Abstract
Aspergillus flavus isolates produce only aflatoxins B1 and B2, while Aspergillus parasiticus and Aspergillus nomius produce aflatoxins B1, B2, G1, and G2. Sequence comparison of the aflatoxin biosynthesis pathway gene cluster upstream from the polyketide synthase gene, pksA, revealed that A. flavus isolates are missing portions of genes (cypA and norB) predicted to encode, respectively, a cytochrome P450 monooxygenase and an aryl alcohol dehydrogenase. Insertional disruption of cypA in A. parasiticus yielded transformants that lack the ability to produce G aflatoxins but not B aflatoxins. The enzyme encoded by cypA has highest amino acid identity to Gibberella zeae Tri4 (38%), a P450 monooxygenase previously shown to be involved in trichodiene epoxidation. The substrate for CypA may be an intermediate formed by oxidative cleavage of the A ring of O-methylsterigmatocystin by OrdA, the P450 monooxygenase required for formation of aflatoxins B1 and B2.
Aflatoxins are toxic and carcinogenic contaminants of foods and feeds that frequently are responsible for health and economic concerns in many countries (3, 34). These metabolites are produced by several species of Aspergillus, including A. flavus and A. pseudotamarii, which produce aflatoxins B1 and B2, and A. parasiticus and A. nomius, which produce aflatoxins B1, B2, G1, and G2 (Fig. 1). The cause for this divergence in the types of aflatoxins produced by different species has not been determined and the ancestral type is unknown. Species that produce only G aflatoxins have not been found. Species producing G aflatoxins could have evolved from an ancestral type which produced only B aflatoxins. Alternatively, B-producing species could have evolved from a B and G aflatoxin-producing ancestral type by a loss-of-function mutation in a gene needed for G but not B aflatoxin production.
FIG. 1.
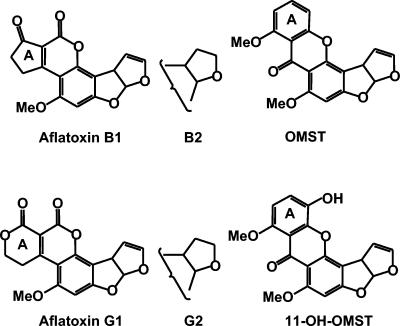
Structures of aflatoxins B1, B2, G1, and G2 and the aflatoxin precursors, OMST and 11-hydroxyOMST (11-OH-OMST).
Aflatoxins are polyketides with characteristic dihydro (B1 and G1)- or tetrahydro (B2 and G2)-bisfuran rings (4, 26) (Fig. 1). The B aflatoxins have a blue fluorescence, while the G aflatoxins have greenish-blue fluorescence when viewed under UV light due to structural differences in the A ring. Production of aflatoxins from malonyl coenzyme A involves a complex biosynthetic pathway, consisting of at least 25 genes (3, 34). These genes are clustered and expression of most of them is regulated by a single Zn2Cys6-type transcription factor, AflR, which is encoded by one of the genes in the cluster (37). Fifteen of the 25 proteins encoded by the cluster are enzymes that catalyze oxidative reactions; of these, six have the structural characteristics of cytochrome P450 monooxygenases (35-37). One of these P450 monooxygenases, OrdA, converts the stable biosynthetic precursor, O-methylsterigmatocystin (OMST) (Fig. 1), to aflatoxins B1 and B2 (28, 38), probably by a two-step process involving initial oxidation of OMST to 11-hydroxy-OMST (29).
How the G aflatoxins are formed has not been determined. From several studies on A. parasiticus, in which the conversion of radiolabeled aflatoxin B1 into G1 was measured, it was suggested that aflatoxin G1 is formed directly from aflatoxin B1 by oxidation of its cyclopentanone ring (16, 21, 25, 30). However, unexpectedly, the B1-to-G1 conversion rate was very low, indicating that a more-complex pathway must be involved. Other studies confirmed that separate pathways lead to the two types of aflatoxins (2, 4, 22, 33). By using cell extracts of A. parasiticus, Yabe et al. (34) found that formation of aflatoxin G1 required another P450 monooxygenase in addition to OrdA. We now report that cypA, an aflatoxin cluster gene upstream of the polyketide synthase gene, encodes the cytochrome P450 monooxygenase required for formation of G aflatoxins.
MATERIALS AND METHODS
Fungal isolates.
The following fungal isolates were used: A. flavus strain L isolates AF13 (ATCC 96044; SRRC 1273) (12) and NRRL 3357 (ATCC 200026; SRRC 167); A. flavus strain S isolate AF70 (ATCC MYA384) (12); A. nomius NRRL13137 (ATCC 15546; SRRC 375), the ex-type isolate (24), and BN008R (ATCC MYA-379), a B and G aflatoxin-producing isolate belonging to an unnamed taxon (15); A. oryzae isolates ATCC 46244 (NRRL 3485; SRRC 493), FRR 2874 (SRRC 2044), and ATCC 12892 (NRRL 1808; SRRC 304); and A. parasiticus isolates NRRL 2999 (ATCC 201461; SRRC 143), ATCC 15517 (SRRC 235), and BN009E (20). For fungal transformations, a niaD− mutant of BN009E was prepared by selection on chlorate-containing medium as previously described (1). Cultures were grown on 5/2 agar (5% V-8 vegetable juice, 2% agar, pH 5.2) in the dark at 31°C or on YES medium (per liter, 60 g of sucrose, 2 g of yeast extract) as previously described (10).
