Abstract
Infections with human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2, respectively) are zoonotic infections. In Africa, the potential exists for additional cross-species transmissions from at least 33 different species of simian immunodeficiency virus (SIV)-infected nonhuman primates (NHPs) through hunting and butchering of these animals for food. Here we describe a highly sensitive and specific enzyme immunoassay (EIA) with chemically modified, multiple antigenic peptides (MAPs) developed for the detection and discrimination of antibodies to SIV genetic lineages. The SIV EIA was developed by using a comprehensive array of MAPs covering two envelope gene regions from all of the SIV lineages for which env sequences were available. Assay sensitivity was evaluated by using 63 plasma or serum samples obtained from primates naturally or experimentally infected with SIVs from 10 genetic lineages. Assay specificity was determined by using 97 known SIV-negative plasma specimens from these same species. Also used in the evaluations were 369 human samples: 198 HIV seronegative, 170 HIV-1 and/or HIV-2 seropositive, and 1 from a human SIVsm infection. Overall assay sensitivity and specificity were 100% with both immunodominant region (IDR) and V3 region MAPs. Although SIV env sequences from talapoin monkeys were not available for specific MAP inclusion, 5 (100%) of 5 SIVtal-infected samples were detected through cross-reactivity with other SIV IDR MAPs used in the assay. The one human SIVsm infection was identified. In conclusion, our SIV MAP EIA proved to be highly sensitive and specific for detecting SIV infections in NHPs and humans. As shown with SIV-infected talapoin monkeys, this assay has the potential to detect previously unidentified SIV strains and should be suitable for sentinel surveillance for potential new cross-species transmissions of SIVs to humans.
Six major lentivirus phylogenetic lineages have been described for African nonhuman primates (NHPs) based on available full-length genome viral sequences: simian immunodeficiency virus (SIV) cpz (SIVcpz) from chimpanzees (Pan troglodytes), SIVsm from sooty mangabeys (Cercocebus atys), SIVagm from four species of African green monkeys (members of the genus Chlorocebus), SIVsyk from Sykes' monkeys (Cercopithecus mitis albogularis), SIVmnd from a mandrill (Mandrillus sphinx), and SIVlhoest from l'Hoest’s monkeys (Cercopithecus lhoesti), SIVsun from sun-tailed monkeys (Cercopithecus solatus), and SIVcol from colobus monkeys (Colobus guereza) (6, 25). In addition, SIVs from other NHPs have been partially characterized (mainly in the pol gene region) and appear to represent additional lineages: SIVrcm, SIVtal, SIVmnd, SIVdrl, SIVdeb, SIVmon, SIVgsn, and SIVmus (9, 19, 28, 29).
Human immunodeficiency virus (HIV) types 1 and 2 (HIV-1 and HIV-2, respectively) are phylogenetically related to the SIVcpz and SIVsm lineages and are thought to be the result of cross-species transmissions from SIV-infected chimpanzees and sooty mangabeys, respectively. While the origin of HIV-1 from SIV-infected chimpanzees is primarily supported by the phylogenetic clustering of HIV-1 and SIVcpz sequences (7, 32), all conditions for HIV-2 as a zoonosis from SIV-infected sooty mangabeys are met: similarity in viral genome organization, phylogenetic relatedness, prevalence in the natural host, geographic coincidence, and plausible route of transmission (17, 33). There is currently no confirmed evidence that SIV strains other than SIVcpz and SIVsm have infected humans, although most grow in human cells in vitro (11).
A number of studies have provided evidence for SIV infection in at least 33 NHPs by using serological assays containing HIV antigens and genetic characterization of whole or partial genomes from 21 SIVs (1, 12, 23, 29). The bush meat trade in sub-Saharan Africa provides the opportunity for human exposures to the numerous SIVs prevalent in different species of feral NHPs (26) through contact with blood and body fluids through hunting and butchering or keeping NHPs as pets. These repeated exposures could result in human infections with SIV strains that may be missed by current HIV testing algorithms, because SIV-specific tests are not available, resulting in the potential to cause new epidemics. This situation is beyond hypothetical, since two laboratory workers have been reported to be infected with SIVs (21).
Due to the lack of SIV-specific tests, serological detection of SIV infections in NHPs has been done by HIV enzyme immunoassay (EIA) and/or Western blot (WB) assays (2, 9, 16, 28, 30, 31, 38) based on the cross-reactivity of SIV antibodies with some HIV antigens. However, these HIV serological assays may have a limited range for detecting divergent SIVs, especially those that have not yet been genetically characterized (24, 26). Furthermore, the common approach of using EIA and WB assays as screening tools for detecting SIV infections is expensive, can be difficult to interpret, and does not allow for differentiation among SIV lineages. PCR-based diagnosis has further limitations, since commonly used “universal” PCR primers do not reliably amplify all SIVs because of the high genetic divergence among SIV lineages. In addition, there are currently no data on the validation of available HIV assays to reliably detect potential SIV infections in humans. The availability of an assay that can be used for the surveillance of SIV infections in humans is critical for detecting potential new cross-species transmissions of these simian retroviruses.
Therefore, it is important to develop and validate serological screening assays that are sensitive and specific enough to detect all known and potentially some currently unknown but antigenically similar SIV lineages. Other synthetic peptide-based SIV assays have been reported, but they used a limited number of peptides from either consensus sequences (24) or specific SIV sequences. Furthermore, these were representative of only four lineages (35) and could fail in the detection of unrecognized or highly divergent SIVs. In addition, these previous assays were based on linear peptides, which have been shown to have a low analytical sensitivity (4, 24, 35, 36), probably due to a low coating efficiency (8).
Here we present a highly sensitive and specific SIV EIA with multiple antigenic peptides (MAPs) (SMAP-EIA) covering the V3 region and the immunodominant region (IDR) of the env gene for the detection and discrimination of six major and at least three partially defined SIV lineages for which env sequences were available. Validation of the assay was performed with plasma or serum samples obtained from known SIV-infected and uninfected species of NHPs representing the lineages from which the assay MAPs were derived and some NHP lineages known to be SIV infected but for which SIV env sequences were not available.
MATERIALS AND METHODS
Peptide synthesis.
