Abstract
Spoligotyping is a major tool for molecular typing of Mycobacterium tuberculosis complex organisms. For epidemiological purposes, strains are considered clonal only when their spoligotyping patterns are identical. We report a change in the spoligotyping profiles of truly isogenic strains (a clinical isolate and a subculture derived in the laboratory) caused by deletion of a direct variable repeat. Without the information about the relationship between them, a link between these strains would have gone unnoticed. Evolutionary events should be taken into account in the interpretation of spoligotyping results and in the design of databases.
The direct variable repeat (DVR) spacer oligonucleotide typing technique (spoligotyping) (17) detects DNA polymorphism within the direct repeat (DR) locus of Mycobacterium tuberculosis complex organisms. The DR locus contains multiple DVRs that consist of well-conserved 36-bp DRs interspersed with spacer sequences (spacers) 34 to 41 bp long (14). The order of the DVRs is strongly conserved in the various isolates (29). Polymorphism in this region appears to comprise mainly the presence or absence of single, discrete DVRs or stretches of contiguous DVRs (13, 29). Differentiation of strains is based on the presence or absence of the spacers in a hybridization pattern (spoligotype), and this has been exploited for the study of the epidemiology of tuberculosis. Although its level of discrimination is much lower than that obtained with the restriction fragment length polymorphism associated with IS6110 in most strains (strains carrying five or more IS6110 copies), the performance of the technique regarding the degree of differentiation and reproducibility is good (17, 18). Furthermore, the polymorphism obtained by spoligotyping of the IS6110 low-copy-number M. tuberculosis complex isolates proved to be superior to the polymorphism obtained by IS6110-associated restriction fragment length polymorphism (2, 10, 11, 18, 34).
This technique has gained widespread acceptance because it is a simple, rapid, and robust method. Results can be expressed in a simple digital format and are easier to compare and store in comparison with those of other available techniques. Thus, it has been extensively applied, alone or in conjunction with other techniques, for tracking epidemics (1, 4, 12, 16, 20, 26, 27, 34); for the description of highly prevalent families such as the Beijing family (30), multidrug-resistant strain W-Beijing (3), and Western Cape F11 (32); and to study global epidemiology (8). A search in June 2004 of the PubMed website of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/entrez) retrieved 186 papers on this topic since 1996. Furthermore, it has allowed the construction of databases with spoligotypes collected worldwide for the comparison of large numbers of strains with simple software, i.e., the database at http://www.pasteur-guadeloupe.fr/tb/spoldb3.htm, Institute Pasteur, Guadeloupe (8), and the specific database on its close relative M. bovis at http://www.Mbovis.org, hosted by the University of Sussex, Falmer, United Kingdom.
Three types of mechanisms have been hypothesized to be responsible for M. tuberculosis complex strain DR polymorphism: homologous recombination between DRs, rearrangements driven by the insertion element IS6110, which is present in the DR region of most M. tuberculosis complex strains (13), and successive deletion of a single DVR or multiple discrete DVRs from an archetypal DR region (29).
We report a change in the spoligotyping profile of M. tuberculosis isogenic strains (a clinical isolate and a subculture) obtained in the laboratory. This change was caused by the deletion of a single spacer and the adjacent DR.
M. tuberculosis strains.
An M. tuberculosis complex isolate (98/426) was cultured from the mediastinal lymph node collected at the post mortem examination of a dog with respiratory distress that did not respond to conventional treatments (31). Granulomatous lesions from the lymph node were homogenized with sterile distilled water and decontaminated with 0.35% hexadecylpyridinium chloride for 30 min (5), centrifuged at 1,068 × g for 30 min, and cultured onto Coletsos and 0.2% (wt/vol) pyruvate-enriched Löwenstein-Jensen media (bioMérieux España and Biomedics, Madrid, Spain). Tubes were incubated at 37°C and checked for growth weekly. The isolate was identified as M. tuberculosis by staining for acid-alcohol fastness, colony morphology, and PCR amplification of the Mycobacterium genus-specific 16S rRNA fragment and MPB70 sequence (35).
Isolate 98/426 was subcultured on Coletsos (Biomedics) from 1998 to 2003 as a part of the culture collection kept in our laboratory. The original isolate, designated isolate a, was subcultured, resulting in the first subculture, designated subculture b. Subsequently, subculture b was subcultured and the same procedure was repeated periodically during the aforementioned period of time. Bacterial growth was recovered from the tubes (although cells were not recovered from every subculture), suspended in sterile purified water, heat inactivated, and kept frozen at −25°C for use as the M. tuberculosis control in routine molecular tests. A series of DNAs was therefore available in chronological order, designated a (clinical isolate, 6/1998), b (first subculture, 6/1999), c (a subculture from 2/2000), d (9/2000), e (1/2001), f (4/2001), g (1/2002), and h (1/2003, last subculture).