Aflatoxin production.
Qualitative analysis of aflatoxin production involved extraction of aflatoxins from YES medium cultures by first adding an equal volume of acetone, waiting 1 h, and then adding 1/10 volume of methylene chloride and 1 volume of water (11). From the acetone-methylene chloride layer (lower layer), 10 μl was spotted onto silica gel thin-layer chromatography (TLC) plates (Baker Si250) and plates were developed in ethyl ether-methanol-water, 96:3:1, to separate aflatoxin B1 from G1 (14). The plates were then visualized under 360-nm UV light.
DNA sequencing.
DNA was isolated from mycelia as previously described (17). For sequencing, DNA fragments from AF13, BN008R, AF70, and NRRL13137 which contained sequence homologous to A. parasiticus norB and cypA (GenBank accession number AY391490) were selected by blot hybridization to pCC1Fos genomic DNA libraries (Agencourt Bioscience Corporation, Beverly, Mass.). Sequence analysis involved shearing the DNA from fosmid clones into 3,000- to 3,500-bp fragments before cloning into vector pGTC (Genome Therapeutics Corporation, Waltham, Mass.) to construct shotgun subclone libraries. Subclones were sequenced (MegaBase 1000 sequencing apparatus; Amersham) using dye-terminator chemistry (Applied Biosystems). Sequencing was performed with sixfold redundancy to achieve a probable sequencing error rate of less than 1 bp per 1,000 bp.
Partial sequences of norB and cypA in other Aspergillus isolates.
A portion of the intergenic region and the 5′ coding sequences of norB and cypA was amplified by PCR using 20 to 50 ng of genomic DNA, 0.25 U of Amplitaq (Applied Biosystems), and 200 pmol of primers AP1729, 5′-GTGCCCAGCATCTTGGTCCACC, and AP3551, 5′-AAGGACTTGATGATTCCTC, at an annealing temperature of 58°C. The oligonucleotide numbers refer to nucleotide positions in the A. parasiticus aflatoxin gene cluster sequence, GenBank accession number AY371490. The PCR products were sequenced using the above primers (Auburn University Genomics and Sequencing Laboratory, Auburn, Ala.). DNA sequence manipulations and alignments were performed with DNAMAN software (Lynnon Biosoft, Vandreuil, Quebec, Canada). Where necessary, alignments were adjusted manually to minimize gaps.
Construction of the cypA disruption vector.
The cypA gene disruption vector was constructed by amplifying a 0.4-kb portion of the 5′ coding region of cypA (pcr1) and a 1.3-kb portion of the 3′ end of cypA and the downstream aflT gene (pcr2) using A. parasiticus NRRL2999 genomic DNA as template. Two primer sets, with their restriction endonuclease sites underlined and the enzymes indicated in parentheses, were as follows: for pcr1, 5′-TATGGTACCTTCTTCTCGAAGCAATACGTC (KpnI) and 5′-TAATTCTAGATACGTCGGCGGTGGC (XbaI); for pcr2, 5′-TATATCTAGACCCCGTTCCCCTTC (XbaI) and 5′-GAGGCGCATGCTACGGATCG (SphI). The XbaI-KpnI fragment from pcr1 and the SphI-XbaI fragment from pcr2 were cloned sequentially into pUC18 by standard recombinant DNA techniques. Then, the 6.7-kb XbaI fragment containing the entire A. parasiticus niaD gene and flanking regions (8) was ligated into the unique XbaI site of the above construct to give the disruption plasmid, pCypDV, whose sequence was checked by restriction analysis. Before transformation, pCypDV was linearized with XhoI and SphI to release the insert portion from the pUC18 vector.
Preparation of cypA disruption mutants.
A. parasiticus BN009E (niaD−) protoplasts for transformations were prepared from approximately 1 g of wet-weight mycelia (germinated from 108 spores) in potato dextrose broth by incubation at 29°C for 3 h with Novozyme234 enzyme mix (A. S. M. Sonnenberg, Applied Plant Research, Mushroom Research Unit, Horst, The Netherlands). The protoplasts were transformed as previously described (9) with 5 μg of XhoI/SphI-digested pCypDV DNA. Transformants were selected on Czapek's agar plates after incubation at 30°C for 7 to 10 days. Approximately 40 transformants per 5 μg of DNA were obtained. Cultures were grown from spores in 5 ml of YES medium for 3 days for analysis of aflatoxins by TLC on silica gel plates. Six transformants whose TLC aflatoxin profile lacked aflatoxin G1 and one transformant which produced both aflatoxins B1 and G1 were selected. To test for the insert position the following oligonucleotides were used in PCRs with DNA obtained from both types of transformants and from the wild-type untransformed fungi: P1 (3518), 5′-CCACTATCAAGCACAATCACCA; P2 (niaD), 5′-CTGTTTCGGACTCTCTTCTG; P3 (4022), 5′-CCACGCGACTGCAAATGGAG; P4 (4762), 5′-CTCGACTGTCGTCTGGTAGG; P5 (niaD), 5′-TCTCTTCCACTGTGCTATCCA; and P6 (6722), 5′-ACATGGAGGCGCCGATGAAG. The numbers in parentheses refer to positions of hybridization of the primers to A. parasiticus aflatoxin biosynthesis cluster DNA, GenBank accession number AY371490. The niaD primers hybridize to the ends of the A. parasiticus niaD insert (accession number U38948).