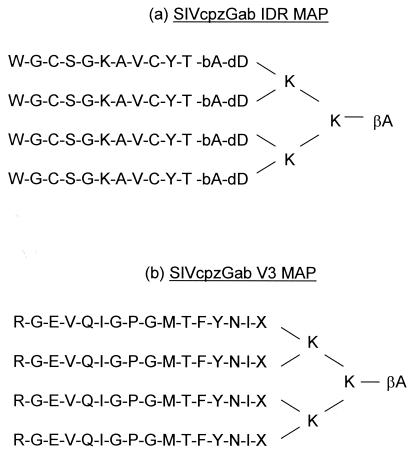
To improve the sensitivity and specificity of our SMAP-EIA relative to previously reported peptide-based assays, we (i) increased the number of SIV lineage-specific peptides for broader sensitivity, (ii) shortened the length of the peptides to increase specificity, and (iii) chemically modified the structure of the peptides to increase analytical sensitivity. Hence, we used a comprehensive array of peptides covering the V3 region of gp120 and the IDR of gp41/36 and representing all NHP lentivirus lineages for which env sequences were available: SIVcpz, SIVsm, SIVagm, SIVsyk, SIVlhoest, SIVmnd, SIVcol, SIVrcm, SIVdeb (Table 1). Given that long peptides could give rise to nonspecific cross-reactivity due to the potential presence of additional epitopes, our peptides were designed to be shorter than those in previous studies (11 and 15 amino acids instead of either 26 and 28 [35] or 44 and 21 [24] amino acids for IDR and V3, respectively). To compensate for a possible reduction in sensitivity from the use of shorter peptides, the peptides were designed and synthesized as MAPs, consisting of four branches of linear peptides held together by lysine molecules. The MAP structure has been shown to significantly increase the analytical sensitivity of peptide-based assays (10, 22) by improving immunogenicity and overcoming the low coating efficiency of peptides on microtiter plates (34, 37). Figure 1 shows the schematic structure of a representative IDR and V3 tetrameric MAP, SIVcpzGab.
TABLE 1.
Amino acid sequences of IDR and V3 region peptides and corresponding SIV strains used in the SMAP-EIA
| Peptide | Sequence | SIV strain(s) |
|---|---|---|
| IDR | WGCSGKAVCYT | SIVcpzGab |
| WGCSGKAICYT | SIVcpzCam | |
| WGCADKVICHT | SIVcpzAnt | |
| WGCAFRQVCHT | SIVsm | |
| WGCAWKQVCHT | SIVagm(1)a | |
| WGCAFKQVCHT | SIVagm(2)a | |
| WGCAFKQICHT | SIVsyk, SIVdeb | |
| WGCQWKQVCHT | SIVsun, SIVlhoest | |
| IGCANMQICRT | SIVcol | |
| FGCAWRQVCHT | SIVrcm | |
| WGCSFSQVCHT | SIVmndl4b | |
| WGCSWAQVCHT | SIVmndGB1b | |
| V3 region | RGEVQIGPGMTFYNI | SIVcpzc |
| VLPVTIMSGLVFHSQ | SIVsm | |
| VLPVTIMAGLVFHSQ | SIVagma | |
| IKNIQLAAGYFLPVI | SIVsyk | |
| EVSTISSTGLLFYYG | SIVsun, SIVlhoest | |
| HRNLNTANGAKFYYE | SIVcol | |
| VKGISLATGVFISLR | SIVrcm | |
| IVSVPSASGLIFYHG | SIVmndb | |
| YRAVHMATGLSFYTT | SIVdeb |
There are many strains of SIVagm, and two IDR sequences were needed to cover the amino acid variations; however, the V3 region sequences were the same.
Two distinct SIV strains are found in mandrills, SIVmnd14 and SIVmndGB1,and the panel includes two separate IDR peptides and a single V3 region peptide.
The V3 region amino acid sequences were identical for all three SIVcpz strains.
FIG. 1.
Schematic structures of MAPs composed of four identical linear peptide molecules from the IDR (a) and the V3 region (b) of the env gene held together by lysine (K) molecules. Non-virus-encoded amino acids β-alanine (bA or βA), d-aspartic acid (dD), and diaminopropionic acid (X) were added as spacers. The amino acid sequences shown are from SIVcpzGab.
All of the peptide sequences used in the assay are shown in Table 1, along with the SIV strains they represent. The MAPs were synthesized by using a solid-phase method, 9-fluorenylmethoxy carbonyl chemistry, according to the manufacturer's protocol, and an automatic synthesizer (model 432A; Applied Biosystems, Inc., Foster City, Calif.). After synthesis, the MAPs were partially purified by reverse-phase high-performance liquid chromatography (Bio-Rad, Richmond, Calif.), lyophilized, and stored desiccated at room temperature until use.
SMAP-EIA.
The peptide-based SMAP-EIA was carried out as previously described with slight modifications (22). Briefly, peptides were first dissolved (10 mg/μl) in dimethyl sulfoxide (Aldrich, Milwaukee, Wis.) and then diluted in cold bicarbonate buffer (0.1 M, pH 9.4) to a final concentration of 0.25 μg/100 μl. The peptide solutions were used to coat (110 μl/well) microtiter plates (Maxisorb; Nalge Nunc International, Rochester, N.Y.) at 4°C overnight. Peptides belonging to the same lineage or with very similar amino acid sequences were combined to a maximum of two SIV MAPs and used to coat the same well. Unbound MAPs were removed by two washes with phosphate-buffered saline (pH 7.5) containing 0.05% Triton X-100 (Sigma Chemical Co., St. Louis, Mo.), and coated plates were stored at −20°C until use. Nonspecific binding sites were blocked with 5% nonfat dry milk in phosphate-buffered saline containing 0.1% Triton X-100 (milk buffer) for 30 min at 37°C just before the assay was performed. Plasma or serum samples were diluted 1:500 in milk buffer, and 100 μl was added to the peptide-coated well and incubated for 1 h at 37°C. Bound antibodies were detected by the addition of 100 μl of a 1:8,000 dilution of goat anti-human immunoglobulin G (heavy and light chains) conjugated to peroxidase (Bio-Rad, Hercules, Calif.) for 30 min at 37°C followed by 100 μl of tetramethylbenzydine-hydrogen peroxidase substrate (BioFX, Owings Mills, Md.) for 10 min at room temperature. Color development was stopped with 1 M sulfuric acid, and the optical density (OD) was measured at 450 nm against a reference wavelength of 630 nm.
NHP evaluation panel.
