Abstract
We systematically investigated the physiological response as well as DNA damage repair and damage tolerance in Shewanella oneidensis MR-1 following UVC, UVB, UVA, and solar light exposure. MR-1 showed the highest UVC sensitivity among Shewanella strains examined, with D37 and D10 values of 5.6 and 16.5% of Escherichia coli K-12 values. Stationary cells did not show an increased UVA resistance compared to exponential-phase cells; instead, they were more sensitive at high UVA dose. UVA-irradiated MR-1 survived better on tryptic soy agar than Luria-Bertani plates regardless of the growth stage. A 20% survival rate of MR-1 was observed following doses of 3.3 J of UVC m−2, 568 J of UVB m−2, 25 kJ of UVA m−2, and 558 J of solar UVB m−2, respectively. Photoreactivation conferred an increased survival rate to MR-1 of as much as 177- to 365-fold, 11- to 23-fold, and 3- to 10-fold following UVC, UVB, and solar light irradiation, respectively. A significant UV mutability to rifampin resistance was detected in both UVC- and UVB-treated samples, with the mutation frequency in the range of 10−5 to 10−6. Unlike in E. coli, the expression levels of the nucleotide excision repair (NER) component genes uvrA, uvrB, and uvrD were not damage inducible in MR-1. Complementation of Pseudomonas aeruginosa UA11079 (uvrA deficient) with uvrA of MR-1 increased the UVC survival of this strain by more than 3 orders of magnitude. Loss of damage inducibility of the NER system appears to contribute to the high sensitivity of this bacterium to UVR as well as to other DNA-damaging agents.
Solar UV radiation (UVR) is lethal and potentially mutagenic to all organisms at species-specific levels. The stratospheric ozone layer absorbs UVC (<290 nm) effectively; however, both UVA (320 to 400 nm) and UVB (290 to 320 nm) wavelengths penetrate to the earth's surface. UVR-induced damage is greatly dependent on the sources of radiation and the time of exposure. Photons of UVB and UVC wavelengths cause direct DNA damage by inducing the formation of DNA photoproducts, such as cyclobutyl pyrimidine dimers (CPD) and pyrimidine (6-4) pyrimidinone (37). The accumulation of DNA photoproducts can be lethal through the blockage of DNA replication and transcription. UVA can cause photodamage to a variety of molecules as well as physiological processes directly or indirectly by inducing the production of reactive oxygen species (5, 6, 17, 53). Distinct differences between far-UV (UVC) and near-UV (UVB and UVA) damage have been observed in both bacteria and bacteriophages (6).
Bacteria are particularly vulnerable to UVR damage due to their small size and unicellular structure. Thus, the possession of mechanisms to repair UVR-induced damage as well as other sheltering strategies, e.g., pigments and sunscreen molecules, are essential contributors to the ecological fitness of organisms that are regularly exposed to solar UVR. Several mechanisms have evolved in bacteria to repair or tolerate UVR-induced DNA damage. Photoreactivation and nucleotide excision repair (NER) are the two primary mechanisms that accurately repair UVR-damaged DNA, whereas mutagenic DNA repair (MDR) is a determinant that increases damage tolerance (11). In addition, many of the genes involved in DNA damage repair are inducible through the SOS response (29). Approximately 30 genes have been reported to belong to the SOS regulon of Escherichia coli (4, 8).
Photoreactivation in bacteria involves a single enzyme called photolyase, which binds to CPDs and, in the presence of light (300 to 500 nm), reverses the dimer to its component monomers (26). CPD photolyases are widely but also sporadically distributed among bacteria, archaea, and eukaryotes (56). NER is present from bacteria to humans and plays a critical role in protecting cells from a variety of DNA-damaging agents since it can recognize a broad range of DNA lesions, including ionizing radiation-induced purine damage, active oxygen species-induced base loss, and UV-induced pyrimidine dimers (41). During the repair process, NER component enzymes hydrolyze two phosphodiester bonds, one on either side of the lesion, to generate an oligonucleotide carrying the damage. The excised oligonucleotide is released from the duplex, and the resulting gap is filled and ligated (28, 42, 43). In E. coli, the NER component genes uvrA, uvrB, and uvrD are subject to SOS regulation (4, 10, 21, 22, 45). UmuDC-mediated MDR functions in translesion synthesis enabling bypass of DNA lesions that would normally block replication by DNA polymerase III (46, 50, 51). Translesion DNA synthesis provides the cell with an additional mechanism of survival, although the process is accompanied by an elevation of the cellular mutation rate (46, 55). Expression of the umuDC operon is regulated by the SOS response in many bacteria (46).