Northern blot analysis.
Expression of norB and cypA was assessed by Northern hybridization of total RNA from 3-day cultures of fungal cultures grown separately on glucose (aflatoxin-conducive) and peptone (aflatoxin-nonconducive) minimal salts medium (32) as previously described (5). DNA fragments used for probes for the Northern blots were obtained by PCR amplification of BN008R DNA with the oligonucleotide primer set 5′-GCTCCCTCTACCCAGTCAAA and 5-GTCCAAGGCAAATCAATACGC for norB and A. parasiticus NRRL2999 DNA with 5′-CGGTTCAATCCCAACGAAGTGCA and 5′-GCTGTCCATGCCTGCGAGAA for cypA. Probes were radiolabeled using a Rediprime II kit and [32P]dCTP (Amersham). Blots on Nytran+ were autoradiographed for 3 days at −70°C.
Nucleotide sequence accession numbers.
GenBank accession numbers for the entire aflatoxin biosynthesis cluster including the norB and cypA genes are as follows: AY510451 (AF13), AY510452 (BN008R), AY510453 (AF70), and AY510454 (NRRL13137). GenBank accession numbers for the partial sequences are AY566564 (NRRL3357), AY566565 (ATCC12892), AY566566 (ATCC46244), and FRR2874 (AY566567).
RESULTS
Comparison of the proximal end of the aflatoxin cluster in A. flavus and B and G aflatoxin-producing Aspergillus species.
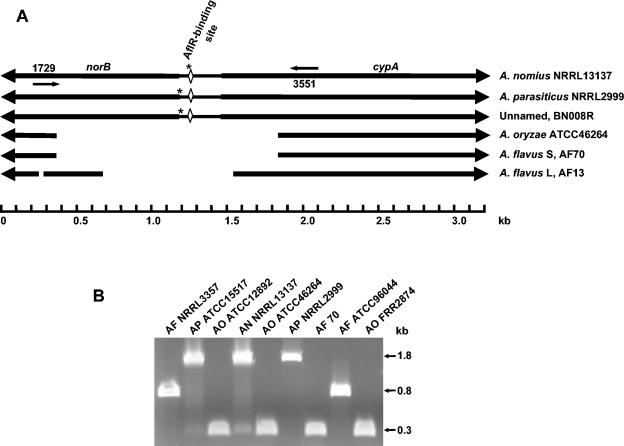
Alignment of sequences of the farthest upstream portion of the aflatoxin biosynthesis gene cluster from isolates of two strains of A. flavus (S and L) and three aflatoxin B- and G-producing Aspergillus species (BN008R, A. nomius, and A. parasiticus) revealed the presence of a 0.8- to 1.5-kb gap in the sequences of the A. flavus isolates. The gap was approximately 0.4 to 0.6 kb from the translational stop codon of norB, which is the putative 5′ terminus of the aflatoxin biosynthesis gene cluster. This deleted region is shown schematically in Fig. 2A. As determined by sequencing, the deletion covers part of the 5′ ends of the coding regions and all of the intergenic region of the two putative aflatoxin biosynthesis genes, norB and cypA, which are bidirectionally transcribed from a 279-bp intergenic region. The deletion is not limited to the above isolates. Based on sequencing (shown schematically in Fig. 2A) and PCR (Fig. 2B), the gap is also present in the sequences of A. oryzae (a domesticated form of A. flavus) and other A. flavus isolates. The length of the gap in the sequences of A. orzyae isolates is identical to that in the sequence of A. flavus AF70, an S strain isolate. The A. flavus L strain isolates, AF13 and NRRL3357, also have a 32-bp deletion in the coding region of norB, 324 bp upstream from the larger deleted region (Fig. 2A). Small insertions or deletions (Fig. 2A) occur in the intergenic regions of norB and cypA from isolates BN008, NRRL2999, and NRRL13137 from the three different aflatoxin B- and G-producing Aspergillus species.
FIG. 2.