Spoligotyping.
PCR amplification and hybridization were performed as described by Kamerbeek et al. (17), with only minor changes to obtain good hybridization patterns under our laboratory conditions. Amplification of the DR locus was performed with heat-treated cell suspensions. Crude supernatants of M. tuberculosis 98/426 and a clinical isolate of M. bovis and purified sterile water were included as controls in every batch of tests. The test was carried out with membranes obtained from Isogen Bioscience BV, Maarssen, The Netherlands.
A change (1 to 0) in spacer 2 in the traditional spoligotyping result (genome spacer 3 according to the numbering system of van Embden et al. [29]) was observed in batches g and h (Fig. 1) in comparison with results that had been obtained with batches a to f. The rest of the spacers remained unchanged. During this period of time, several membranes had been used; therefore, these results were corroborated in the same membrane.
FIG. 1.
Spoligotyping pattern of original strain 98/426 (a) and the pattern that resulted from batch g, showing the difference in the results obtained for spacer 2.
Detection of the DVR deletion.
To study the reason for this change in the spoligotyping profile, the following primers targeting adjacent spacers were designed to amplify a fragment of the 5′ region of the DR locus: sp1 (forward) (5′ AAC CAT AGA GGG TCG CCG 3′) and sp4 (reverse) (5′ GAT GAT TGG TCG GCG TAT 3′), which are based on the known sequences of spacers 1 and 4, respectively. DNA amplifications were performed in 25-μl reaction volumes containing 2.5 μl of 10× standard reaction buffer including 2 mM MgCl2 (Biotools, B&M Labs), 200 mM each deoxynucleoside triphosphate (dNTP mix; Biotools), 0.5 U of DNA polymerase (Biotools), and 50 ng of each oligonucleotide primer (Roche Molecular Biochemicals, Roche Diagnostics). Amplification was performed with 35 cycles of 30 s at 94°C, 2 min at 65°C, and 3 min at 72°C; the initial denaturation and final extension steps were extended for 10 min. Sterile water was included as a negative control. Amplification products were checked on a 2% agarose gel and examined under UV light after staining with ethidium bromide. The amplification products obtained with original isolate a (1998) and DNA batch h (2003) were purified with the QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany) and sequenced with the DyeDeoxy (dRhodamine) Terminator cycle sequencing kit in an automatic ABI Prism 373 DNA sequencer (Applied Biosystems) (CIB Sequencing Facilities, Madrid, Spain).
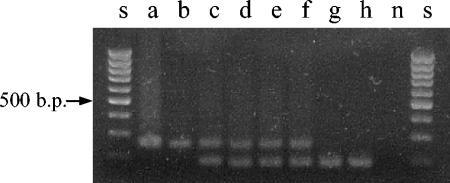
When DNA was amplified by PCR with primers sp1and sp4, a product of the expected 259 bp was observed in original isolate a and subculture b; two products (a 259-bp product and a smaller fragment) were detected in DNA batches c to f, while batches g and h yielded only the smaller PCR product (Fig. 2). The sequences from the amplification products of subcultures a and h, 259 and 187 nucleotides, respectively, were aligned with the M. tuberculosis DR cluster DNA (accession no. Z48304; Kamerbeek et al. [17]) from the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank). Sequence analysis showed a deletion of spacer 2 and an adjacent DR, but from the sequencing data it cannot be deduced whether the previous or the subsequent one was lost (Fig. 3).
FIG. 2.
PCR amplification of DNA batches a to h of M. tuberculosis strain 98/426 with primers sp1 (forward) and sp4 (reverse). Lanes s, 100-bp ladder (Biotools, B&M Labs) used as a DNA molecular size marker; lane n, negative control. The 259-bp band indicates the presence of the original sequence of the DR; the 187-bp band corresponds to the sequence without DVR 2 (deleted).
FIG. 3.
Alignment of the sequences of a fragment of the 5′ region of the DR locus. The first nucleotide corresponds to position 872 (accession no. Z48304). Annealing sites for the primers are underlined, and the DRs are shaded. Dashes indicate deletions. The true positions of the DR and the adjacent T (in italics) from subculture h cannot be deduced from the sequencing data (bases 916 to 951 and 952 and bases 988 to1023 and 1024, respectively).
