; β-HCH, 5 mg/liter; γ-HCH, 7 and 0.7 mg/liter; and δ-HCH, 20 mg/liter.
Lin gene expression after exposure of strain B90A to other HCH isomers.
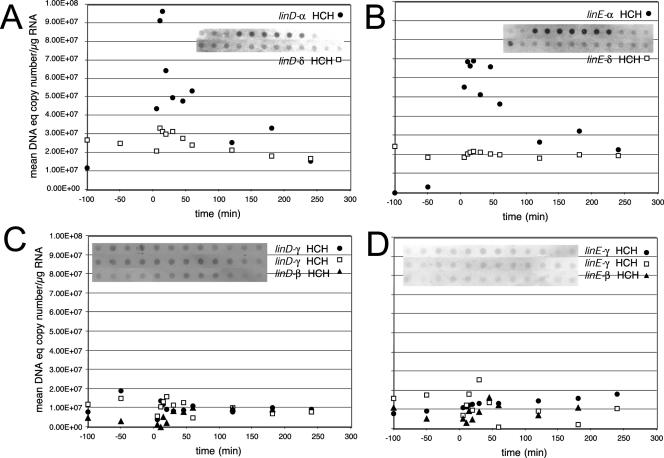
When the cells were exposed to a pulse of α-HCH (2 mg/liter), a similar response was seen as for γ-HCH (Fig. 5). The transcript levels of all lin genes previously observed to be constitutively expressed were again similar irrespective of α-HCH application to the chemostat (data not shown) and the same as after exposure to γ-HCH (i.e., around 2 ×108 mean DNA equivalent copies per μg of RNA). Induction of linE and linD also took place with α-HCH and with an induction factor of about 10-fold, although the measured linD transcript was slightly less abundant than with γ-HCH (maximum 108 mean DNA equivalent copies per μg of RNA). Induction and subsequent disappearance of the linD and linE transcripts followed the measured α-HCH concentration in the chemostat (Fig. 4). This strongly suggests that α-HCH is metabolized by S. paucimobilis B90A cells via a pathway similar to that of γ-HCH, giving rise to similar metabolites, which can induce the linE promoter (21). In contrast, no induction of linD and linE was observed in RNA isolated from cells exposed to β- and δ-HCH isomers (Fig. 5), although their concentrations were of the same order as those of α- and γ-HCH and both compounds were at least partially degraded by the cells (Fig. 4). This was surprising but indicates that β- and δ-HCH are either partially degraded by LinA, LinB, and LinC activity without formation of the metabolite necessary to achieve linED induction or are degraded via a different metabolic pathway not involving the lin gene products. The disappearance of β- and δ-HCH in the chemostat was rather different. Whereas β-HCH did not seem to be touched by the cells for about 1 h but then began disappearing at a higher rate (Fig. 4), δ-HCH was only partially degraded for about 1 h, after which degradation stopped completely. This could be an indication for induction of a different degradation pathway in the case of β-HCH. The apparent complete stop of further δ-HCH degradation after 1 h might have been due to the accumulation of a toxic intermediate. After exposure to both β- and δ-HCH, the cells accumulated a metabolite in the culture medium, which was detectable by a gas chromatograph equipped with an electron capture detector but is not further identified here (data not shown). It could be that the higher concentration of δ-HCH (20 mg/liter) resulted in higher concentrations of a δ-HCH intermediate, which was toxic for the cells, similar to observations of 2,4-dichlorophenol in 2,4-D degradation (19). In other ongoing studies in our laboratories, we have observed that S. paucimobilis cultures exposed to β- and δ-HCH release only half of all chlorine atoms of HCH as chloride (C. Holliger, unpublished data), which supports the idea that the limited degradation of β- and δ-HCH in chemostat cultures of strain B90A did not lead to formation of the (di)chlorohydroquinones necessary for induction of the linE promoter.
FIG. 5.
