Abstract
The role of the rRNA gene copy number as a central component of bacterial life histories was studied by using strains of Escherichia coli in which one or two of the seven rRNA operons (rrnA and/or rrnB) were deleted. The relative fitness of these strains was determined in competition experiments in both batch and chemostat cultures. In batch cultures, the decrease in relative fitness corresponded to the number of rRNA operons deleted, which could be accounted for completely by increased lag times and decreased growth rates. The magnitude of the deleterious effect varied with the environment in which fitness was measured: the negative consequences of rRNA operon deletions increased under culture conditions permitting more-rapid growth. The rRNA operon deletion strains were not more effective competitors under the regimen of constant, limited resources provided in chemostat cultures. Enhanced fitness in chemostat cultures would have suggested a simple tradeoff in which deletion strains grew faster (due to more efficient resource utilization) under resource limitation. The contributions of growth rate, lag time, Ks, and death rate to the fitness of each strain were verified through mathematical simulation of competition experiments. These data support the hypothesis that multiple rRNA operons are a component of bacterial life history and that they confer a selective advantage permitting microbes to respond quickly and grow rapidly in environments characterized by fluctuations in resource availability.
An organism's life history is its lifetime pattern of growth, differentiation, storage of resources, and reproduction (6). Tradeoffs for the optimization of one life history trait at the expense of another have led to the evolution of a magnificent array of ecological strategies in plants and animals. That is why the identification and investigation of life histories remain a cornerstone of contemporary ecology. Since natural selection favors individuals that are better able to survive and leave viable progeny, regardless of whether they are multicellular or single-celled, it should be valuable to consider the life histories of microbes.
One readily quantifiable feature of bacteria that has been proposed as a component of bacterial life histories is the number of rRNA genes (25, 26, 39). This proposal is based on the understanding that multiple copies of rRNA genes allow for increased rates of rRNA synthesis, leading to more rapid synthesis of ribosomes and ultimately conferring the potential for a quicker response to an influx of resources and rapid growth (13). This characteristic fits the classical definition of a component of life history and may also exhibit a tradeoff, namely, a metabolic cost for retention of multiple copies of rRNA genes associated with basal levels of transcription of these genes. This proposed expense would be particularly important in environments characterized by slow and constant flux of resources, where there is little selection for shorter lag times or rapid growth.
We have investigated the potential selective forces behind the occurrence of multiple rRNA gene copies in bacteria, and we report the results of experimental tests of the hypothesis that multiple copies of rRNA genes are a central component of life history and confer an advantage in environments defined by fluctuations in resource availability. Previous work addressed this hypothesis by illustrating the connection between the ecological strategies and rRNA gene copy numbers of a phylogenetically diverse collection of bacteria (25). In the present study, the number of rRNA genes was altered experimentally in an evolved lineage of Escherichia coli so as to investigate any direct link between the life history of an organism and its rRNA gene copy number.
MATERIALS AND METHODS
Growth media and culture conditions.
For cloning and genetic manipulations, cultures were grown in Luria-Bertani liquid medium (LB), with 1.5% (wt/vol) Bacto agar (Becton Dickinson and Company, Franklin Lakes, N.J.) added for a solid medium. For selection of sucrose-resistant strains, sucrose was added at a final concentration of 6% (wt/vol) to LB and NaCl was omitted (LBsuc) (8). Kanamycin (50 mg/liter) and chloramphenicol (30 mg/liter) were added to media where noted. All competition cultures were grown in Davis-Mingoli broth (DM) supplemented with 2.0 μg of thiamine hydrochloride and 0.1, 25, or 1,000 mg of glucose per liter (e.g., DM+25) (28). Tetrazolium arabinose (TA) agar medium was used to differentiate Ara+ and Ara− phenotypes, which appear as white and red colonies, respectively (29).
For all experiments, cultures were first grown overnight in LB inoculated from −80°C glycerol stocks: these cultures were then used to inoculate a DM conditioning culture, which was incubated with shaking at 225 rpm and 37°C for exactly 24 h. The inoculum for the conditioning cultures represented approximately 1/100 of the population density achieved at the specific glucose concentration of the test medium to be used. This conditioning culture was then used to inoculate fresh DM (1/100) for the experiment performed.
Batch cultures for competition experiments, conditioning cultures, and cultures for the measurement of individual growth parameters were grown in 10 ml of DM in a 50-ml Erlenmeyer flask or in 50 ml of DM in a 250-ml Erlenmeyer flask shaken at 225 rpm at 37°C. Competition experiments in chemostats were carried out in a system of vessels designed like those described previously (9, 11). The chemostat vessels were incubated in a 37°C water bath, and sterile DM + 25 was delivered to each vessel by a variable-speed peristaltic pump to achieve a dilution rate of 0.11 or 0.81 per h. A vacuum was applied to each culture vessel through a glass tube that maintained the culture volume at approximately 65 ml. The vacuum was also used to draw air through a ≤0.30-μm-pore-size HEPA filter (HEPA-VENT; Whatman Inc., Clifton, N.J.) attached to each medium inflow tube, providing sterile aeration and mixing.
General cloning procedures.
Restriction endonuclease digestions were carried out according to the manufacturer's protocols (New England Biolabs, Beverly, Mass.). If necessary, specific restriction fragments were isolated on an agarose gel and purified (QIAEX II gel extraction kit; QIAGEN, Valencia, Calif.). A commercially available, chemically competent strain of E. coli (One Shot TOP10; Invitrogen, Carlsbad, Calif.) was used for most cloning according to the manufacturer's protocols. When E. coli strain D308 was transformed, cells were first made chemically competent by using a modification of the protocol of Chung et al. (12, 39).
PCR conditions.