We suppose that the deletion event happened in a single cell that subsequently multiplied successfully. Both populations coexisted during a period of time, and the spoligotyping pattern detected in the X-ray film is in the superimposed images of both profiles, with spacer 2. Finally, only the spoligotyping profile from the deleted strain can be detected. We cannot explain the mechanism causing the mutated population to overgrow the parental strain. It is tempting to speculate that the deletion gave the strain a growth advantage. The DVR belongs to the clustered regularly interspaced short palindromic repeats (CRISPR) family of sequences that are widely distributed among prokaryotic species (15, 22). A comparison of genes that flank the CRISPR loci has shown homology among four genes designated CRISPR-associated genes (cas); two of them, cas1 and cas2, have been found in the M. tuberculosis complex (15). However, except for their involvement in replicon partitioning in the archaeon Haloferax mediterranei (21), it is unclear whether the CRISPR loci and cas genes have a biological function (15, 22). Because the purpose of the subsequent subculturing was to obtain a large amount of mycobacterial cells, it was performed by loading the loops generously before inoculating the new medium, but we cannot dismiss the possibility of random selection of a single colony of the mutated strain.
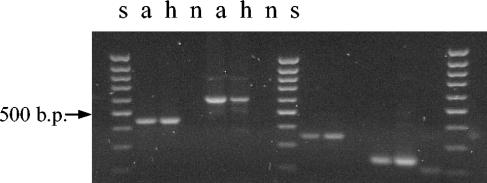
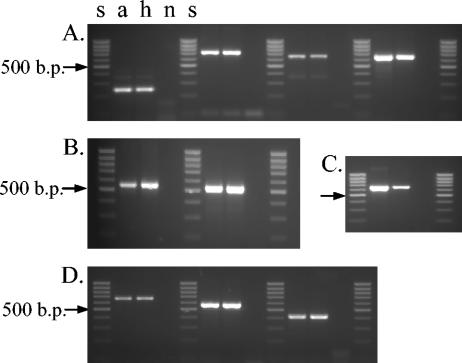
Contamination of the cultures was ruled out, as great precautions were always taken when subculturing this M. tuberculosis strain. The few other M. tuberculosis strains that we have isolated in our laboratory have different spoligotypes (data not shown). The original isolate, a, and the last DNA batch, h, were also studied by mycobacterial interspersed repetitive unit (MIRU)-variable-number tandem repeat (VNTR) analysis aimed at loci QUB11a (positions 2163333 to 2163531), QUB26 (positions 4052968 to 4053547) (25), ETR-A (positions 2165204 to 2165611), ETR-B (positions 2461280 to 2461550) (9), 4, 26, 40, 10, 16, 31, 2, 23, 39, and 27 (28), which showed identical profiles for these 14 loci (Fig. 4 and 5).
FIG. 4.
MIRU-VNTR analysis of batches a and h of M. tuberculosis strain 98/426 with the primers described by Skuce et al. (25). Amplification products were checked on a 2.5% agarose gel, run for 120 min at 45 V, and examined under UV light after staining with ethidium bromide. From left to right, loci QUB-11a, QUB-26, ETR-A, and ETR-B are shown. Lanes s, 100-bp ladder (Biotools, B&M Labs) used as a DNA molecular size marker; lane n, negative control.
FIG. 5.
MIRU-VNTR analysis of batches a and h of M. tuberculosis strain 98/426 with the primers described by Supply et al. (28). Amplification products were checked on a 2.5% agarose gel, run for 90 min at 45 V, and examined under UV light after staining with ethidium bromide. Panels: A, loci 4, 16, 10, and 31 (left to right); B, loci 26 and 40; C, locus 39; D, loci 2, 23, and 27. Lanes s, 100-bp ladder (Biotools, B&M Labs) used as a DNA molecular size marker; lane n, negative control.
Spoligotyping has rapidly become an accepted standard technique for molecular epidemiology of M. tuberculosis complex infections. DR spacer markers are stable enough for spoligotyping to be used as a tool for molecular epidemiology; thus, for epidemiological purposes, strains are considered clonal only when their spoligotyping profiles are identical. Little is know about the microevolutionary events associated with the DR locus and their influence on the interpretation of spoligotyping. The pace of the “molecular clock” associated with this genetic marker seems to be extremely slow, as multiple M. tuberculosis isolates from the same patients corresponding to relapses or infections at different sites showed identical spoligotypes (6, 10, 11, 23, 26).