The infection status of the primates included in the NHP evaluation panel had been determined previously by commercial HIV-1 and HIV-2 serological assays following a commonly used algorithm whereby the samples were first screened by an EIA (Genetics Systems HIV-1/HIV-2 peptide EIA; Bio-Rad, Hercules, Calif.). Seroreactive samples were confirmed by HIV WB analysis (Cambridge Biotech, Rockville, Md. [HIV-1], and Genelabs Diagnostics, Redwood City, Calif. [HIV-2]) based on the presence of at least one gag band and one env band. In some cases, seroreactive samples were further confirmed by PCR with a variety of primers (2, 3, 18, 29). Samples with seronegative EIA results were considered to be from uninfected animals. Table 2 shows the evaluation panel, which consisted of 63 serum or plasma samples from various NHPs either naturally or experimentally infected and 97 samples from uninfected NHPs.
TABLE 2.
SIV-infected and uninfected NHP panel
| Species | Common name | Virus | No. of samples that were SIV
|
|
|---|---|---|---|---|
| Positive | Negative | |||
| Cercocebus atys | Sooty mangabey | SIVsm | 7 | 7 |
| Macaca species | Macaque | SIVsm | 11 | 8 |
| Cercocebus torquatus | Red-capped mangabey | SIVrcm | 2 | 15 |
| Chlorocebus pygerythrus | Vervet monkey | SIVagmVer | 3 | 3 |
| Chlorocebus tantalus | Tantalus monkey | SIVagmTan | 4 | 4 |
| Chlorocebus sabaeus | Green monkey | SIVagmSab | 3 | |
| Macaca species | Macaque | SIVagmVer | 1 | |
| Cercopithecus albogularis | Sykes' monkey | SIVsyk | 4 | 4 |
| Cercopithecus l'hoesti | L'Hoest’s monkey | SIVlhoest | 1 | |
| Macaca species | Macaque | SIVlhoest | 1 | |
| SIVsun | 1 | |||
| Cercopithecus neglectus | DeBrazza's monkey | SIVdeb | 3 | 1 |
| Colobus guereza | Guereza colobus | SIVcol | 4 | 32 |
| Mandrillus sphinx | Mandrill | SIVmnd | 13 | 8 |
| Mandrillus leucophaeus | Drill | SIVdrl | 1 | 3 |
| Miopithecus talapoin | Talapoin monkey | SIVtal | 5 | 5 |
| Pan troglodytes troglodytes | Central African chimpanzee | SIVcpz | 2 | 3 |
| Pan troglodytes schweinfurthii | East African chimpanzee | SIVcpz | 1 | |
| Total | 63 | 97 | ||
Of the 63 SIV-positive samples that were used to determine the sensitivity of our assay, 7 samples were from sooty mangabeys and 11 samples were from macaques (5 rhesus and 6 stump-tailed macaques) experimentally infected with SIVsm; 7 samples were from African green monkeys (4 tantalus and 3 vervet monkeys) and 1 macaque infected with SIVagm; 4 samples were from Sykes' monkeys infected with SIVsyk; 3 samples were from chimpanzees infected with SIVcpz (SIVcpzGab, SIVcpzAnt, and SIVcpzUS); 2 samples were from macaques experimentally infected with SIVlhoest or SIVsun; 4 samples were from colobus monkeys infected with SIVcol; 13 samples were from mandrills and 1 sample was from a drill infected with SIVmnd and SIVdrl, respectively (18); 2 samples were from red-capped mangabeys infected with SIVrcm; 5 samples were from talapoin monkeys infected with SIVtal; and 3 samples were from DeBrazza's monkeys infected with SIVdeb.
The specificity of the SMAP-EIA was calculated by using the test results for the 97 SIV-negative serum or plasma samples obtained from 7 sooty mangabeys, 8 macaques (6 stump-tailed and 2 pigtailed macaques), 10 African green monkeys (4 tantalus, 3 vervet, and 3 sabaeus monkeys), 3 Central African chimpanzees, 4 Sykes' monkeys, 1 l'Hoest’s monkey, 8 mandrills, 3 drills, 32 colobus monkeys, 5 talapoin monkeys, 15 red-capped mangabeys, and 1 DeBrazza's monkey. Seventeen of these SIV-negative NHPs were found to be infected with simian foamy viruses (SFVs), as determined by an in-house SFV WB test (20), and samples from these NHPs were also used to evaluate the specificity of the SMAP-EIA.
Human evaluation panel.
Since the SMAP-EIA is ultimately intended for surveillance of SIV-like infections in human populations, we obtained and tested a plasma sample from a human with a confirmed SIV infection following occupational exposure to SIVsm (21). To assess the specificity of the SMAP-EIA for prospective application in human testing, 198 HIV-seronegative and human T-cell leukemia virus (HTLV)-seronegative serum samples from U.S. blood donors were also tested. Seronegative human serum samples from the United States were chosen for evaluation because of the potential for close contact between humans and NHPs in African populations.
To evaluate the potential for the cross-reactivity of HIV-1 and HIV-2 antibodies with SIV MAPs, we included in our evaluation panel 170 HIV-1-seropositive serum samples from 70 persons infected with non-B HIV-1 group M subtypes from Africa (A [n = 27], C [n = 1], D [n = 14], AE [n = 1], F [n = 7], G [n = 18], H [n = 1], and J [n = 1]), 50 persons with HIV-1 subtype B from the United States, 4 persons with HIV-1 group O from Cameroon, 41 persons with HIV-2 from Nigeria and the Ivory Coast, and 5 persons with dual HIV-1 and HIV-2 infections from Nigeria.
In addition to HIV, we also checked the specificity of the SMAP-EIA by testing plasma samples from persons infected with other etiological agents common to African populations. These included 18 plasma samples from malaria patients infected with one of the four main species of Plasmodium (P. falciparum [n = 4], P. malariae [n = 2], P. ovale [n = 5], and P. vivax [n = 7]), 44 serum samples from HTLV type 1- or 2-positive persons, 3 plasma samples from persons with strong antibody titers to measles virus, and 3 plasma samples from SFV-infected persons working with primates (15).
Data analyses and cutoff determinations.