DNA fragments of <4 kb were amplified by using 0.25 U of Taq DNA polymerase (Biolase; Intermountain Scientific Corporation, Kaysville, Utah) with 1× reaction buffer, 2 mM MgCl2, 2.5 mM each deoxynucleoside triphosphate, 50 to 100 ng of template DNA, and 0.5 μM forward and reverse primers (Table 1) in a total volume of 50 μl. Reactions were carried out in a PTC100 thermal cycler (MJ Research Inc., South San Francisco, Calif.) with the following incubation conditions: 94°C for 3 min; 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s; and 72°C for 10 min. When larger DNA fragments (>4 kb) were to be amplified, the proofreading DNA polymerase (rTth) was used (GeneAmp XL PCR kit; Perkin-Elmer Applied Biosystems, Foster City, Calif.). Reactions were set up according to the manufacturer's protocols except that colony material was sometimes used as the template. The reactions were carried out under the following conditions: 94°C for 3 min; 30 cycles of 94°C for 30 s, 60°C for 10 min, and 72°C for 12 min; and 72°C for 10 min.
TABLE 1.
PCR primers used in this study
| Name | Sequencea (5′-3′) |
|---|---|
| UpA-Forward | GCG AAT TAC ATA TGC CCT CAC GCC ATC CTC TTT TAT |
| UpA-Reverse | AAG GTA CCC TGA CCG CGC ATT TTT TAT TCT |
| DnA-Forward | AAG GTA CCC CAT CGC TCA ACG GAT AAA A |
| DnA-Reverse | CTA CTC TAG ACG TCG CAT CCG GCA TTT TTT T |
| UpB-Forward | CCG AAT TAC ATA TGA CCG TGC TGG TGT TTG AC |
| UpB-Reverse | GAG GGA TCC GCA ACA TTC AAC CAA ATC A |
| DnB-Forward | CGA GGA TCC CCA TCG CTC AAC GGA TAA AA |
| DnB-Reverse | CTA CTC TAG ACC TGA TGC AAA AAC GAG GCT AGT TTA |
Recognition sequences (underlined) of restriction endonucleases NdeI, (UpA-Forward, UpB-Forward); KpnI (UpA-Reverse, DnA-Forward), XbaI (DnA-Reverse, DnB-Reverse), and BamHI (UpB-Reverse, DnB-Forward) are shown within additional nucleotide sequence (boldfaced) at the 5′ end of each primer.
Deletion of rRNA operons.
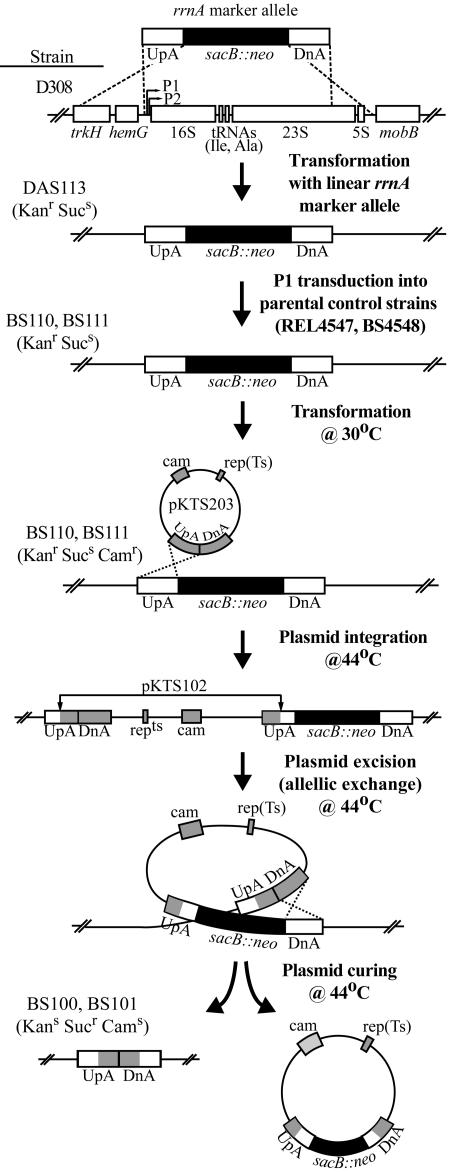
rRNA operons were deleted from a well-studied strain of E. coli (REL4547 [43]) that is unable to utilize the sugar arabinose (Ara−) and had evolved for 10,000 generations in batch cultures of glucose minimal medium, and from an otherwise isogenic strain (BS4548) carrying a spontaneous mutation (Ara+). The first step in the deletion of each rRNA operon was the construction of “intermediate” strains, in which the wild-type rRNA operon was replaced with an operon-specific marker allele (a sacB::neo cassette; see below), leaving no homology upstream of the marker to any of the other rRNA operons (Fig. 1). The use of site-specific sequence upstream of the targeted rRNA operon prevented the cell from repairing the mutation through recombination with one of the other rRNA operons (discussed in reference 20).
FIG. 1.
Outline of procedure used to delete the rRNA operon rrnA from the chromosome of E. coli. Homologous recombination events are depicted as crossed dotted lines, which encompass regions of homology; plasmid-derived regions of DNA are depicted in grey.