Induction of linE- and linD-specific mRNAs after addition of different HCHs. At time zero a single dose of HCH was added to the chemostat culture of S. paucimobilis B90A. (A and B) α-HCH (filled symbols) and δ-HCH (open symbols) at concentrations of 2 and 20 mg per liter, respectively. (C and D) γ-HCH (0.7 mg/liter) and β-HCH (5 mg/liter). The corresponding autoradiogram used for scanning is included within the graphs. The mRNA amounts are represented as the mean DNA equivalent copy number (by comparing to a DNA standard, as explained in Materials and Methods) per microgram of RNA loaded in each spot. Induction experiments with 0.7 mg of γ-HCH per liter were carried out twice; other experiments were carried out once.
Induction of linD and linE in response to a low γ-HCH concentration.
Finally, it was determined whether linD and linE would still be induced at a 10-fold-lower γ-HCH concentration (0.7 mg/liter), which is indicative of what may happen at the low micromolar concentrations more often found in the environment. Although linD and linE genes were induced by γ-HCH at a concentration of 7 mg/liter (25 μM), no measurable change in mRNA was observed upon exposing the cells to a concentration of 0.7 mg/liter (2.5 μM). This was reproducibly observed (Fig. 5C and D), but since quantitation of individual spots resulted in standard deviations of up to 20%, very low mRNA bursts may have remained undetectable. Interestingly, γ-HCH was still degraded by the cells (Fig. 4), although we cannot conclude whether full mineralization took place on the basis of the gas chromatograph-electron capture detector measurements.
There is good evidence from other experimental systems to believe that such a low concentration (2.5 μM) might be at or below a threshold for induction of the linD and linE genes and similarly for other catabolic genes in general. We conclude this from experimental studies on catabolic promoter response with reporter gene fusions, which independently discovered the lowest detectable induction to take place in a concentration range of 1 to 10 μM. For example, by using promoter fusions to the luxAB bacterial luciferase genes, the lowest response from the tfdDI promoter in R. eutropha JMP134 to 2,4-D was found to occur at 2 μM (11). Similarly, the lowest inducer concentration for the hbpC promoter in Pseudomonas azealica was 9 μM 2-hydroxybiphenyl (13), for the Po promoter in Pseudomonas putida was 3.2 μM phenol (30), and for the todX promoter of P. putida F1 was 1 μM toluene (2). It might be that a threshold for specific gene activation occurs at low substrate concentrations (around 1 μM), because when most of the substrate is channeled through the metabolic pathway, only a small fraction of inducer is left to be detected by the regulatory protein. For example, a previous study calculated a diffusion rate of uncharged 2,4-D through the cytoplasmic membrane of R. eutropha at a 1 μM outside concentration in the order of 100 molecules per s per cell (18), which, in fact, was mostly driven by the rate of 2,4-D metabolism.
Orchestrating expression of the lindane degradation pathway.
In conclusion, the results of this work have shown that expression of the lin genes behaves rather differently than what is usually observed for catabolic pathways of, e.g., Beta- and Gammaproteobacteria. Probably due to the absence of regulatable promoters and a scattering of the different genes involved in the pathway, most of the lin genes are expressed to a constitutive level (i.e., linA1 and/or linA2, linB, linC, and linX). Only the linE and linD genes, as observed before for S. paucimobilis UT26 (21), were inducibly expressed. The scattering of pathway genes is more commonly found in sphingomonads, for example, for the pentachlorophenol degradation pathway in Sphingobium chlorophenolicum ATCC 39723 (7), for the genes involved in the dioxin degradation pathway in Sphingomonas sp. strain RW1 (3), or for the catabolic plasmid of Sphingomonas aromaticivorans (26), and may reflect a recent acquisition of gene sequences (in which case regulatable expression may evolve in the end) or simply be a more common property for this class of bacteria. Constitutively expressing part of the lin genes does not provide an apparent physiological disadvantage for the cells, except perhaps at the highest δ-HCH concentration, where a toxicity effect cannot be excluded. The present genetic organization is sufficiently advantageous, and Sphingomonas strains with related lin genes and expression characteristics have been isolated from all over the world (e.g., from Japan [29], India [27], and France [31]). On the contrary, it might be that by providing constitutive expression, the cells escape at least part of the phenomena of apparent noninduction at low substrate concentrations. An absence of measurable gene induction does not necessarily mean that complete degradation is brought to a halt. S. paucimobilis B90A cells could rely on small amounts of LinD and LinE enzymes that are present because of low constitutive gene expression. However, it is currently not known whether S. paucimobilis cells gain any energy, carbon, or reducing equivalents by cometabolizing γ- or α-HCH partially, without inducing the lower pathway, or whether they will still completely metabolize HCH at concentrations below 2.5 μM in the environment. Mineralization studies with radiolabeled γ-HCH and with γ-HCH concentrations in the microgram-per-liter range are on their way. Pseudomonas sp. strain B13 metabolized 3-chlorobenzoate even in the nanomolar range but with completely different uptake and metabolic rates prevailing at the nanomolar range than at the micromolar range (32). Although this is a very relevant environmental question, it remains unclear which biological phenomena are taking place at very low pollutant concentrations (micromolar and lower) (6).