The data were analyzed by using Analyze-it for Microsoft Excel and the statistical package for the social sciences (SPSS; SPSS, Inc., Chicago, Ill.). The crude ODs for each SIV lineage panel of samples were summarized by using the SPSS box-plot technique, which is a graphical representation of the median and the 25th and 75th percentiles (vertical boxes with error bars). Possible outliers are represented by asterisks and ovals depending on their interquartile ranges from the quartiles.
All 97 SIV-negative NHP samples were tested against MAPs from each lineage, and the cutoff value for a particular MAP(s) was calculated as the mean of all of the ODs plus 4 standard deviations. Hence, a sample with an OD/cutoff ratio of ≥1 for any MAP was considered seropositive, while a sample with an OD/cutoff ratio of <1 was considered seronegative. The performance of the SMAP-EIA was evaluated by calculating the sensitivity and the specificity of MAPs from each SIV lineage.
RESULTS
Assay cutoff determinations for NHP serum or plasma.
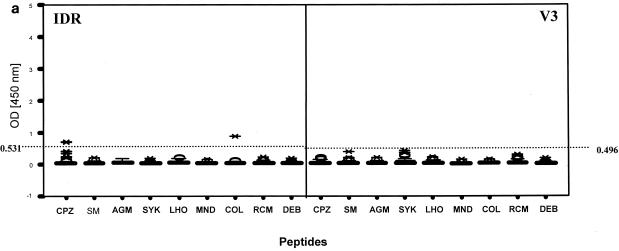
Based on the observed seroreactivity of the 97 SIV-negative primate serum samples (Fig. 2a), the cutoff values for different SIV MAPs were as follows for IDR: 0.122 (SIVmnd), 0.150 (SIVsyk), 0.150 (SIVdeb), 0.177 (SIVsm), 0.201 (SIVrcm), 0.241 (SIVagm), 0.265 (SIVlhoest or SIVsun), 0.420 (SIVcol), and 0.531 (SIVcpz). For the V3 region, the cutoff values were as follows: 0.050 (SIVdeb), 0.082 (SIVlhoest or SIVsun), 0.102 (SIVsm), 0.129 (SIVagm), 0.133 (SIVmnd), 0.252 (SIVcpz), 0.284 (SIVrcm), 0.492 (SIVcol), and 0.496 (SIVsyk).
FIG. 2.
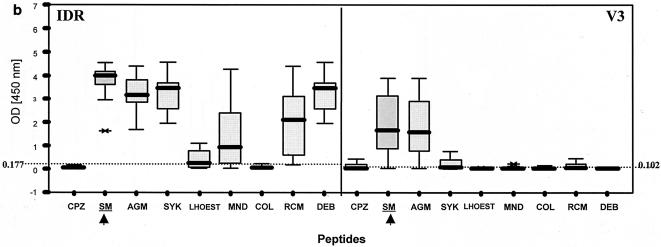
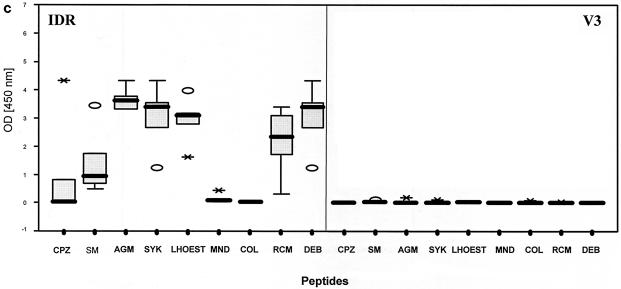
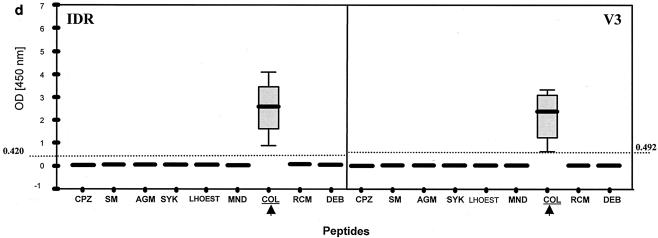
Patterns of reactivity of serum and plasma samples from SIV-infected and uninfected NHPs with SIV MAPs. Samples were from SIV-negative NHPs (n = 97) (a), SIVsm from naturally infected sooty mangabeys (n = 7) and experimentally infected macaques (n = 11) (b), SIVtal from naturally infected talapoin monkeys (n = 5) (c), and SIVcol from naturally infected colobus monkeys (n = 4) (d). The left portion of each graph shows SMAP-EIA data for the IDR; the right portion shows data for the V3 region component of the assay. The vertical boxes with error bars represent the 25th to 75th percentiles of the seroreactivity OD of each MAP. The horizontal dotted lines represent the indicated cutoff values for the homologous MAPs for the NHP species-specific samples (b and d) or the highest cutoff values for the negative (or seronegative) NHP samples against all MAPs (a). The asterisks and open ovals represent outliers. The homologous MAP(s) for each of the NHP samples is indicated by an arrow.
Assay specificity.
Based on the cutoff value for each SIV MAP, we examined the seroreactivity of samples from all of the uninfected NHPs to determine the specificity of the SMAP-EIA. None of the 97 SIV-negative NHP serum samples showed any reactivity with the homologous SIV IDR and V3 MAPs (Fig. 2a). However, 2 (13.3%) of 15 red-capped mangabey samples showed weak, nonspecific reactivity (both with an OD/cutoff ratio of 1.3) with the heterologous SIVcpz IDR MAP. Similarly, 1 (5.5%) of 18 sooty mangabey samples showed reactivity (OD/cutoff ratio of 2.1) with the heterologous SIVcol IDR MAP. Thus, although the homologous IDR SMAP-EIA had a specificity of 100%, the overall assay specificity of the IDR SMAP-EIA was 96.9% when heterologous antibody reactivity was included. The V3 SMAP-EIA specificity was 100% with homologous SIV-negative NHP serum samples and SIV MAPs (Table 3). The high specificity of the IDR and V3 peptides was also demonstrated by the absence of reactivity from 17 SFV-infected NHP samples that were included in the SIV-negative panel.
TABLE 3.