The rrnA marker allele (Fig. 1) contained the region (UpA) between position 369 of trkH and 350 nucleotides upstream of the rrnA 16S gene, which was PCR amplified from the REL4547 chromosome by using the UpA primers (Table 1); a sacB::neo cassette cleaved from pIB279 (8) with BamHI; and the region (DnA) between position 2416 of the rrnA 23S gene and 30 nucleotides upstream from the mobB stop codon, which was PCR amplified from the E. coli K-12 rrnA operon located on pC1 (3) by using the DnA primers (Table 1). The rrnB-specific marker allele contained the region (UpB) between position 78 of the murI gene and 358 nucleotides upstream of the rrnB 16S gene, which was PCR amplified from the REL4547 chromosome by using the UpB primers (Table 1); the sacB::neo cassette cleaved from pIB279 with BamHI; and the region (DnB) between position 2415 of the rrnB 23S gene and 9 nucleotides upstream of murB, which was PCR amplified from the REL4547 chromosome by using the DnB primers (Table 1). The rrnA and rrnB marker alleles, cleaved from pRA113 and pRB222 with NdeI and XbaI, were used to transform a recD mutant strain of E. coli (D308) (36). Transformants (Kanr Sucs) had replaced the chromosomal rRNA operon with the marker allele through recombination. The marker allele was moved from the chromosome of these transformants (DAS113 for rrnA; DBS222 for rrnB) to the parental strains (REL4547; BS4548) or ΔrrnB strains (BS200; BS201) by using generalized transduction with P1vir (35). Transductants (Kanr Sucs) with the marker allele in place of the original rRNA operon were referred to as “intermediate” strains (BS110 and BS111, marker replacing rrnA; BS210 and BS211, marker allele replacing rrnB; BS310 and BS311, marker allele replacing rrnA in ΔrrnB strains).
Intermediate strains were transformed at the permissive temperature (30°C) with pKTS102 (Fig. 1) or pKTS203, both of which were derived from pMAK705 [rep(Ts) Camr] (18), and contained a truncated rrnA (UpA:DnA) or rrnB (UpB:DnB) operon. Growth of transformants (Kanr Sucs Camr) at the nonpermissive temperature (44°C) promoted the integration of the plasmid into the chromosome through recombination (Fig. 1). Subsequent growth in LB without antibiotics at 44°C favored cells that had undergone a second recombination event (excision of the plasmid from the chromosome) and lost the plasmid (8). An allelic exchange occurred if this second recombination was within the region of homology opposite from the site of integration (as shown in Fig. 1). The resulting rRNA operon deletion strains (Kans Sucr Cams), which had undergone this allelic exchange and lost the plasmid containing the marker allele, were selected by their growth on LBsuc at 30°C.
The genotypes of the parental and deletion strains were verified by size analysis of PCR products from affected genomic regions and Southern hybridization of PvuII-digested genomic DNA with a digoxigenin-dUTP-labeled DNA probe for a conserved region of the 16S gene (positions 8 to 536) (as described in references 20, 25, and 39).
Relative fitness assays.
For batch culture competition experiments, relative fitness was determined as reported in reference 20 by a method described by Lenski et al. (28), by the following equations:
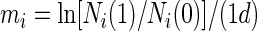 |
(1) |
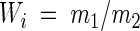 |
(2) |
where Ni(0) is the initial population density, Ni(1) is the population density after 1 day (d), and mi is the Malthusian parameter for that strain. The relative fitness (Wi) of each rRNA operon deletion strain is defined by equation 2 as the ratio of its Malthusian parameter (m1) to that of the parental strain (m2). Statistical analyses of the relative fitness measured for each strain included analysis of variance and planned comparisons among means (38).
The relative fitness of each deletion strain was measured in chemostat culture competitions against the parental strain by monitoring the change in density of each population as a function of total population doublings. The fitness of each strain under chemostat conditions was also assessed by comparison of calculated J parameters. The J parameter, a weighted value of Ks (see below), represents the subsistence or “break-even” concentration of the limiting resource for each strain under chemostat conditions at equilibrium, and the steady-state concentration of the limiting resource when each strain is grown alone. The J parameter was calculated by the following equation:
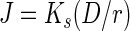 |
(3) |
where Ks is the Monod half-saturation constant (see below), D is the chemostat dilution rate (per hour), and r, the intrinsic rate of increase of a particular species, is calculated as (μmax − D) > 0, where μmax is the maximal growth rate (19). J parameters were calculated for each strain at the slow (0.11 h−1) and fast (0.81 h−1) dilution rates used in chemostat competition experiments. Chemostat competition experiments were initiated from individual DM+25 chemostat cultures at equilibrium (no change in population density for three volume changes) for each competitor. Equal volumes of each equilibrium chemostat culture were transferred to largely fill empty replicate chemostat vessels, thus reducing the time needed to reestablish equilibrium of the total population density. Relative population densities were estimated by dilution and plating onto TA agar medium.
Measurement of individual components of fitness.
The time required for each strain to reach exponential growth after inoculation (lag time) was estimated by subtraction. The cultures used for these measurements were treated identically to those in batch culture competition experiments, except that each inoculation was staggered by 15-min intervals to ensure precise timing. Immediately after inoculation and mixing (t = 0), and exactly 4 h later (t = 4), population density was estimated by dilution and plating onto LB agar plates. The time interval of 4 h was determined empirically to be within mid-exponential growth for all strains.
The theoretical time required to reach the population density at t = 4 from the initial population density, at the maximal specific growth rate determined for each strain (described below), was subtracted from the actual time required (4 h). The μmax of each strain was estimated by calculating the slope of the linear regression of ln-transformed population density values (optical densities at 600 nm) versus time during exponential growth in DM+1000. DM + 1000 was used to obtain sufficient optical densities for accurate measurement with a Lambda 3 UV-Vis spectrophotometer (Perkin-Elmer Applied Biosystems).
Ks represents the concentration of a growth-limiting substrate at which a culture grows at half the maximal specific growth rate (30) and is defined by the following equation:
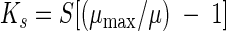 |
(4) |
where S is the concentration of the substrate and μ is the specific growth rate at [S]. The specific growth rate was estimated for each strain in a manner similar to that described above for μmax, except that cultures were grown in DM+0.1, which contains glucose at a concentration very near the Ks for the parental strain (42), and population densities were estimated by dilution and plating on LB agar. Ks values represent the mean of estimates calculated for three replicate cultures of each strain.