Shewanella oneidensis MR-1, a γ-proteobacterium, was originally isolated from the sediment of Oneida Lake in New York State (35). Extensive studies have been carried out on this bacterium due to its respiratory versatility: it can reduce a variety of compounds, including toxic metals and radionuclides (30, 31). This unique feature offers potential for bioremediation by immobilization of soluble metal species at contaminated sites. To succeed, MR-1 has to tolerate toxic levels of pollutants and exposure to ionizing or solar radiation. Recently, the genome sequence of MR-1 has been completed (15). It consists of a 4,969,803-bp chromosome with 4,758 predicted open reading frames (ORFs) and a 161,613-bp plasmid with 173 predicted ORFs. Three prophages, lambdaSo (51,857 bp), MuSo1 (34,551bp), and MuSo2 (35,666 bp), are present on the MR-1 chromosome (15). Compared to E. coli K-12, MR-1 has most of the genes involved in repairing DNA damage and defending oxidative stress. Knowledge on bacterial UV resistance and repairing mechanisms has come predominantly from E. coli. Limited knowledge on molecular and physiological responses to UVR is available for environmentally relevant bacteria. Here, we report the responses of MR-1 following UVC, UVB, UVA, and natural sunlight exposures. We found that MR-1 was uniformly sensitive to all wavelengths of UVR. We also evaluated the contribution of photoreactivation, NER, and mutagenic repair to the survival of MR-1 following UVR exposures. An inefficiently expressed NER system in MR-1 appears to contribute to its high sensitivity to both UVB and UVC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, plasmids, and PCR primers used in this study are listed in Table 1. E. coli and Pseudomonas aeruginosa strains were grown in Luria-Bertani (LB) medium (pH 7.2) at 37°C. All Shewanella strains were grown at 30°C in tryptic soy broth, except for Shewanella algae, which was grown in a modified marine broth (5 g of peptone, 2 g of yeast, and 17 g of sea salts in 1 liter; pH 7.2). For gene expression experiments, S. oneidensis MR-1 was grown in Davis medium (Difco) supplemented with 15 mM lactic acid. Ampicillin (100 μg ml−1) was used to grow E. coli carrying plasmids pJJK20, pJB321, pXQ01, and pXQ03, whereas carbenicillin (200 μg ml−1) was used to grow P. aeruginosa carrying the plasmids described above.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s)a or nucleotide sequenceb | Source, reference, or relative position of primer |
|---|---|---|
| Bacterial strains | ||
| E. coli DH10B | Plasmid-free strain used for cloning | 13 |
| E. coli K12 | Reference strain | DSM498 |
| S. oneidensis MR-1 | Lake Oneida, N.Y., sediment | 35 |
| S. oneidensis DLM7 | Lower Green Bay sediment, Lake Michigan, Wis. | 52 |
| S. oneidensis MR-4 | Black sea water column, 5 m | 52 |
| S. putrefaciens 200 | Crude oil pipeline, Canada | ATCC |
| S. algae | Red algae, Japan | ATCC |
| P. aeruginosa PAO1 | Prototrophic, no UV resistance plasmid present | A. M. Chakrabarty |
| P. aeruginosa UA11079 | Same as PAO1, but Rifr, uvrA::ΩHg (Hgr), plasmid-free strain | 39 |
| Plasmids | ||
| pJJK20 | Apr, E. coli umuDC promoter source | 24 |
| pJB321 | Cbr, broad-host-range cloning vector | 3 |
| pRK2013 | Helper plasmid for triparental matings | 9 |
| pXQ01 | Apr, cloned MR-1 uvrA coding sequence as NdeI-BamHI in pJJK20 | This study |
| pXQ03 | Apr, umuDC promoter and MR-1 uvrA coding sequence from pXQ01 as SphI-BamHI in pJB321 | This study |
| Primersc | ||
| uvrA NdeI 5′ | GATCCATATGGATAAGATTGAAATACGCGGTGC | ATG start codon |
| uvrA BamHI 3′ | GATCGGATCCCTACTGCTGTTTGGTTAGC | CTA stop codon |
| uvrA1F | TAACGGTCGTAAGGGTGAGC | 465 |
| uvrA1R | GAGAGTTCGAGTGCGGTTTC | 683 |
| uvrA2F | GCTTATTCACCTGCGTGACA | 1898 |
| uvrA2R | GTATAAGTGGCGGGGTTTGA | 2114 |
| uvrB1F | CACCATCGCCAATGTGATAG | 144 |
| uvrB1R | CAATCAGCACCACATCCTTG | 421 |
| uvrB2F | TAGTGCCTAAAGGGGTGGTG | 1754 |
| uvrB2R | CCCTTAAGCGTTTCACCTCA | 2002 |
| uvrD1F | AAGACCAAGGATTACGGCCT | 449 |
| uvrD1R | ATCCAAGCGTATTGAATGGC | 698 |
| uvrD2F | CAAGAGCTCACGTTATGGCA | 1297 |
| uvrD2R | CTCTTCAGGCATTTCGAAGG | 1572 |
Phenotype resistance (r) abbreviations are: Ap, ampicillin; Cb, carbenicillin; Hg, mercury; Rif, rifampin.
For primer oligonucleotide sequences, the restriction sites incorporated in primers are underlined. CATATG, NdeI; GGATCC, BamHI.
Primers were designed using putative gene sequences of S. oneidensis MR-1.
Molecular techniques.