Sensitivity and specificity of SMAP-EIA with the NHP evaluation panel
| SIV lineage | SIV-positive group
|
SIV-negative group
|
||||||
|---|---|---|---|---|---|---|---|---|
| No. of samples tested | SMAP-EIA | No. of positive samplesa | Sensitivity (%) | No. of samples tested | SMAP-EIA | No. of negative samplesa | Specificity (%) | |
| SIVsm | 18 | IDR | 18 | 100 | 15 | IDR | 14 | 93.3 |
| 18 | V3 | 17 | 94.4 | 15 | V3 | 15 | 100 | |
| SIVagm | 8 | IDR | 8 | 100 | 10 | IDR | 10 | 100 |
| 8 | V3 | 5 | 62.5 | 10 | V3 | 10 | 100 | |
| SIVsyk | 4 | IDR | 4 | 100 | 4 | IDR | 4 | 100 |
| 4 | V3 | 3 | 75.0 | 4 | V3 | 4 | 100 | |
| SIVcpz | 3 | IDR | 3 | 100 | 3 | IDR | 3 | 100 |
| 3 | V3 | 2 | 66.6 | 3 | V3 | 3 | 100 | |
| SIVlhoest or SIVsun | 2 | IDR | 2 | 100 | 1 | IDR | 1 | 100 |
| 2 | V3 | 2 | 100 | 1 | V3 | 1 | 100 | |
| SIVcol | 4 | IDR | 4 | 100 | 32 | IDR | 32 | 100 |
| 4 | V3 | 4 | 100 | 32 | V3 | 32 | 100 | |
| SIVmnd and SIVdrl | 13 | IDR | 13 | 100 | 11 | IDR | 11 | 100 |
| 13 | V3 | 13 | 100 | 11 | V3 | 11 | 100 | |
| SIVrcm | 2 | IDR | 2 | 100 | 15 | IDR | 13 | 86.6 |
| 2 | V3 | 2 | 100 | 15 | V3 | 15 | 100 | |
| SIVdeb | 1 | IDR | 1 | 100 | 1 | IDR | 1 | 100 |
| 1 | V3 | 1 | 100 | 1 | V3 | 1 | 100 | |
| SIVtalb | 5 | IDR | 5 | 100 | 5 | IDR | 5 | 100 |
| 5 | V3 | 0 | NA | 5 | V3 | 5 | 100 | |
| Overall | 60 | IDR | 60 | 100 | 97 | IDR | 94 | 96.9 |
| 55b | V3 | 49 | 89 | 97 | V3 | 97 | 100 | |
Based on the seroreactivity of each NHP serum or plasma sample against the homologous SIV-specific MAP.
SIVtal-specific peptides were not included in the assay because the envelope gene of this virus has not been sequenced. Strong IDR reactivity to any SIV IDR peptide was considered to be indicative of a positive SIVtal serum or plasma. No V3 reactivity was observed. NA, not applicable.
Sensitivity of the SMAP-EIA.
In general, the SIV-seropositive NHP samples showed broad cross-reactivity with the IDR peptides and more specific V3 peptide reactivity. Figure 2b shows the seroreactivity pattern for SIVsm-infected animals, which is representative of the general pattern observed for plasma or serum samples from most SIV lineages except for SIVcol. Typically, antibodies from SIV-infected monkeys reacted with most SIV IDR MAPs, except for that from SIVcol, with the highest IDR seroreactivity generally being observed with the homologous SIV MAP. In contrast, V3 seroreactivity usually was limited to the homologous SIV MAP and, in a few instances, to one other closely related SIV MAP.
Each SIV-seropositive NHP serum or plasma sample was tested against all IDR and V3 MAPs. The sensitivity of the IDR peptides was found to be 100% with all 63 SIV-positive NHP samples for which corresponding peptides were represented in the assay (Table 3). One sample from an SIV-infected Sykes' monkey was not detected by the homologous SIVsyk IDR peptide; however, this sample was reactive with the SIVsm IDR peptide and with the homologous SIVsyk V3 peptide. It is encouraging to note that all five SIV-positive talapoin monkey samples included in the validation testing were detected by our assay even though SIVtal MAPs were not included; the five SIVtal-positive talapoin monkey samples showed cross-reactivity with all SIV IDR MAPs except for those from SIVmnd and SIVcol (Fig. 2c). However, the five talapoin monkey samples were not reactive with any of the V3 peptides, most likely due to the amount of divergence of SIVtal from other SIVs in this variable gene region. The overall sensitivity of the V3 SMAP-EIA was 89.0% because six samples (three SIVagm, one SIVsm, one SIVcpz, and one SIVsyk) had ODs below the established cutoff. However, all six samples were reactive with the IDR peptides. Thus, when the results for both the IDR and the V3 peptides were combined, the overall SMAP-EIA sensitivity was 100%; this result suggests that a testing algorithm that includes both IDR and V3 components should be used.
Of three SIVcpz-seropositive plasma or serum samples (one from Pan troglodytes schweinfurthii and two from P. troglodytes troglodytes), two showed strong monospecific seroreactivity with the homologous IDR MAP, while the SIVcpzUS-infected sample showed strong homologous SIVcpz IDR MAP seroreactivity and weak cross-reactivity with the SIVagm, SIVlhoest, SIVcol, and SIVrcm IDR MAPs. Two of the three chimpanzee samples showed monospecific reactivity with the homologous SIVcpz V3 MAP, while one did not react with any of the V3 peptides. This lack of V3 reactivity might have been due to low antibody titers in this sample, since the IDR seroreactivity was also relatively low; alternatively, the virus from this chimpanzee was significantly more divergent. Nonetheless, all SIVcpz infections were detected by both the IDR and the V3 components of the assay.
SIVsm- and SIVagm-infected samples showed stronger seroreactivity with homologous SIV IDR MAPs than with heterologous SIV IDR MAPs. No seroreactivity with the SIVcol IDR MAP was seen. More specific seroreactivity was observed with the V3 component of the SMAP-EIA, but antibody cross-reactivity was observed between SIVsm and SIVagm MAPs. However, the highest ODs were seen with homologous V3 MAPs. Three SIVsyk-infected samples reacted similarly with the IDR component, while one SIVsyk-infected sample reacted solely with the heterologous SIVsm MAP. Three (75%) of four samples from SIV-infected Sykes' monkeys showed monospecific reactivity with the homologous V3 MAP, and one had no seroreactivity. Thirteen samples from monkeys infected with SIVlhoest or SIVsun, SIVmnd or SIVdrl, SIVrcm, and SIVdeb showed typical cross-reactivity with the IDR component, yet V3 sensitivity for these specimens was 100%. In contrast to SIV-seropositive samples from the other SIV lineages, all four SIV-positive colobus monkey samples reacted only with the homologous SIVcol IDR and V3 peptides (Fig. 2d). It is important to note that while other SIV-positive samples showed antibody cross-reactivity with many heterologous IDR MAPs, no cross-reactivity was seen with the SIVcol IDR MAP. Conversely, serum samples from SIVcol-infected monkeys reacted only with the homologous MAP, most likely reflecting a higher diversity of SIVcol than of SIV lineages in other NHPs. Interestingly, two mandrill samples and two DeBrazza's monkey samples that were HIV-1 or HIV-2 EIA seronegative but HIV-2 WB assay positive all tested positive in the SMAP-EIA.