The death rate of each strain during stationary phase was represented by the rate of decrease in population density between 12 and 24 h, measured as described by Vasi et al. (42). Population densities of three replicate cultures were estimated every hour after inoculation between 12 and 24 h by dilution and plating onto LB agar. The mean CFU per liter for each culture at each time point was ln transformed and plotted versus time. This ln-transformed regression was assumed to be linear, and the inverse slope was recorded as the death rate (i.e., a positive death rate represented a decrease in population density). Population densities continued to be monitored periodically for each strain up to 197 h after inoculation.
Data analyses.
Analysis of variance was used to test for significant differences in the means (P < 0.05) for relative fitness in batch culture (n = 24), lag time (n = 8), μmax (n = 12), and Ks (n = 3), followed by planned comparisons of the sum of squares to determine which means were significantly different from each other (38).
Modeling competition experiments.
A model designed to simulate the competition of two populations for a single limited resource was programmed using object-oriented software (STELLA, version 5.1.1; High Performance Systems, Hanover, N.H.) (39). The model is a three-compartment system that defines the interaction between the state variables of a limiting resource (glucose, expressed in grams per liter) and two competing populations (population 1 and population 2, expressed in cells per liter), in which the concentration of the limited resource in the culture vessel is the central state variable of the system. The dynamics of the limited resource are determined by the initial concentration (in grams per liter) and the utilization of the resource by each population (in grams of the resource per cell). The growth (cells per liter per hour) divided by the yield (cells per gram of resource) of each population determined the rate of removal of the resource from the system due to utilization (grams of the resource per liter per hour) by each population. The model was programmed to also allow for continuous or semicontinuous addition of resources and washout of cells to simulate chemostat culture competitions.
The dynamics of each population's density are controlled by growth and death. The rate of growth of each population follows the Monod equation, which defines the relationship between the specific growth rate of a population and to the concentration of a limiting resource (30). In addition to the parameters described above, lag time is programmed to delay the growth of a population at the beginning of a simulation, for the duration of the lag time (in hours). Since no significant death rate was measured during the 24-h competition experiments, this value was set to zero. All simulations were run by using Euler's method of derivation with a step time of 0.00625 h (Stella Technical Documentation; High Performance Systems, Inc.).
RESULTS
Deletion of rRNA operons rrnA and rrnB.
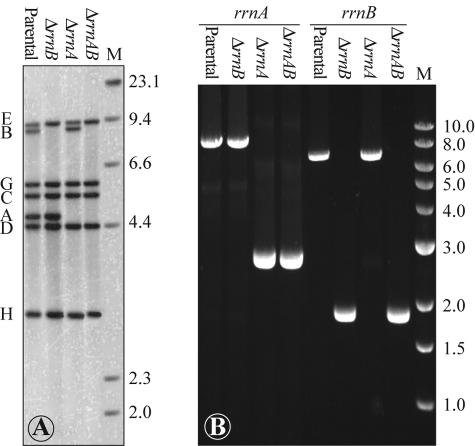
Strains of E. coli in which one or two (rrnA, rrnB, or both rrnA and rrnB) of the seven rRNA operons were deleted from the chromosome were constructed. Each deletion included removal of the tandem promoters for the operon, the 16S gene, the internally transcribed spacer (ITS) region, and a majority of the 23S gene (Fig. 1). Southern hybridization of PvuII-digested genomic DNA with a 16S ribosomal DNA-specific probe showed the loss of fragments corresponding to each deleted rRNA operon (Fig. 2A), and PCR of affected regions of the chromosome resulted in products that were 4.8 kb smaller (Fig. 2B), verifying the genotype of each rRNA deletion strain.
FIG. 2.
(A) Southern hybridization of PvuII-digested genomic DNA from parental, ΔrrnB, ΔrrnA, and ΔrrnAB strains. Locations of hybridized bands corresponding to the 16S rRNA gene from each rRNA operon (bands A through E, G, and H) are indicated for the parental strain. (B) PCR-amplified rrnA and rrnB regions from each strain. PCR products corresponding to intact rrnA and rrnB operons are approximately 7.4 and 6.7 kb long, whereas deleted rrnA and rrnB operons are much smaller (2.6 and 1.9 kb, respectively). Sizes of markers in lanes M are given (in kilobases) to the right of each panel.
Fitness effect of rRNA operon deletion(s).
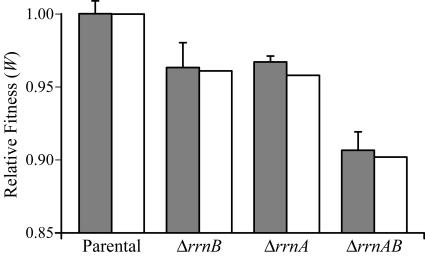
In batch culture competition experiments, the fitness of strains with one rRNA operon deleted (ΔrrnA or ΔrrnB) decreased relative to that of the parental strain by 3 to 4% (P < 0.025), while strains with two rRNA operons deleted experienced a 9% decrease in fitness (P < 0.025) (Fig. 3; Table 2). The mean relative fitness values did not differ between the ΔrrnA and ΔrrnB strains, in which a single rRNA operon was deleted (P > 0.10). In reciprocal experiments, the arabinose marker proved to be neutral, and therefore Ara+ and Ara− strains with the same rRNA operon genotype were treated as identical.
FIG. 3.
Effects of rRNA operon deletions on relative fitness (W) in batch culture. The mean relative fitness of the parental strain (Ara+ versus Ara−) and of rRNA operon deletion strains (ΔrrnB, ΔrrnA, and ΔrrnAB) versus the parental strain was measured in direct (grey) and simulated (white) batch culture competition experiments. Error bars, standard deviation of the mean (n = 8).
TABLE 2.