Characteristics of the norB-cypA region in different Aspergillus species. (A) Schematic diagram of the norB-cypA sequences of different aflatoxin biosynthesis gene cluster homologs. Thick arrows indicate coding regions and direction of transcription of norB and cypA. Gaps represent deletions of 32 and 854 bp in A. flavus NRRL3357 and AF13, and 1516 bp in A. flavus AF70 and A. oryzae ATCC12892, ATCC46244, and FRR2874 when the sequences are compared to the sequence of A. parasiticus in this region. Additional smaller deletions or insertions are marked by asterisks (11 bp in A. parasiticus NRRL2999 at bp 1166, 13 bp in BN008R, and 4 bp in A. nomius NRRL13137). The positions of oligonucleotide primers AP1729 and AP3551 are indicated by small arrows. (B) Agarose gel (1.0%) electrophoresis of PCR fragments obtained by amplification of different Aspergillus DNAs with primers AP1729 and AP3551. Abbreviations: AF, A. flavus; AP, A. parasiticus; AO, A. oryzae; AN, A. nomius.
Disruption of cypA.
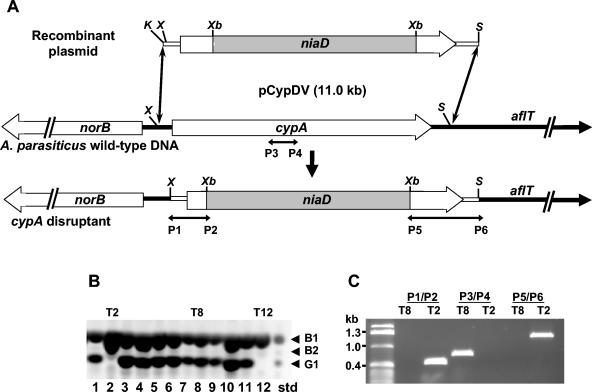
To test the involvement of cypA in aflatoxin G formation, we prepared a construct to insertionally disrupt the gene in A. parasiticus. The construct that was used had a 0.4-kb portion of the intergenic region and the 5′ coding sequence of cypA on one side of an niaD cassette and a 1.3-kb portion of the 3′ coding sequence of cypA and part of the downstream aflT gene on the other side of the cassette (Fig. 3A). The plasmid contains unique restriction enzyme sites for XhoI and SphI which allowed release of the insert portion from the vector to facilitate homologous recombination.
FIG. 3.
Preparation and characterization of cypA disruptants. (A) Schematic of the cypA disruption plasmid, pCypDV, and the expected product obtained by homologous recombination. Directions of transcription of norB, cypA, and aflT are shown as thick arrows. The insert niaD cassette DNA (shaded box) was obtained as a XbaI fragment from pSL82 (7). P1 to P6 are approximate annealing sites for the oligonucleotide primers used to test for insertion of the XhoI/SphI-digested plasmid fragment in transformants. K, KpnI; X, XhoI; S, SphI; Xb, XbaI. (B) A representative silica gel TLC profile of aflatoxins produced by pCypDV transformants. Locations of authentic aflatoxin standards are indicated on the right. Aflatoxins were visualized under 366-nm light and the negative image is shown. T2 and T12 are transformants that do not produce G aflatoxins; T8 produces both B and G aflatoxins. (C) Bands on a 1.0% agarose gel after electrophoresis of PCR products obtained with oligonucleotide primer pairs P1-P2, P3-P4, and P5-P6 used to check cypA disruption transformants. The positions of the primers are shown in panel A.
Transformation of A. parasiticus BN009E with the XhoI/SphI-digested plasmid gave six transformants which produced only aflatoxin B1 and a minor amount of aflatoxin B2 when grown on aflatoxin-conducive medium. The thin-layer chromatography profile for 12 transformants including two of the putative cypA knockout transformants (T2 and T12) is shown in Fig. 3B. To demonstrate that the gene was insertionally disrupted in one of the putative cypA knockout transformants, T2, we compared the PCR bands obtained using DNAs from this transformant and from a transformant which still produced both B and G aflatoxins (T8). Primer pairs P1-P2 and P5-P6 were designed to amplify between the niaD cassette and a region slightly outside of the portion of cypA contained in the disruption vector (Fig. 3A). PCR of transformant T8 with these primer pairs resulted in no bands, whereas the same PCR with transformant T2 resulted in bands of the predicted size. PCR with the P3-P4 primer pair designed to amplify a portion of cypA that was expected to be replaced by the niaD cassette in knockout transformants gave bands only with transformant T8 (Fig. 3C).
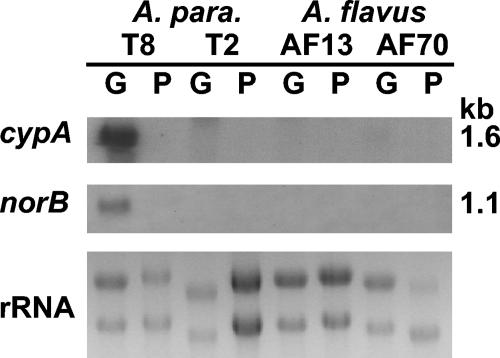
The results of blot hybridization of total RNA with DNA probes generated by PCR of portions of the coding regions of norB and cypA show that these genes were expressed by the transformant T8 under conducive conditions for aflatoxin production, but not by the cypA knockout transformant T2 (Fig. 4). In the nonconducive medium containing peptone minimal salts, neither gene was expressed for any of the fungal isolates. No hybridization bands for either gene were observed in blots of A. flavus RNA, indicating that neither of these genes is expressed in these isolates. This result was expected since the promoter regions and parts of the 5′ ends of the coding regions of both genes are missing.