Cutoff calculations for human serum or plasma.
In order to check for any background antibody cross-reactivity when the SMAP-EIA is used for the detection of SIV-like strains in humans and to determine a cutoff value that could be used in screening humans for infection with SIV, we tested a total of 198 HIV- and HTLV-negative U.S. blood donor specimens. As before, the cutoff values for the IDR component were calculated as the mean OD plus 4 standard deviations: 0.104 (SIVcpz), 0.213 (SIVsyk), 0.213 (SIVdeb), 0.239 (SIVrcm), 0.273 (SIVmnd), 0.274 (SIVsm), 0.357 (SIVlhoest), 0.371 (SIVagm), and 0.472 (SIVcol). For the V3 component, the cutoff values were as follows: 0.067 (SIVagm), 0.072 (SIVdeb), 0.079 (SIVmnd), 0.087 (SIVlhoest), 0.088 (SIVsm), 0.095 (SIVrcm), 0.103 (SIVcpz), 0.115 (SIVsyk), and 0.161 (SIVcol).
Specificity of the SMAP-EIA.
Evaluation of 198 HIV- and HTLV-negative U.S. blood donor samples with the calculated cutoff values revealed nine samples with low seroreactivity, slightly above the cutoff value of the IDR component of the SMAP-EIA. In contrast, all 198 U.S. blood donor samples were below the V3 component cutoff value. Therefore, the specificities for the IDR and V3 SMAP-EIAs were 95.45 and 100%, respectively. However, since one U.S. blood donor sample was reactive in the IDR SMAP-EIA, with an OD of 0.866, we recommend using a more stringent OD cutoff of ≥1.00 for screening human samples for exposure to SIVs with the SMAP-EIA. As such, the assay will be more specific but possibly less sensitive.
To determine whether antibodies to HIV cross-react with SIV MAPs, 120 serum samples from infections with HIV-1 group M subtypes, 4 samples from infections with HIV-1 group O, 41 samples from infections with HIV-2, and 5 samples from dual HIV-1 and HIV-2 infections were tested with our SMAP-EIA. All samples from HIV-1 group M and group O infections cross-reacted primarily with the SIVcpz IDR MAP; only a few cross-reacted with SIV MAPs from other lineages, such as SIVlhoest, SIVdeb, and SIVagm (data not shown). The seroreactivity of the HIV-1-positive samples with V3 MAPs was reduced and directed only to the SIVcpz MAP. As expected, all 41 HIV-2-positive samples reacted primarily with the SIVsm IDR MAP, since these viruses share identical amino acid sequences in this region. The V3 peptide assay reactivity of the HIV-2-positive samples was also comparable to that of the SIVsm-positive samples. The five HIV-1- and HIV-2-positive samples showed reactivities reflecting the presence of heterotypic infections. While HIV-1-positive serum or plasma samples cross-reacted with SIV MAPs, plasma or serum samples from all SIV-infected NHPs did not cross-react with any HIV-1 peptides (data not shown).
Of 44 specimens from HTLV-infected persons, only 1 demonstrated any seroreactivity with an SIV MAP, the SIVcol IDR MAP (data not shown). One of the 18 malaria-positive plasma specimens showed weak reactivity with the SIVcpz IDR MAP. No SMAP-EIA reactivity was observed with any of the three measles virus-infected or three SFV-infected human samples.
Sensitivity of the SMAP-EIA for detecting SIV infections in humans.
To further evaluate the ability of our assay to detect SIV infections in humans, we analyzed one plasma sample from a person who was reported to be infected with SIVsm following an occupational exposure (21). This sample was reactive with both the SIVsm IDR and the SIVsm V3 MAPs.
DISCUSSION
We developed and validated a novel SMAP-EIA for the detection of SIV antibodies with a panel of MAPs covering nine highly divergent SIV lineages. We showed that this assay is highly sensitive and specific for detecting infections with SIVs of all nine lineages and in an SIV-infected person following occupational exposure to SIVsm (21). Samples from two SIVmnd-infected mandrills and two SIVdeb-infected DeBrazza's monkeys were seronegative in a commercially available HIV-1 or HIV-2 peptide EIA but were seroreactive with their homologous IDR and V3 MAPs in our SMAP-EIA and were confirmed to be seropositive by an HIV-2 WB assay. These results show that this new assay is more sensitive than standard HIV EIAs for detecting SIV infections in NHPs and offers a new screening tool for the surveillance of SIV infections in humans.
The IDR component of our SMAP-EIA is highly sensitive but less specific than the V3 component and can be used as a screening tool in serosurveillance for SIV or SIV-like infections in humans and NHPs. The IDR of gp41/36 is highly conserved among all known SIV lineages, and SIV antibodies can cross-react with several SIV MAPs. This property can be highly advantageous during serosurveillance for potentially new, unrecognized SIV-infected NHPs or for recognizing additional cross-species transmissions of SIV strains into humans. No SIVtal env sequences were available in the published databases for designing SIVtal MAPs. However, we found that sera from SIV-infected talapoin monkeys contained anti-SIVtal antibodies that cross-reacted with heterologous SIV MAPs used in our IDR SMAP-EIA. The V3 component of the SMAP-EIA is very specific but less sensitive than the IDR component, most likely due to the high genetic variability in the V3 portion of the envelope, and can be used for confirmation of IDR seroreactivity and identification of the infecting SIV lineage.