Fitness for each strain in batch and chemostat competition experiments
| Strain | Relative fitness (W) in batch culturea |
J in chemostat cultureb with a:
|
|
|---|---|---|---|
| Slow dilution rate | Fast dilution rate | ||
| Parental | 1.000 (±0.009) | 0.0141 | 0.434 |
| ΔrrnB | 0.963 (±0.017) | 0.0137 | 0.473 |
| ΔrrnA | 0.967 (±0.004) | 0.0189 | 0.652 |
| ΔrrnAB | 0.907 (±0.013) | 0.0186 | 0.671 |
Relative fitness of Ara+ versus Ara− strains for the parental strain, or of ΔrrnB, ΔrrnA, and ΔrrnAB strains versus the parental strain in glucose minimal medium (DM + 25). Values in parentheses are standard deviations of the mean (n = 24).
J, subsistence concentration for the limiting resource (Glu; calculated in micrograms per milliliter) in chemostats with slow (0.11 h−1) and fast (0.81 h−1) dilution rates.
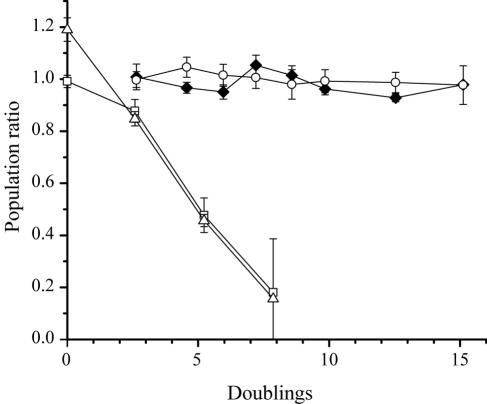
In chemostat culture competition experiments, the deletion of rrnA or rrnB had a differential effect on fitness, which did not correlate with the number of rrn operons deleted. Deletion of the rrnB operon in ΔrrnB strains had very little effect on fitness in chemostats, whereas deletion of rrnA in ΔrrnA and ΔrrnAB strains resulted in a significant decrease in fitness relative to that of the parental strain (Fig. 4). Compared to the parental strain, the relatively small differences in J for ΔrrnB strains (≤8%) were consistent with their coexistence with the parental strain in chemostat competition experiments over 14 doublings. The substantially larger differences in J for ΔrrnA and ΔrrnAB strains (≥24%) compared to the parental strain accurately predicted the outcome of those chemostat competition experiments, in which ΔrrnA and ΔrrnAB strains were undetectable after as few as 8 doublings (Table 2; Fig. 4).
FIG. 4.
Effects of rRNA operon deletions on relative fitness in chemostat culture (dilution rate, 0.11 h−1). The ratios of population densities (rrn deletion strain/parental strain) for ΔrrnA (squares), ΔrrnB (circles), ΔrrnAB (triangles), and parental strains (filled diamonds) (Ara+ versus Ara−) are plotted as a function of the number of doublings. Error bars, standard errors for four replicate competition cultures.
Effects of rRNA operon deletions on individual components of fitness.
The mean values for the lag time, μmax, and Ks for each strain are given in Table 3. The effects on lag time and maximal growth rate were directly related with the number of rRNA operons in each strain. Only the mean lag time of ΔrrnAB strains, which was about 20% (12 min) longer, was significantly different from that of the parental strain (P < 0.05). The parental strains, therefore, had a head start of about one-third of a doubling over cells with only five rRNA operons in competition experiments. Maximal growth rates of the rRNA deletion strains were slightly lower than that of the parental strain and also correlated with the number of deleted rRNA operons (P < 0.10 for six rrn operons; P < 0.05 for five rrn operons).
TABLE 3.
Growth parameters for each straina
| Strain | Lag time (h)b | μmax (h−1)c | Ks (μg/ml)d) |
|---|---|---|---|
| Parental | 1.43 (±0.039) | 1.03 (±0.011) | 0.118 (±0.0035) |
| ΔrrnB | 1.48 (±0.091) | 1.00 (±0.012) | 0.111 (±0.0025) |
| ΔrrnA | 1.48 (±0.076) | 1.00 (±0.016) | 0.153 (±0.0047) |
| ΔrrnAB | 1.71 (±0.070) | 0.99 (±0.014) | 0.149 (±0.0116) |
Values are means of measurements made in DM + 1000 for lag time (n = 8) and μmax (n = 12) and in DM + 0.1 for Ks (n = 3). Standard errors of the means are given in parentheses.
Only the mean for the ΔrrnAB strain is significantly different from all others (P < 0.05).
Means for the ΔrrnA and ΔrrnB strains are significantly different from that of the parental strain (P value between 0.05 and 0.10), and the mean for the ΔrrnAB strain is significantly different from all others (P < 0.05).
Based on specific growth rates for each strain on DM + 0.1 (0.47 for the parental strain, 0.49 for the ΔrrnB strain, 0.40 for the ΔrrnA strain, and 0.41 for the ΔrrnAB strain. Means for the ΔrrnA and ΔrrnAB strains are not different from each other but are different from those for the ΔrrnB and parental strains (P < 0.05), which are not different from each other.
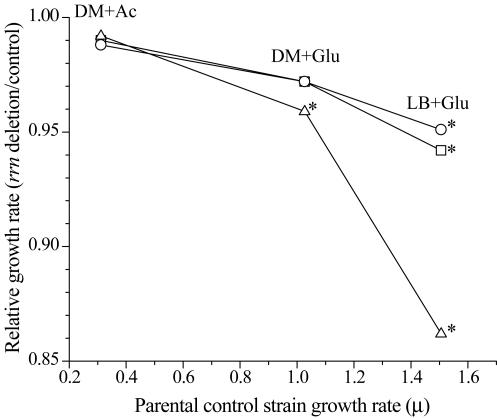
The deleterious effect of having fewer rRNA operons was magnified as strains were grown in media permitting more-rapid growth (Fig. 5). In the richest medium tested (LB+Glu), all deletion strains had growth rates that were significantly different from that of the parental strain (μ = 1.51 h−1) (P < 0.05). Furthermore, the decrease in relative growth rates correlated with the number of rRNA operons deleted: deletion of both rrnA and rrnB had a consistently greater impact on the growth rate than deletion of either rrnA or rrnB alone. In media providing for the slowest growth tested, there was no significant difference (P > 0.05) in growth rates between any of the rRNA deletion strains and the parental strain (μ = 0.31 h−1 for all).