Some recent studies have focused on the processes involved in the genetic changes that may produce polymorphism in the DR region by comparison of M. tuberculosis strains that have some features in common or isolates from a strain family, although they are not strictly isogenic. Occasional insertion element-driven polymorphisms have been described: IS6110-mediated deletion in the DR region leading to the loss of large blocks of DVRs adjacently situated (24), insertion of IS6110 into the DR sequence that results in asymmetrical disruption that does not serve as a PCR target (19, 24, 29, 33), and insertion of IS6110 into the spacer sequence that prevents hybridization (33). Loss of an intact, discrete DVR or a set of contiguous DVRs has also been described in presumably isogenic pairs of strains or related strains (7, 26, 29, 33).
To our knowledge, this is the first report of a change in the DR region in truly isogenic strains (subculture derived in the laboratory). This change has been caused by deletion of a DVR, yielding a different spoligotyping pattern. Our observation corroborates the hypotheses suggested by Van Embden et al. (29) of deletion of a single DVR or multiple discrete DVRs as a mechanism underlying M. tuberculosis strain-to-strain variation.
These two strains would not have been grouped together in the same cluster under the current criteria for interpretation of spoligotyping data. This means that a link between these isogenic strains would have gone unnoticed. It is likely that these deletion events also happen in vivo, although different molecular clocks may exist. Bringing our observation into the perspective of epidemiology highlights two aspects: (i) the information derived from spoligotyping must always be integrated with detailed and precise information such as that provided by classical epidemiology and contact tracing and (ii) the need to take into account the evolutionary events in the DR region for the interpretation of spoligotyping results and for the design of databases.
Acknowledgments
A. Aranaz is the recipient of a fellowship from the Ramon y Cajal Program (Spanish Ministry of Science and Technology). This work was supported by project AGL 2001-2029 of the Spanish Ministry of Science and Technology and by the Spanish Ministry of Agriculture, Fisheries and Food (MAPA).
We thank the staff of the SADNA (CIB, Madrid, Spain) for sequencing. We are grateful to Matthew Guilmour for careful revision of the manuscript.
REFERENCES
- 1.Banu, S., S. V. Gordon, S. Palmer, R. Islam, S. Ahmed, K. M. Alam, S. T. Cole, and R. Brosch. 2004. Genotypic analysis of Mycobacterium tuberculosis in Bangladesh and prevalence of the Beijing strain. J. Clin. Microbiol. 42:674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, J., A. B. Andersen, K. Kremer, and H. Miörner. 1999. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J. Clin. Microbiol. 37:2602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, Z. Liu, S. L. Moghazeh, B Shopsin, B. Tempalski, J. Driscol, R. Frothingham, J. M. Muser, P. Alcabes, and B. N. Kreiswirth. 1999. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA 282:2321-2327. [DOI] [PubMed] [Google Scholar]
- 4.Caminero, J. A., M. J. Pena, M. I. Campos-Herrero, J. C. Rodríguez, I. García, P. Cabrera, C. Lafoz, S. Samper, H. Takiff, O. Afonso, J. M. Pavón, M. J. Torres, D. van Soolingen, D. A. Enarson, and C. Martín. 2001. Epidemiological evidence of the Beijing genotype on Gran Canaria island. Am. J. Respir. Crit. Care Med. 164:1165-1170. [DOI] [PubMed] [Google Scholar]
- 5.Corner, L. A., and A. C. Trajstman. 1988. An evaluation of 1-hexadecyl-pyridinium chloride as a decontaminant in the primary isolation of Mycobacterium bovis from bovine lesions. Vet. Microbiol. 18:127-134. [DOI] [PubMed] [Google Scholar]
- 6.Driscoll, J. R., M. A. McGarry, and H. W. Taber. 1999. DNA typing of a nonviable culture of Mycobacterium tuberculosis in a homeless shelter outbreak. J. Clin. Microbiol. 37:274-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, Z., N. Morrison, B. Watt, C. Doig, and K. J. Forbes. 1998. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J. Bacteriol. 180:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valétude, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Moström, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rosetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 10.Goguet de la Salmonière, Y. O., H. L. Li, G. Torrea, A. Bunschoten, J. van Embden, and B. Gicquel. 1997. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 35:2210-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goyal, M., N. A. Saunders, J. D. A. van Embden, D. B. Young, and R. J. Shaw. 1997. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J. Clin. Microbiol. 35:647-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal, M., S. Lawn, B. Afful, J. W. Acheampong, G. Griffin, and R. Shaw. 1999. Spoligotyping in molecular epidemiology of tuberculosis in Ghana. J. Infect. 38:171-175. [DOI] [PubMed] [Google Scholar]