Acknowledgments
The authors thank Christoph Werlen and Alexandra Bähler, EAWAG, Dübendorf, Switzerland, for helping with chemostat cultivation.
Part of this work was supported by grants under the Indo-Swiss Collaboration in Biotechnology Program.
REFERENCES
- 1.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. [PubMed] [Google Scholar]
- 2.Applegate, B., C. Kelly, L. Lackey, J. McPherson, S. Kehrmeyer, F.-M. Menn, P. Bienkowski, and G. Sayler. 1997. Pseudomonas putida B2: a tod-lux bioluminescent reporter for toluene and trichloroethylene co-metabolism. J. Ind. Microbiol. 18:4-9. [DOI] [PubMed] [Google Scholar]
- 3.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann, B., M. Snozzi, A. J. B. Zehnder, and J. R. van der Meer. 1996. Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J. Bacteriol. 178:4367-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braga, A. M., T. Krauss, C. R. Reis dos Santo, and P. Mesquita de Souza. 2002. PCDD/F-contamination in a hexachlorocyclohexane waste site in Rio de Janeiro, Brazil. Chemosphere 46:1329-1333. [DOI] [PubMed] [Google Scholar]
- 6.Button, D. K. 1998. Nutrient uptake by microorganisms according to kinetic parameters from theory as related to cytoarchitecture. Microbiol. Mol. Biol. Rev. 62:636-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daunert, S., G. Barrett, J. S. Feliciano, R. S. Shetty, S. Shrestha, and W. Smith-Spencer. 2000. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem. Rev. 100:2705-2738. [DOI] [PubMed] [Google Scholar]
- 9.Dogra, C., V. Raina, R. Pal, M. Suar, S. Lal, K.-H. Gartemann, C. Holliger, J. R. van der Meer, and R. Lal. 2004. Organization of lin genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J. Bacteriol. 186:2225-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin, F. C. H., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring-cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay, A. G., J. F. Rice, B. M. Applegate, N. G. Bright, and G. S. Sayler. 2000. A bioluminescent whole-cell reporter for detection of 2,4-dichlorophenoxyacetic acid and 2,4-dichlorophenol in soil. Appl. Environ. Microbiol. 66:4589-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaspers, M. C., A. Schmid, M. H. Sturme, D. A. Goslings, H. P. Kohler, and J. R. van der Meer. 2001. Transcriptional organization and dynamic expression of the hbpCAD genes, which encode the first three enzymes for 2-hydroxybiphenyl degradation in Pseudomonas azelaica HBP1. J. Bacteriol. 183:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaspers, M. C., W. A. Suske, A. Schmid, D. A. Goslings, H. P. Kohler, and J. R. van der Meer. 2000. HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J. Bacteriol. 182:405-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johri, A. K., M. Dua, D. Tuteja, R. Saxena, D. M. Saxena, and R. Lal. 1998. Degradation of α-, β-, γ-, and δ-hexachlorocyclohexane by Sphingomonas paucimobilis. Biotechnol. Lett. 20:885-887. [Google Scholar]
- 15.Kumari, R., S. Subudhi, M. Suar, G. Dhingra, V. Raina, C. Dogra, S. Lal, C. Holliger, J. R. van der Meer, and R. Lal. 2002. Cloning and characterization of lin genes responsible for the degradation of hexachlorocyclohexane isomers in Sphingomonas paucimobilis strain B90. Appl. Environ. Microbiol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutz, F. W., P. H. Wood, and D. P. Bottimore. 1991. Organochlorine pesticides and polychlorinated biphenyls in human adipose tissue. Rev. Environ. Contam. Toxicol. 120:1-82. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, C. M., J. H. J. Leveau, A. J. B. Zehnder, and J. R. van der Meer. 2000. Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 182:4165-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leveau, J. H., A. J. B. Zehnder, and J. R. van der Meer. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 180:2237-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leveau, J. H. J., F. König, H.-P. Füchslin, C. Werlen, and J. R. van der Meer. 1999. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol. Microbiol. 33:396-406. [DOI] [PubMed] [Google Scholar]