FIG. 5.
The decreases in relative growth rates (rrn deletion strain/parental strain) for ΔrrnA (squares), ΔrrnB (circles), and ΔrrnAB (triangles) strains are plotted as a function of the growth rate of the parental strain on the same medium (DM+Ac, DM+Glu, LB+Glu). Data points marked with an asterisk represent growth rates significantly different from that of the parental strain (P < 0.05).
Unlike lag times and maximal growth rates, the Ks for each strain was not correlated with the number of rRNA operons deleted (Table 3). Instead, the increases in Ks for ΔrrnA and ΔrrnAB strains correlated with the deletion of the rrnA operon (P < 0.05). None of the strains had an appreciable death rate between 12 and 24 h after inoculation, with no significant change in population densities until after 48 h, when the number of viable cells dropped rapidly in all strains.
Model simulations of competition experiments.
Competition experiment simulations were conducted using the individual parameters (lag time, μmax, Ks, and death rate) measured for each strain. Empirically determined values for initial population densities (2.0 × 108 cells liter−1) and cell yields (1012 cells g−1) were also used to parameterize the model. Relative fitness values determined from simulations of batch and chemostat competition experiments were very close to empirical values. Simulated batch culture competitions predicted relative fitness values (ΔrrnA, 0.958; ΔrrnB, 0.961; ΔrrnAB, 0.902) that were within 1 to 2% of empirical values (Fig. 3; Table 2). In chemostat competition simulations, the population densities of deletion strains followed dynamics similar to those indicated by empirical results, as shown by the values for J in Table 2. Furthermore, the chemostat simulations with data corroborated the mean Ks values measured for ΔrrnA and ΔrrnAB strains (Table 3).
DISCUSSION
Resource availability is arguably one of the largest selective forces acting upon populations of microbes (see, e.g., references 9 and 28). The ability of microbes to respond effectively to transient increases in resource levels is, therefore, a life history trait that would have a profound impact on their evolution. Understanding the evolution and physiology behind this life history trait would advance our capacity to predict their behavior in natural and in managed ecosystems.
Correlative data exists which links the number of rRNA operons on a bacterial chromosome with an organism's capacity for response to resource availability. Strains of the genus Mycobacterium, for example, can be roughly divided between slow growers (doubling time, ≥16 h) and fast-growers (doubling time, 3 to 4 h), having either one or two rRNA operons per chromosome, respectively (7, 34). There are also numerous examples of bacteria for which the number of rRNA operons on the chromosome coincides with their presumed life history. Bacteria such as the bioluminescent symbiont of the Caribbean flashlight fish Kryptophanaron alfredi (43), members of the aphid endosymbiotic genus Buchnera (37, 41), parasitic mycoplasmas (1), and the marine oligotroph Sphingomonas sp. strain RB2256 (17) have one or two rRNA operons. These bacteria exist in relatively stable environments, tend to have slow growth rates, and respond slowly to fluctuations in resource availability. Bacteria with a relatively high number of rRNA operons on their chromosomes include E. coli with 7 (5), Vibrio spp. with 7 to 10 (21, 24, 33), and Bacillus subtillus with 10 rRNA operons (27); these organisms have relatively fast growth rates and would be expected to respond more quickly to fluctuations in resource availability. The hypothesis that the number of rRNA operons is reflective of an organism's ecological strategy is also supported by the correlation between rRNA operon copy number and the rate of colony appearance for soil isolates (25). Bacterial isolates that formed visible colonies in a few days had an average of five rRNA operons, whereas isolates that formed colonies up to 2 weeks later had only one or two rRNA operons on their chromosomes.
Despite the considerable predictive power associated with having a readily assessed genomic marker (rRNA operon copy number) with which to infer an organism's ecological strategy, there are few direct measurements of the influence of rRNA operon copy number on an organism's life history. Previous alterations of rRNA operons in E. coli included strains in which plasmid-borne rRNA operons were introduced (as in references 23 and 40), where rRNA genes were truncated or interrupted with antibiotic resistance genes but could still be expressed from intact promoters (2-4, 13), or in which adjacent genes on the chromosome were affected (16). Potential secondary effects in each of these manipulated strains confound attempts to measure the effect of rRNA operon copy number on physiological traits.
To assess the consequences of variation in rRNA operon copy number, we developed a strategy for deleting rRNA operons in order to create strains of E. coli with one or two rRNA operons (including promoters) deleted that were otherwise isogenic (Fig. 1). rRNA operons rrnA and rrnB were targeted for deletion in order to minimize perturbation of the tRNA gene pool. The tRNAs present in the internally transcribed spacers of rrnA and rrnB are replicated in other rRNA operons, and neither of these operons contains distally located tRNA genes (31). rrnA and rrnB are also located adjacent to each other on the chromosome, reducing differences in gene dosage that could occur in cells with multiple DNA replication loops (10, 15). The construction of these strains enabled a direct measurement of the fitness effects associated with varying the rRNA operon copy number.
Results from batch culture competition experiments support the hypothesis that multiple rRNA operons are an advantage under culture conditions defined by fluctuating resource availability (Fig. 3; Table 2). As the number of rRNA operons was decreased from 7 to either 6 or 5 copies per chromosome, the ability of these cells to compete with the parental strain for the same limiting resource (glucose) decreased. Although differences between individual components of fitness for each strain were not always statistically significant, the cumulative changes in lag time and growth rate were sufficient to account for 98 to 99% of the relative fitness of the strains in simulated competition experiments modeled on Monod kinetics (Fig. 3).