Commercial SIV EIAs and WB assays are not currently available for the detection of SIV infections in either NHPs or occupationally exposed persons; such infections generally are determined with HIV EIAs and WB assays (29). Although SIV antibodies cross-react with some HIV antigens, especially the major Gag proteins (13, 14, 23, 27), not all SIV infections have been detected by commercial HIV assays. Some SIV-infected monkeys whose samples were nonreactive with HIV antigens were found to be infected only by PCR amplification with various generic primers and DNA sequencing (29). SIV peptide-based assays were used previously in the serological testing of primate lentiviruses (24, 35), but these studies used linear peptides that have limitations in sensitivity, most likely due to a low microtiter plate coating efficiency (8). Our use of MAPs increases the analytical sensitivity of the assay (22). Even if two of the four chains of a MAP become bound to the well of the microtiter plate, there are still at least two chains that are readily antibody accessible. Another limitation of previous studies was the use of peptides representing only a few representative SIV lineages or representing consensus peptides. Our assay used all of the currently known SIV lineages for which env sequences were available at the time of testing, and as new SIV sequences from NHPs become available, additional MAPs could be incorporated into the assay. In fact, three new SIV whole-genome sequences representing mona monkeys (Cercopithecus mona), mustached monkeys (Cercopithecus cephus), and greater spot-nosed monkeys (Cercopithecus nictitans) were recently published (5). Additional MAPs representing these newly sequenced SIVs will be incorporated into the SMAP-EIA for serosurveillance of SIV-like infections among African populations, zoo workers, animal handlers, and laboratory researchers working with SIV strains.
The recognition of AIDS as a zoonosis from SIV-infected NHPs (12), the ability of SIVs from multiple lineages to infect human macrophages (11), and the continued exposure of persons to SIV-infected NHP blood and body fluids via hunting and butchering, the keeping of NHPs as pets, or following occupational exposure all raise concerns about the seeding of new lentivirus epidemics that may go unchecked because current serological assays may not be able to detect such potential emerging infectious diseases. The development of highly sensitive and specific serological assays, such as the SMAP-EIA, for the surveillance of SIV infections in humans is urgently needed at this time due to risk from invasion of previously protected ecological niches. For instance, commercial logging companies in sub-Saharan Africa are building new roads into the remote portions of the rain forest, previously inaccessible to most of the population. Additionally, truck drivers provide transportation of NHP bush meat from these sites back to the urban markets in the major cities, increasing the magnitude of human exposure to SIVs and providing the population density and transmission opportunities necessary to seed new lentivirus epidemics.
Acknowledgments
We are grateful to the following persons for providing the valuable primate samples used in the evaluation of the assay: Brigitte Beer, Vanessa Hirsch, and Larry Arthur from NIH; Zena Tooze from CERCOPAN, Cross River State, Nigeria; Martine Peeters from IRD, Montpellier, France; Francois Villinger from Emory University; JoAnn Yee and Nick Lerche from the University of California at Davis; and many U.S. zoo veterinary staff members.
Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the U.S. Public Health Service, or the Centers for Disease Control and Prevention.
REFERENCES
- 1.Apetrei, C., D. L. Robertson, and P. A. Marx. 2004. The history of SIVs and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front. Biosci. 9:225-254. [DOI] [PubMed] [Google Scholar]
- 2.Beer, B. E., E. Bailes, R. Goeken, G. Dapolito, C. Coulibaly, S. G. Norley, R. Kurth, J. P. Gautier, A. Gautier-Hion, D. Vallet, P. M. Sharp, and V. M. Hirsch. 1999. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73:7734-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. St. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brattegaard, K., D. Soroh, F. Zadi, H. Digbeu, K. M. Vetter, and K. M. De Cock. 1995. Insensitivity of a synthetic peptide-based test (Pepti-LAV 1-2) for the diagnosis of HIV infection in African children. AIDS 9:656-657. [DOI] [PubMed] [Google Scholar]
- 5.Courgnaud, V., B. Abela, X. Pourrut, E. Mpoudi-Ngole, S. Lout, E. Delaporte, and M. Peeters. 2003. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different Cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J. Virol. 77:12523-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. Hahn, A. M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 8.Geerligs, H. J., W. J. Weijer, W. Bloemhoff, G. W. Welling, and S. Welling-Wester. 1988. The influence of pH and ionic strength on the coating of peptides of herpes simplex virus type 1 in an enzyme-linked immunosorbent assay. J. Immunol. Methods 106:239-244. [DOI] [PubMed] [Google Scholar]
- 9.Georges-Courbot, M. C., C. Y. Lu, M. Makuwa, P. Telfer, R. Onanga, G. Dubreuil, Z. Chen, S. M. Smith, A. Georges, F. Gao, B. H. Hahn, and P. A. Marx. 1998. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J. Virol. 72:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomara, M. J., S. Riedemann, I. Vega, H. Ibarra, G. Ercilla, and I. Haro. 2000. Use of linear and multiple antigenic peptides in the immunodiagnosis of acute hepatitis A virus infection. J. Immunol. Methods 234:23-34. [DOI] [PubMed] [Google Scholar]
- 11.Grimm, T. A., B. E. Beer, V. M. Hirsch, and K. A. Clouse. 2003. Simian immunodeficiency viruses from multiple lineages infect human macrophages: implications for cross-species transmission. J. Acquir. Immune Defic. Syndr. 32:362-369. [DOI] [PubMed] [Google Scholar]
- 12.Hahn, B. H., G. M. Shaw, K. E. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-617. [DOI] [PubMed] [Google Scholar]
- 13.Hayami, M., E. Ido, and T. Miura. 1994. Survey of simian immunodeficiency virus among nonhuman primate populations. Curr. Top. Microbiol. Immunol. 188:1-20. [DOI] [PubMed] [Google Scholar]