- 20.Miyauchi, K., Y. Adachi, Y. Nagata, and M. Takagi. 1999. Cloning and sequencing of a novel meta-cleavage dioxygenase gene whose product is involved in degradation of γ-hexachlorocyclohexane in Sphingomonas paucimobilis. J. Bacteriol. 181:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyauchi, K., H. S. Lee, M. Fukuda, M. Takagi, and Y. Nagata. 2002. Cloning and characterization of linR, involved in regulation of the downstream pathway for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 68:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagata, Y., K. Miyauchi, and M. Takagi. 1999. Complete analysis of genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J. Ind. Microbiol. Biotechnol. 23:380-390. [DOI] [PubMed] [Google Scholar]
- 23.Nagata, Y., T. Nariya, R. Ohtomo, M. Fukuda, K. Yano, and M. Takagi. 1993. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 175:6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagata, Y., R. Ohtomo, K. Miyauchi, M. Fukuda, K. Yano, and M. Takagi. 1994. Cloning and sequencing of a 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase gene involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 176:3117-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira, R. M., O. M. Brilhante, J. C. Moreira, and A. C. Miranda. 1995. Hexachlorocyclohexane contamination in urban areas of the south eastern region of Brazil. Rev. Saude Publica 29:228-233. (In Portuguese.) [DOI] [PubMed] [Google Scholar]
- 26.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahu, S. K., K. K. Patnaik, M. Sharmila, and N. Sethunathan. 1990. Degradation of α-, β-, and γ-hexachlorocyclohexane by a soil bacterium under aerobic conditions. Appl. Environ. Microbiol. 56:3620-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Senoo, K., and H. Wada. 1989. Isolation and identification of an aerobic γ-HCH decomposing bacterium from soil. Soil Plant Nutr. 35:79-87. [Google Scholar]
- 30.Shingler, V., and T. Moore. 1994. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J. Bacteriol. 176:1555-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas, J. C., F. Berger, M. Jacquier, D. Bernikkon, F. Baud-Grasset, N. Truffault, P. Normand, T. M. Vogel, and P. Simonet. 1996. Isolation and characterization of a novel γ-hexachlorocyclohexane-degrading bacterium. J. Bacteriol. 178:6049-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tros, M. E., G. Schraa, and A. J. B. Zehnder. 1996. Transformation of low concentrations of 3-chlorobenzoate by Pseudomonas sp. strain B13: kinetics and residual concentrations. Appl. Environ. Microbiol. 62:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, K., D. A. Vallero, and R. G. Lewis. 1999. Factors influencing the distribution of lindane and other hexachlorocyclohexanes in the environment. Environ. Sci. Technol. 33:4373-4378. [Google Scholar]
- 34.Ware, G. W. 1989. The pesticide book, 3rd ed. Thomas Publications, Fresno, Calif.
- 35.Windholz, M., S. Budavari, L. Y. Stroumtsos, and M. N. Fertig (ed.). 1976. The Merck index, 9th ed. Merck & Co., Inc., Rahway, N.J.
- 36.Zylstra, G. J., W. R. McCombie, D. T. Gibson, and B. A. Finette. 1988. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl. Environ. Microbiol. 54:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]