The observation that deletion of rrnA or rrnB had a relatively small (1 to 2%) impact on the growth rate suggests that the mutant strains are able to compensate, in large part, for these deletions. If there were no means for compensation, deletion of any one of the seven rRNA operons would result in a 14% decrease in growth rate. The most likely mechanism to account for that compensation is the recognized capacity for increased expression from the remaining intact operons (13). However, as expression of rRNA operons approaches maximal levels, the capacity to compensate for deletions should decrease. To test for this compensatory capacity at different expression levels, the growth rate of each of the strains was varied. As predicted, deletion strains were able to compensate completely for the deletion of either rrnA or rrnB or both at low growth rates, but as expression of the rRNA operons was increased at higher growth rates, the deleterious effect of the deletions became increasingly pronounced (Fig. 5). Although the decrease in maximal growth rate that coincided with fewer rRNA operons might appear to be relatively small, sensitivity analyses of our batch culture competition model corroborated those by Vasi et al., which demonstrate that relative fitness is extremely sensitive to even minor changes in growth rates under batch culture conditions (42).
While it may be tempting to assert that the advantage conferred by multiple rRNA genes is simply the capacity for higher growth rates, deletion of rrnA or rrnB in E. coli had a similar impact on a second component of fitness—lag time. In nature, the capacity for rapid response to resources (i.e., shorter lag time) may be more important than maximal growth rate, particularly in environments with fluctuating resource availability. Lag times measured in these experiments may be more informative than measurements of “shift-up” times in strains of E. coli with inactivated rRNA operons (13). While “shift-up” times reflect the amount of time needed to shift from one exponential-growth rate to a higher growth rate, lag times include the time necessary to initiate growth from a nongrowing physiological state—a common condition for microbes in nature.
The results from competition experiments in the chemostat did not reveal a selective advantage for the knockout strains that might have resulted from faster growth due to more efficient utilization of resources (Fig. 4). There was, however, a differential effect on fitness in chemostat culture with the deletion of the rrnA versus the rrnB operon. Deletion of the rrnA operon resulted in a substantial decrease in fitness, whereas no difference in fitness was found for the deletion of the rrnB operon. Although we do not know the cause of this differential effect, it might be the first significant evidence of differential properties between rRNA operons in E. coli (14), and it directly correlates with differences in measured Ks and therefore with estimates of J in these strains (Table 3). The J parameter represents the lowest concentration of a limiting resource where the specific growth rate of an organism is equal to the dilution rate, and it can be used to accurately predict the outcome of chemostat competition experiments (19). J parameter values for the ΔrrnB strain were within 8% of those for the parental strains, which is consistent with the coexistence of these two strains in chemostat culture competition experiments (Fig. 4). The more substantial differences in J (>24%) between rrnA deletion strains and the parental strain were consistent with the much lower relative fitness of ΔrrnA strains in chemostats. Perhaps it is not surprising that strains with fewer rRNA operons were unable to make more efficient use of the limiting resources, considering that these experiments were carried out on a line of E. coli with cell machinery that had evolved for 10,000 generations to take advantage of periodic fluctuations in resource availability (42). The large effect of deleting the rrnA operon in ΔrrnA and ΔrrnAB strains, relative to the parental strain or the ΔrrnB strain, was unexpected and remains unresolved.
It is necessary to consider other possible explanations for the experimental results reported here. The fact that genomic DNA from an intermediate construct in E. coli K-12 was used during generalized transduction with phage P1vir raises the possibility that DNA other than the targeted rRNA operons could have been incorporated into the recipient strain and could have been responsible for the observed changes in phenotype. We view this as an unlikely source of the variation, because the E. coli recipient strain and strain K-12 are closely related within one subgroup of E. coli (22), and there is only a single known region of intraspecific sequence divergence (the phn operon) located within 100 kb of the rrnA and rrnB operons. We also note that a similar decrease in growth rate was observed when the rrnB operon was inactivated in the same parental strain of E. coli by the insertion of the sacB::neo marker allele into the 16S gene (20). Since the effect on relative fitness of the inactivation of the rrnB operon was indistinguishable from that of BS200 and BS201, used in the present study, we conclude that changes in growth rate and lag time are attributable to deletion of the rRNA operons.
It is also not possible to distinguish the effect of the deletion of rRNA genes from the deletion of tRNA genes that are found in the ITS in rrnA and rrnB operons. The rrnA ITS region contains the two tRNA genes tRNAAla and tRNAIle, whereas the rrnB ITS region contains the tRNAGlu gene (32). Although the tRNA genes found in the ITS regions of rrnA and rrnB are repeated in the other rRNA operons found elsewhere on the chromosome, the pools of the respective tRNAs may have been perturbed in these strains.
In summary, the research reported here supports the hypothesis that the rRNA operon copy number is a component of bacterial life histories. The ecological strategies of bacteria with many rRNA operons most likely include quick response and fast growth upon an influx of resources, as opposed to a strategy of more efficient utilization of limited resources. We are continuing to investigate the mechanisms behind a potential tradeoff for rRNA operon copy numbers in microbes in order to better understand the role that this trait plays in evolution and life history in the microbial world.
Acknowledgments
This research was supported by a grant from the National Science Foundation (IBN 9875254), awarded to T.M.S.
We thank Richard E. Lenski for providing essential guidance on the measurement of relative fitness in batch culture and the parental strain REL4547. We also thank Les Dethlefsen for many valuable conversations regarding fitness.
REFERENCES
- 1.Amikam, D., G. Glaser, and S. Razin. 1984. Mycoplasmas (Mollicutes) have a low number of rRNA genes. J. Bacteriol. 158:376-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ammons, D., J. Rampersad, and G. E. Fox. 1999. 5S rRNA gene deletions cause an unexpectedly high fitness loss in Escherichia coli. Nucleic Acids Res. 27:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai, T., C. Condon, J. Voulgaris, D. Zaporojets, B. H. Shen, M. Al-Omar, C. Squires, and C. L. Squires. 1999. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181:3803-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asai, T., D. Zaporojets, C. Squires, and C. L. Squires. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. USA 96:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachellier, S., E. Gilson, M. Hofnung, and C. W. Hill. 1996. Repeated sequences, p. 2012-2040. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, D.C.