FIG. 4.
Northern hybridization analysis of A. parasiticus and A. flavus RNAs with probes for the genes norB and cypA. A. parasiticus BN009E transformant T8, the cypA-knockout transformant, T2, A. flavus AF13, and A. flavus AF70 were grown on peptone minimal salts (P) and glucose minimal salts (G) medium for 3 days and 20 μg of total RNA was separated on a 0.4 M formaldehyde-1.2% agarose gel and transferred to a Nytran+ membrane for probing with radiolabeled norA and cypA probes. The approximate sizes of the bands are shown on the right side of the blots.
Homologies of NorB and CypA to proteins in the GenBank database.
Based on protein-protein BLAST searches of the GenBank database, the sequence of the predicted protein encoded by the first gene in the cluster, norB, is highly homologous to sequences for fungal aryl alcohol dehydrogenases (COG0667; E value of e−112 for the highest homology sequence in the database). A gene coding for a similar type of protein (NorA) is present in the aflatoxin pathway cluster approximately 38 kb further downstream. The amino acid identity of NorB with NorA is 50% (E value = e−100). A NorA and NorB homolog, StcV, is present in the sterigmatocystin gene cluster of A. nidulans (E value = 3e−94). The function of such putative aryl alcohol dehydrogenases in either aflatoxin or sterigmatocystin biosynthesis has not been determined (6, 34, 37).
Similar BLAST searches of the GenBank database with the predicted protein encoded by cypA revealed the highest homology to Tri4 (38% amino acid identity; E value, 5e−83), one of the P450 monooxygenases involved in trichothecene biosynthesis in Fusarium species (23). Alignment of the predicted amino acid sequences of CypA from A. parasiticus, A. nomius, BN008R, and Gibberella zeae Tri4 is shown in Fig. 5. The deduced amino acid sequence of CypA shows the presence of conserved regions characteristic of P450 enzymes (31). These include a heme-binding loop beginning at amino acid (aa) 447 in the L helix, a conserved P450 E-X-X-R motif at aa 355 to 358 in the K helix, and a protein transfer groove A/G-G-X-D/E-T-T/S at aa 297 to 302 in the I helix. The presence of two adjacent Arg residues 17 residues from the C-terminal side of the putative heme-binding loop (Fig. 5) is characteristic of flavin-adenine dinucleotide-utilizing reductases, and in this regard distinguishes CypA from Tri4. The deduced protein sequence of CypA also has a membrane-binding motif (P-X-P) and adjacent cluster of basic residue(s) near the N terminus, characteristic of membrane-bound P450 proteins. CypA has approximately 20 to 32% overall amino acid homology to other fungal P450 enzymes and falls in the pfam00067.9 family of P450 monooxygenases (Table 1). Because of its homology to Tri4, CypA has been given the P450 designation CYP58B1 (David R. Nelson, personal communication).
FIG. 5.
Alignment of CypA protein sequences from isolates of three Aspergillus species that produce both B and G aflatoxins and Tri4 from G. zeae, GenBank accession number AAM48924. Locations of presumed consensus P450 catalytic and other domains are shown as bars under the sequence. Shaded regions represent locations of amino acid identity in the sequences. Basic amino acids adjacent to the P-X-P motif and the heme-binding loop are indicated by arrows. The different helix domains are indicated by letters and brackets above the sequence.
TABLE 1.
Comparison of heme-binding loop sequences and amino acid identity of fungal cytochrome P450 proteins
| CYP no. | Sequencea | Name | Protein identity (%)b | Putative oxidative functionc | Organism | Accession no. |
|---|---|---|---|---|---|---|
| CYP58B1 | FSKGSRHCLGINFALAEIYL | CYPA | Epoxidase | Aspergillus parasiticus | AAS66021 | |
| CYP58A | FSQGSRQCIGYTMAFAEMYL | TRI4 | 38.2 | Epoxidase | Gibberella zeae | AAM48924 |
| CYP60A | FTKGSRSCLGQNLSMAEISL | Unnamed | 32.1 | Unknown | Neurospora crassa | XP_330833 |
| CYP60B | FSIGPRNCIGRQLAYVEMRL | AVNA | 22.4 | Hydroxylase | Aspergillus parasiticus | AAB52228.1 |
| CYP60B | WSVGLRNCIGRNLAYAEVRL | VERB | 20.2 | Desaturase | Aspergillus parasiticus | AAD50366 |
| CYP62 | FGQGARQCLGIHLGWMQLRL | CYPX | 21.4 | Unknown | Aspergillus parasiticus | AAF26280 |
| CYP57A2 | FGAGSRSCIGKNISILEMSK | PDA6-1 | 21.7 | Demethylase | Nectria haematococca | P38364 |
| CYP53 | FSTGPRACVGRNVAEMELLV | BPHA | 19.3 | Hydroxylase | Aspergillus niger | P17549 |
| CYP59 | FEFGPRSCIGQTLAMLELRI | VERA | 16.6 | Oxidase | Aspergillus parasiticus | AAS66015 |
| CYP64 | FGFGRRICPGRFVTDEKLFL | ORDA | 12.5 | Hydroxylase | Aspergillus parasiticus | AAC49710 |
The consensus sequence for the heme-binding loop is F-X-X-G-X-R-X-C-X-G.