- 14.Hayami, M., Y. Ohta, T. Hattori, H. Nakamura, K. Takatsuki, A. Kashiwa, K. Nozawa, I. Miyoshi, T. Ishida, and Y. Tanioka. 1985. Detection of antibodies to human T-lymphotropic virus type III in various non-human primates. Jpn. J. Exp. Med. 55:251-255. [PubMed] [Google Scholar]
- 15.Heneine, W., W. M. Switzer, P. Sandstrom, J. Brown, S. Vedapuri, C. A. Schable, A. S. Khan, N. W. Lerche, M. Schweizer, D. Neumann-Haefelin, L. E. Chapman, and T. M. Folks. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 4:403-407. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch, V. M., B. J. Campbell, E. Bailes, R. Goeken, C. Brown, W. R. Elkins, M. Axthelm, M. Murphey-Corb, and P. M. Sharp. 1999. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J. Virol. 73:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch, V. M., R. A. Olmsted, M. Murphey-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 18.Hu, J., W. M. Switzer, B. T. Foley, D. L. Robertson, R. M. Goeken, B. T. Korber, V. M. Hirsch, and B. E. Beer. 2003. 2003. Characterization and comparison of recombinant simian immunodeficiency virus from drill (Mandrillus leucophaeus) and mandrill (Mandrillus sphinx) isolates. J. Virol. 77:4867-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 20.Hussain, A. I., V. Shanmugam, V. B. Bhullar, B. E. Beer, D. Vallet, A. Gautier-Hion, N. D. Wolfe, W. B. Karesh, A. M. Kilbourn, Z. Tooze, W. Heneine, and W. M. Switzer. 2003. Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses. Virology 309:248-257. [DOI] [PubMed] [Google Scholar]
- 21.Khabbaz, R. F., W. Heneine, J. R. George, B. Parekh, T. Rowe, T. Woods, W. M. Switzer, H. M. McClure, M. Murphey-Corb, and T. M. Folks. 1994. Brief report: infection of a laboratory worker with simian immunodeficiency virus. N. Engl. J. Med. 330:172-177. [DOI] [PubMed] [Google Scholar]
- 22.Kim, P., and C. P. Pau. 2001. Comparing tandem repeats and multiple antigenic peptides as the antigens to detect antibodies by enzyme immunoassay. J. Immunol. Methods 257:51-54. [DOI] [PubMed] [Google Scholar]
- 23.Lowenstine, L. J., N. C. Pedersen, J. Higgins, K. C. Pallis, A. Uyeda, P. Marx, N. W. Lerche, R. J. Munn, and M. B. Gardner. 1986. Seroepidemiologic survey of captive Old-World primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (Cercocebus atys). Int. J. Cancer 38:563-574. [DOI] [PubMed] [Google Scholar]
- 24.Masciotra, S., D. L. Rudolph, G. van der Groen, C. Yang, and R. B. Lal. 2000. Serological detection of infection with diverse human and simian immunodeficiency viruses using consensus env peptides. Clin. Diagn. Lab. Immunol. 7:706-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller, M. C., N. K. Saksena, E. Nerrienet, C. Chappey, V. M. Herve, J. P. Durand, P. Legal-Campodonico, M. C. Lang, J. P. Digoutte, and A. J. Georges. 1993. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J. Virol. 67:1227-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicol, I., D. Messinger, P. Dubouch, J. Bernard, I. Desportes, R. Jouffre, R. Snart, P. Nara, R. C. Gallo, and D. Zagury. 1989. Use of Old World monkeys for acquired immunodeficiency syndrome research. J. Med. Primatol. 18:227-236. [PubMed] [Google Scholar]
- 27.Ohta, Y., T. Masuda, H. Tsujimoto, K. Ishikawa, T. Kodama, S. Morikawa, M. Nakai, S. Honjo, and M. Hayami. 1988. Isolation of simian immunodeficiency virus from African green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int. J. Cancer 41:115-122. [DOI] [PubMed] [Google Scholar]
- 28.Osterhaus, A. D., N. Pedersen, G. van Amerongen, M. T. Frankenhuis, M. Marthas, E. Reay, T. M. Rose, J. Pamungkas, and M. L. Bosch. 1999. Isolation and partial characterization of a lentivirus from talapoin monkeys (Myopithecus talapoin). Virology 260:116-124. [DOI] [PubMed] [Google Scholar]
- 29.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peeters, M., W. Janssens, K. Fransen, J. Brandful, L. Heyndrickx, K. Koffi, E. Delaporte, P. Piot, G. M. Gershy-Damet, and G. van der Groen. 1994. Isolation of simian immunodeficiency viruses from two sooty mangabeys in Cote d'Ivoire: virological and genetic characterization and relationship to other HIV type 2 and SIVsm/mac strains. AIDS Res. Hum. Retrovir. 10:1289-1294. [DOI] [PubMed] [Google Scholar]
- 31.Peeters, M., K. Fransen, E. Delaporte, M. Van den Haesevelde, G. M. Gershy-Damet, L. Kestens, G. van der Groen, and P. Piot. 1992. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS 6:447-451. [DOI] [PubMed] [Google Scholar]
- 32.Sharp, P. M., E. Bailes, R. R. Chauduri, C. M. Rodenburg, M. O. Santiago, and B. H. Hahn. 2001. The origins of AIDS viruses: where and when? Philos. Trans. R. Soc. Lond. B 356:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp, P. M., D. L. Robertson, and B. H. Hahn. 1995. Cross-species transmission and recombination of ‘AIDS’ viruses. Philos. Trans. R. Soc. Lond. B 349:41-47. [DOI] [PubMed] [Google Scholar]
- 34.Shin, S. Y., M. K. Lee, S. Y. Kim, and K. S. Hahm. 1996. Use of multiple antigenic peptides as coating antigens in detection of antibody. Mol. Cell 6:169-175. [Google Scholar]
- 35.Simon, F., S. Souquiere, F. Damond, A. Kfutwah, M. Makuwa, E. Leroy, P. Rouquet, J. L. Berthier, J. Rigoulet, A. Lecu, P. T. Telfer, I. Pandrea, J. C. Plantier, F. Barre-Sinoussi, P. Roques, M. C. Muller-Trutwin, and C. Apetrei. 2001. Synthetic peptide strategy for the detection of and discrimination among highly divergent primate lentiviruses. AIDS Res. Hum. Retrovir. 17:937-952. [DOI] [PubMed] [Google Scholar]
- 36.Simon, F., T. D. Ly, A. Baillou-Beaufils, V. Fauveau, J. De Saint-Martin, I. Loussert-Ajaka, M. L. Chaix, S. Saragosti, A. M. Courouce, and D. Ingrand. 1994. Sensitivity of screening kits for anti-HIV-1 subtype O antibodies. AIDS 11:1628-1629. [DOI] [PubMed] [Google Scholar]
- 37.Tam, J. P. 1988. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 85:5409-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsujimoto, H., A. Hasegawa, N. Maki, M. Fukasawa, T. Miura, S. Speidel, R. W. Cooper, E. N. Moriyama, T. Gojobori, and M. Hayami. 1989. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature 341:539-541. [DOI] [PubMed] [Google Scholar]