- 13.Groenen, P. M. A., A. E. Bunschotten, D. van Soolingen, and J. D. A. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis: applications for strain differentiation by a novel method. Mol. Microbiol. 105:1057-1065. [DOI] [PubMed] [Google Scholar]
- 14.Hermans, P. W. M., D. van Soolingen, E. M. Bik, P. E. W. de Haas, J. W. Dale, and J. D. A. van Embden. 1991. The insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen R., J. D. A. van Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565-1575. [DOI] [PubMed] [Google Scholar]
- 16.Källenius, G., T. Koivula, S. Ghebremichael, S. E. Hoffner, R. Norberg, E. Svensson, F. Dias, B. Marklund, and S. B. Svenson. 1999. Evolution and clonal traits of Mycobacterium tuberculosis in Guinea-Bissau. J. Clin. Microbiol. 37:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. M. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Muser, and J. D. A. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legrand, E., I. Filliol, C. Sola, and N. Rastogi. Use of spoligotyping to study the evolution of the direct repeat locus by IS6110 transposition in Mycobacterium tuberculosis. J. Clin. Microbiol. 39:1595-1599. [DOI] [PMC free article] [PubMed]
- 20.Martínez, J. I., F. S. Chaves, A. A. Arce, M. S. Alonso, E. M. Palenque, F. H. Jaen, M. M. M. Teresa, M G. Gor, and A. N. Rodríguez. 2000. Recent transmission of tuberculosis in Madrid (Spain): usefulness of molecular techniques. Med. Clin. 115:241-245. [PubMed] [Google Scholar]
- 21.Mojica, F. J., C. Ferrer, G. Juez, and F. Rodríguez-Valera. 1995. Long stretches of short tandem repeats are present in the largest replicons of the archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 17:85-93. [DOI] [PubMed] [Google Scholar]
- 22.Mojica, F. J., C. Diez-Villaseñor, E. Soria, and G. Juez. 2000. Biological significance of a family of regularly spaced repeats in the genome of archaea, bacteria and mitochondria. Mol. Microbiol. 36:244-246. [DOI] [PubMed] [Google Scholar]
- 23.Niemann, S., E. Richter, and S. Rusch-Gerdes. 1999. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J. Clin. Microbiol. 37:409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sampson, S. L., R. M. Warren, M. Richardson, T. C. Victor, A. M. Jordaan, G. D. van der Spuy, and P. D. van Helden. 2003. IS6110-mediated deletion polymorphism in the direct repeat region of clinical isolates of Mycobacterium tuberculosis. J. Bacteriol. 185:2856-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skuce, R., T. P. McCorry, J. F. McCarroll, S. M. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 26.Soini, H., X. Pan, A. Amin, E. A. Graviss, A. Siddiqui, and J. M. Musser. 2000. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J. Clin. Microbiol. 38:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sola, C., A. Devallois, L. Horgen, J. Maïsetti, I. Filliol, E. Legrand, and N. Rastogi. 1999. Tuberculosis in the Caribbean: using spacer oligonucleotide typing to understand strain origin and transmission. Emerg. Infect. Dis. 5:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Embden, J. D. A., T. van Gorkom, K. Kremer, R. Jansen, B. A. M. van der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vela, A. I., I. Simarro, L. de Juan, A. Aranaz, N. Montero, A. Mateos, G. Suárez, L. Domínguez, R. de los Ríos, R. Martín, S. Moreno-Alcalde, A. García-Nieto, C. Martín-Scapa, E. Navas-Elorza, E. Gómez-Mampaso, and N. Gamarra. 2001. A propósito de un caso de tuberculosis canina. Prof. Vet. 49:48-53. [Google Scholar]
- 32.Victor, T. C., P. E. de Haas, A. M. Jordaan, G. D. van der Spuy, M. Richardson, D. van Soolingen, P. D. van Helden, and R. Warren. 2004. Molecular characteristics and global spread of Mycobacterium tuberculosiswith a Western Cape F11 genotype. J. Clin. Microbiol. 42:769-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, S. M., S. Gross, and F. Drobniewski. 1998. Evaluation of strategies for molecular fingerprinting for use in the routine work of a Mycobacterium reference unit. J. Clin. Microbiol. 36:3385-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilton, S., and D. Cousins. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1:209-273. [DOI] [PubMed] [Google Scholar]