Homologies were obtained by alignment using DNAMAN software.
The oxidative function of the cytochrome P450 enzymes has been ascribed on the basis of gene knockout studies.
DISCUSSION
We have shown for the first time why A. flavus isolates do not produce G aflatoxins. The lack of G aflatoxin production is a consequence of a 0.8- to 1.5-kb deletion near the 5′ end of the gene cluster. This deletion includes all of the intergenic region and portions of the 5′ ends of the coding sequences of norB and cypA, the first two genes at the end of the aflatoxin biosynthesis gene cluster proximal to the polyketide synthesis gene. Therefore, because the deletion removes the promoter regions and translational start sites of these two putative oxidative enzyme-encoding genes, neither gene should be transcribed, as was found by Northern blot analysis of RNAs from A. flavus isolates grown under conducive conditions for aflatoxin formation (Fig. 4). Since both of these genes contain an AflR-binding site in their shared intergenic region, their expression is probably regulated by the Zn2Cys6-transcriptional activator, AflR, in a similar fashion to those of the other better-studied aflatoxin and sterigmatocystin pathway genes (9, 18, 19). Disruption in A. parasiticus of one of these genes, cypA, which encodes a cytochrome P450 monooxygenase, prevented accumulation of G aflatoxins. Taken together, these results demonstrate that cypA is required for aflatoxin G formation.
The second gene, norB, which is also not expressed in A. flavus isolates because of the deletion, is predicted to encode an aryl alcohol dehydrogenase which is highly homologous to an enzyme encoded by another gene in the aflatoxin cluster, norA. The function of norA in aflatoxin biosynthesis has not been determined. Knockout of norA in A. parasiticus did not appear to affect aflatoxin production (J. C. Cary, unpublished results). Because the proteins encoded by these genes have about 68% amino acid similarity, it is possible that they can functionally complement one another. This could explain why a phenotype was not observed when only one of these genes was inactivated.
The present results are consistent with the findings of Yabe et al. (34), who inferred by inhibition studies with cell-free A. parasiticus extracts that two different cytochrome P450 enzymes are required for aflatoxin G1 formation. They identified one of these enzymes as OrdA, which was known to be required for B aflatoxin formation (28, 39), but did not mention any candidates for the second enzyme. Because OMST was converted to aflatoxin B1 by yeast cells expressing the ordA gene, previous investigators suggested that OrdA was the only enzyme required for formation of the B aflatoxins (29). Conversion of OMST to aflatoxin B1 requires at least two separate oxidation steps. Udwary et al. (29) proposed that OrdA performs two sequential oxidations of the xanthone A ring of OMST, the first generating the intermediate, 11-hydroxyOMST (Fig. 1, 11-OH-OMST), and the second cleaving the A ring to give an open-chain butenyl carboxylic acid intermediate (structure 1, Fig. 6). It is not known if the subsequent rearrangement of the open-chain intermediate to form aflatoxin B1 requires other enzymes, but if so these enzymes must be available in yeast. Since no A. parasiticus mutants have been isolated which only produce G-type toxins (33), at least one of the enzymes required for B-type aflatoxin formation is probably also required for G-type toxin formation (34).
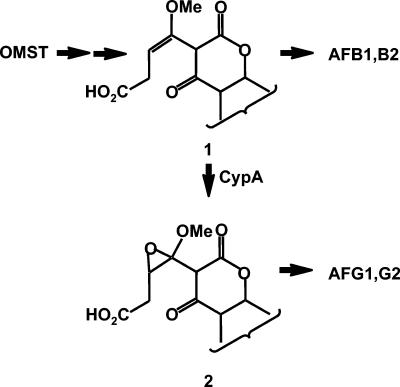
FIG. 6.
Scheme showing the possible conversion of OMST to aflatoxins B1 and G1 (the steps would be similar for the conversion of dihydro-OMST to aflatoxin B2 and G2). The conversion of OMST to aflatoxin B1 is predicted to involve formation of an A-ring-opened intermediate, structure 1, as suggested by Udwary et al. (29). Oxidation of this intermediate by CypA is predicted to give the epoxide which is subsequently converted to the G aflatoxins.
The substrate for CypA oxidation that is required to produce the G aflatoxins has not been identified. Because of its amino acid similarity to Tri4, like Tri4, CypA may be capable of oxidizing an unactivated double bond (23). It is possible that the unactivated double bond of structure 1 in Fig. 6 is oxidized to the epoxide of structure 2, which then could rearrange to aflatoxin G1. Other oxidoreductases in the aflatoxin pathway gene cluster that are predicted to be encoded by genes whose function in aflatoxin biosynthesis is still unknown (for example OrdB, NorA, or NorB) could be involved in the rearrangement of either structure 1 or 2 to B and G aflatoxins, respectively (6, 36, 38).