- 6.Begon, M., J. L. Harper, and C. R. Townsend. 1998. Ecology, 3rd ed. Blackwell Sciences International, Oxford, United Kingdom.
- 7.Bercovier, H., O. Kafri, and S. Sela. 1986. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem. Biophys. Res. Commun. 136:1136-1141. [DOI] [PubMed] [Google Scholar]
- 8.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 9.Bohannan, B. J. M., and R. E. Lenski. 1997. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology 78:2303-2315. [Google Scholar]
- 10.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition, p. 1561-1562. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, D.C.
- 11.Chao, L., B. R. Levin, and F. M. Stewart. 1977. Complex community in a simple habitat: experimental-study with bacteria and phage. Ecology 58:369-378. [Google Scholar]
- 12.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condon, C., D. Liveris, C. Squires, I. Schwartz, and C. L. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condon, C., J. Philips, Z. Y. Fu, C. Squires, and C. L. Squires. 1992. Comparison of the expression of the 7 ribosomal RNA operons in Escherichia coli. EMBO J. 11:4175-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellwood, M., and M. Nomura. 1982. Chromosomal locations of the genes for ribosomal RNA in Escherichia coli K-12. J. Bacteriol. 149:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellwood, M., and M. Nomura. 1980. Deletion of a ribosomal ribonucleic acid operon in Escherichia coli. J. Bacteriol. 143:1077-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, S. R., and S. P. Hubbell. 1980. Single-nutrient microbial competition: qualitative agreement between experimental and theoretically forecast outcomes. Science 207:1491-1493. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto, J. G., B. S. Stevenson, and T. M. Schmidt. 2003. Rates and consequences of recombination between rRNA operons. J. Bacteriol. 185:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Y. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinks-Robertson, S., R. L. Gourse, and M. Nomura. 1983. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell 33:865-876. [DOI] [PubMed] [Google Scholar]
- 24.Khetawat, G., R. K. Bhadra, S. Nandi, and J. Das. 1999. Resurgent Vibrio cholerae O139: rearrangement of cholera toxin genetic elements and amplification of rrn operon. Infect. Immun. 67:148-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Dusterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S. Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Henaut, H. Hilbert, S. Holsappel, S. Hosono, M. F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. KlaerrBlanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S. M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. Oreilly, K. Ogawa, A. Ogiwara, B. Oudega, S. H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 28.Lenski, R. E., M. R. Rose, S. C. Simpson, and S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315-1341. [Google Scholar]
- 29.Levin, B. R., F. M. Stewart, and L. Chao. 1977. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111:3-24. [Google Scholar]
- 30.Monod, J. 1949. The growth of bacterial cultures. Annu. Rev. Microbiol. 3:371-394. [Google Scholar]
- 31.Morgan, E. A., T. Ikemura, L. Lindahl, A. M. Fallon, and M. Nomura. 1978. Some rRNA operons in Escherichia coli have tRNA genes at their distal ends. Cell 13:335-344. [DOI] [PubMed] [Google Scholar]
- 32.Morgan, E. A., T. Ikemura, and M. Nomura. 1977. Identification of spacer tRNA genes in individual rRNA transcription units of Escherichia coli. Proc. Natl. Acad. Sci. USA 74:2710-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nandi, S., G. Khetawat, S. Sengupta, R. Majumder, S. Kar, R. K. Bhadra, S. Roychoudhury, and J. Das. 1997. Rearrangements in the genomes of Vibrio cholerae strains belonging to different serovars and biovars. Int. J. Syst. Bacteriol. 47:858-862. [DOI] [PubMed] [Google Scholar]
- 34.Pongrapeeporn, K. S., J. Jearanaisilavong, A. Chaiprasert, S. Pattanakitsakul, P. Yenchitsomanus, and S. Panyim. 1996. Rapid-growing mycobacteria possess at least two copies of rRNA gene. Asia Pac. J. Mol. Biol. 4:43-47. [Google Scholar]
- 35.Provence, D. L., and R. Curtiss III. 1994. Gene transfer in gram-negative bacteria, p. 332-334. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 36.Russell, C. B., and F. W. Dahlquist. 1989. Exchange of chromosomal and plasmid alleles in Escherichia coli by selection for loss of a dominant antibiotic sensitivity marker. J. Bacteriol. 171:2614-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 38.Sokal, R. R., and F. J. Rohlf. 1995. Biometry: the principles and practice of statistics in biological research, 3rd ed. W. H. Freeman and Company, New York, N.Y.
- 39.Stevenson, B. S. 2000. Life history implications of ribosomal RNA gene copy number in Escherichia coli. Doctoral dissertation. Michigan State University, East Lansing.
- 40.Stevenson, B. S., and T. M. Schmidt. 1998. Growth rate-dependent accumulation of RNA from plasmid-borne rRNA operons in Escherichia coli. J. Bacteriol. 180:1970-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. E. Andersson. 2002. Fifty million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 42.Vasi, F., M. Travisano, and R. E. Lenski. 1994. Long-term experimental evolution in Escherichia coli. 2. Changes in life-history traits during adaptation to a seasonal environment. Am. Nat. 144:432-456. [Google Scholar]
- 43.Wolfe, C. J., and M. G. Haygood. 1993. Bioluminescent symbionts of the Caribbean flashlight fish (Kryptophanaron alfredi) have a single rRNA operon. Mol. Mar. Biol. Biotechnol. 2:188-197. [PubMed] [Google Scholar]