The presence of large sequence deletions in the aflatoxin cluster of A. flavus may shed some light on the phylogeny of the different aflatoxin-producing species in Aspergillus section Flavi. We suggest that B aflatoxin-producing species diverged from B and G toxin-producing species. This hypothesis is in agreement with previous suggestions that A. nomius diverged from a common ancestor prior to the divergence of A. flavus (20, 27). A. flavus is a highly diverse asexual species composed of many genetically isolated vegetative compatibility groups and several morphotypes or strains. The loss-of-function deletion is conserved among the A. flavus isolates examined in the present work including both the L and S morphotypes (13). The deletion is also present in isolates of A. oryzae, an atoxigenic domesticated offshoot of A. flavus. The variability in size of the deletion may provide further insight into the relationships among different A. flavus and A. oryzae isolates.
Acknowledgments
We thank Beverly G. Montalbano and Les L. Scharfenstein for excellent technical assistance. We also thank David R. Nelson, University of Tennessee, Memphis, for his kindness in checking the CypA sequence and assisting with nomenclature.
REFERENCES
- 1.Bayman, P., and P. J. Cotty. 1991. Improved media for selecting nitrate-nonutilizing mutants in Aspergillus flavus. Mycologia 83:311-316. [Google Scholar]
- 2.Bhatnagar, D., T. E. Cleveland, and D. G. Kingston. 1991. Enzymological evidence for separate pathways for aflatoxin B1 and B2 biosynthesis. Biochemistry 30:4343-4350. [DOI] [PubMed] [Google Scholar]
- 3.Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. 2003. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 61:83-93. [DOI] [PubMed] [Google Scholar]
- 4.Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. 1992. Oxidation-reduction reactions in biosynthesis of secondary metabolites, p. 255-285. In D. Bhatnagar, E. B. Lillehoj, and D. K. Arora (ed.), Mycotoxins in ecological systems, vol. 10. Marcel Dekker, New York, N.Y.
- 5.Cary, J. W., J. M. Dyer, K. C. Ehrlich, M. S. Wright, S.-H. Liang, and J. E. Linz. 2002. Molecular and functional characterization of a second copy of the aflatoxin regulatory gene, aflR-2, from Aspergillus parasiticus. Biochim. Biophys. Acta 1576:316-323. [DOI] [PubMed] [Google Scholar]
- 6.Cary, J. W., M. Wright, D. Bhatnagar, R. Lee, and F. S. Chu. 1996. Molecular characterization of an Aspergillus parasiticus gene, norA, located on the aflatoxin biosynthesis gene cluster. Appl. Environ. Microbiol. 62:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, P. K., J. W. Cary, D. Bhatnagar, T. E. Cleveland, J. W. Bennett, J. E. Linz, C. P. Woloshuk, and G. A. Payne. 1993. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:3273-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, P.-K., K. C. Ehrlich, J. E. Linz, D. Bhatnagar, T. E. Cleveland, and J. W. Bennett. 1996. Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr. Genet. 30:68-75. [DOI] [PubMed] [Google Scholar]
- 9.Chang, P.-K., K. C. Ehrlich, J. Yu, D. Bhatnagar, and T. E. Cleveland. 1995. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl. Environ. Microbiol. 61:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotty, P. J. 1994. Comparison of four media for the isolation of Aspergillus flavus group fungi. Mycopathologia 125:157-162. [DOI] [PubMed] [Google Scholar]
- 11.Cotty, P. J. 1988. Simple fluorescence method for rapid estimation of aflatoxin levels in a solid culture medium. Appl. Environ. Microbiol. 54:274-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotty, P. J. 1989. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79:808-814. [Google Scholar]
- 13.Cotty, P. J., D. S. Bayman, D. S. Egel, and K. S. Elias. 1994. Agriculture, aflatoxins and Aspergillus, p. 1-27. In K. Powell (ed.), The genus Aspergillus. Plenum Press, New York, N.Y.
- 14.Cotty, P. J., and D. Bhatnagar. 1994. Variability among atoxigenic Aspergillus flavus strains in ability to prevent aflatoxin contamination and production of aflatoxin biosynthetic pathway enzymes. Appl. Environ. Microbiol. 60:2248-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotty, P. J., and K. F. Cardwell. 1999. Divergence of West African and North American communities of Aspergillus section Flavi. Appl. Environ. Microbiol. 65:2264-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutton, M. F. 1988. Enzymes and aflatoxin biosynthesis. Microbiol. Rev. 52:274-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich, K. C., J. W. Cary, and B. G. Montalbano. 1999. Characterization of the promoter for the gene encoding the aflatoxin biosynthetic pathway regulatory protein AFLR. Biochim. Biophys. Acta 1444:412-417. [DOI] [PubMed] [Google Scholar]
- 18.Ehrlich, K. C., B. G. Montalbano, and J. W. Cary. 1999. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 230:249-257. [DOI] [PubMed] [Google Scholar]
- 19.Ehrlich, K. C., B. G. Montalbano, J. W. Cary, and P. J. Cotty. 2001. Promoter elements in the aflatoxin pathway polyketide synthase gene. Biochim. Biophys. Acta 1576:171-175. [DOI] [PubMed] [Google Scholar]
- 20.Ehrlich, K. C., B. G. Montalbano, and P. J. Cotty. 2003. Sequence comparison of aflR from different Aspergillus species provides evidence for variability in regulation of aflatoxin production. Fungal Genet. Biol. 38:63-74. [DOI] [PubMed] [Google Scholar]
- 21.Heathcote, J. G., M. F. Dutton, and J. R. Hibbert. 1976. Biosynthesis of aflatoxins. Part II. Chem. Ind. (London) 20:270-272. [Google Scholar]
- 22.Henderberg, A., J. W. Bennett, and L. S. Lee. 1988. Biosynthetic origin of aflatoxin G1: confirmation of sterigmatocystin and lack of confirmation of aflatoxin B1 as precursors. J. Gen. Microbiol. 134:661-667. [DOI] [PubMed] [Google Scholar]
- 23.Hohn, T. M., A. E. Desjardins, and S. P. McCormick. 1995. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet. 248:95-102. [DOI] [PubMed] [Google Scholar]
- 24.Kurtzman, C. P., B. W. Horn, and C. W. Hesseltine. 1987. Aspergillus nomius, a new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamarii. Antonie Leeuwenhoek 53:147-158. [DOI] [PubMed] [Google Scholar]
- 25.Maggon, K. K., and T. A. Venkitasubramaian. 1973. Metabolism of aflatoxin B1 and aflatoxin G1 by Aspergillus parasticus. Experientia 29:1210-1212. [DOI] [PubMed] [Google Scholar]
- 26.Minto, R. E., and C. A. Townsend. 1997. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 97:2537-2555. [DOI] [PubMed] [Google Scholar]
- 27.Peterson, S. W. 1997. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis, p. 323-356. In R. A. Samson and J. I. Pitt (ed.), Classification of Penicillium and Aspergillus: integration of modern taxonomic methods. Harwood Publishers, Reading, United Kingdom.
- 28.Prieto, R., and C. P. Woloshuk. 1997. ord1, an oxidoreductase gene responsible for conversion of O-methylsterigmatocystin to aflatoxin in Aspergillus flavus. Appl. Environ. Microbiol. 63:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Udwary, D. W., L. K. Casillas, and C. A. Townsend. 2002. Synthesis of 11-hydroxyl O-methylsterigmatocystin and the role of a cytochrome P-450 in the final step of aflatoxin biosynthesis. J. Am. Chem. Soc. 124:5294-5303. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe, C. M., and C. A. Townsend. 1996. Incorporation of molecular oxygen in aflatoxin B1 biosynthesis. J. Org. Chem. 61:1990-1993. [Google Scholar]
- 31.Werck-Reichhart, D., and R. Feyereisen. 2000. Cytochromes P450: a success story. Genome Biol. [Online.] http://genomebiology.com/2000/I/6/reviews/3003.1. [DOI] [PMC free article] [PubMed]
- 32.Woloshuk, C. P., and G. A. Payne. 1994. The alcohol dehydrogenase gene adh1 is induced in Aspergillus flavus grown on medium conducive to aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:670-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yabe, K., H. Nakamura, Y. Ando, N. Terakado, H. Nakajima, and T. Hamasaki. 1988. Isolation and characterization of Aspergillus parasiticus mutants with impaired aflatoxin production by a novel tip culture method. Appl. Environ. Microbiol. 54:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yabe, K., M. Nakamura, and T. Hamasaki. 1999. Enzymatic formation of G-group aflatoxins and biosynthetic relationship between G- and B-group aflatoxins. Appl. Environ. Microbiol. 65:3867-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, J., D. Bhatnagar, and K. C. Ehrlich. 2002. Aflatoxin biosynthesis. Rev. Iberoam. Micol. 19:191-200. [PubMed] [Google Scholar]
- 36.Yu, J., P. K. Chang, D. Bhatnagar, and T. E. Cleveland. 2000. Genes encoding cytochrome P450 and monooxygenase enzymes define one end of the aflatoxin pathway gene cluster in Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53:583-590. [DOI] [PubMed] [Google Scholar]
- 37.Yu, J., P.-K. Chang, J. W. Cary, D. Bhatnagar, and T. E. Cleveland. 1997. avnA, a gene encoding a cytochrome P-450 monooxygenase, is involved in the conversion of averantin to averufin in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 63:1349-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, J., P.-K. Chang, K. C. Ehrlich, J. W. Cary, D. Bhatnagar, T. E. Cleveland, G. A. Payne, J. E. Linz, C. P. Woloshuk, and J. W. Bennett. 2004. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70:1253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, J., P. K. Chang, K. C. Ehrlich, J. W. Cary, B. Montalbano, J. M. Dyer, D. Bhatnagar, and T. E. Cleveland. 1998. Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl. Environ. Microbiol. 64:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]