Genomic and plasmid DNA isolation, restriction digestion, gel purification, ligation, and transformation were performed using standard techniques (40). PCR primers (Table 1) were designed using the Primer 3 program (http://www.broad.mit.edu/cgi-bin/primer/primer3.cgi/) and synthesized at the Genomic Technology Center of Michigan State University.
UV irradiation, photoreactivation, and MDR assays.
UVA, UVB, and UVC assays were performed using previously described methods (47, 49). The UVA, UVB, and UVC sources used were XX-15L, XX-15 M, and XX-15 lamps (UVP Products; San Gabriel, Calif.), respectively. The energy output of each lamp was monitored with a UV-X radiometer (UVP Products) fitted with the appropriate sensor. The UVB lamp was filtered through cellulose diacetate (Kodacel; Eastman Kodak, Rochester, N.Y.) to eliminate stray UVC wavelengths. During irradiation, cell suspensions were mixed continuously to avoid shading effects. In experiments comparing the UVA sensitivity at different physiological stages, cells grown in Davis medium to exponential phase were used directly for UVA treatment, whereas stationary-phase cells were diluted with Davis medium to an optical density at 600 nm of about 0.2 (the density at mid-exponential phase in Davis medium). Photoreactivation assays and MDR assays were conducted as described previously (24, 25).
Solar radiation sensitivity assays.
Solar radiation sensitivity assays were conducted by exposing cell suspensions to ambient solar radiation. The suspensions were maintained in sterile boxes constructed of 64-mm-thick Acrolyte OP-4 plastic (Professional Plastics, Austin, Tex.). The Acrolyte OP-4 plastic transmits greater than 90% of the total radiation throughout the UVA and UVB wavelengths (Acrolyte OP-4 technical data sheet; Cyro Industries, Arlington, N.J.). Replicate boxes were maintained on ice on a rocking platform during the exposures. Solar UVB radiation was measured with a UVB detector (SED240/UVB-1/W) attached to an IL-1700 research radiometer (International Light, Newburyport, Mass.). UVB radiation was measured every second and the readings were integrated over the exposure period, yielding a quantitative output in joules per square meter. At appropriate time points, the boxes were temporarily shaded from sunlight exposure, and two samples (5 ml) were taken. One sample was plated in the dark, and the other was plated following a photoreactivation treatment as described previously (25).
Transcriptional analysis of NER by using a cDNA microarray.
S. oneidensis MR-1 whole-genome DNA arrays were produced by Liyou Wu and Jizhong Zhou at Oak Ridge National Laboratory (Oak Ridge, Tenn.). Mid-exponential-phase cells (80 ml) grown in Davis medium were split into two parts: one was used for UVR treatments and the other was used as controls. The exposure doses were 3.3 J m−2 for UVC, 568 J m−2 for UVB, and 25 kJ m−2 for UVA, which yielded about a 20% survival rate. After irradiation, cells were transferred to a 100-ml flask and incubated at 30°C on a shaker (200 rpm). An aliquot of cells (12 ml) was transferred to a centrifuge tube after 5, 20, and 60 min of incubation and concentrated by centrifugation at 4°C. The cell pellet was resuspended in 2 ml of supernatant and mixed with 4 ml of bacterial RNA protection reagent (QIAGEN, Valencia, Calif.). The cell suspension was kept at room temperature until all the samples were collected (within 2 h). Cells were then pelleted and stored at −80°C until RNA extraction. Controls were treated in the same way except for the UVR treatment. Total RNA was isolated using a QIAGEN RNeasy mini kit (QIAGEN), digested with RNase-free DNase I (Invitrogen, Carlsbad, Calif.) at 25°C for 30 min, extracted with phenol, phenol-chloroform (1:1), and chloroform, and stored in ethanol at −80°C until use. Both PCR and gel electrophoresis were used to confirm the complete digestion of any contaminating DNA. We confirmed both RNA purity and quality by the 260-to-280-nm absorbance ratio and gel electrophoresis before the reverse transcription reaction. Prehybridization and RNA labeling were performed as described by Schroeder et al. (44) with a 2:3 ratio of 5-(3-aminoallyl)-dUTP and dTTP. Hybridization and washing were carried out as described by Hegde et al. (14). The array was scanned using an Axon 4000B scanner (Axon Instruments, Inc., Union City, Calif.). The data were imported into GeneSpring (Silicon Genetics, Redwood City, Calif.) for analysis. Data were normalized both per chip and per gene (Lowess method). Spots with less than 55% of pixels greater than background plus two standard deviations were not included in the data analysis (34).
Functional analysis of uvrA in P. aeruginosa strain UA11079.
The uvrA gene (SO4030; GenBank accession no. NP_719560) from S. oneidensis MR-1 was amplified from 50 ng of genomic DNA using primers uvrA NdeI 5′ and uvrA BamHI 3′ (Table 1) and cloned into pJJK20 (25), creating plasmid pXQ01 (Table 1). A 3.6-kb SphI and BamHI fragment from pXQ01 containing the 0.75-kb umuDC promoter from E. coli and the 2.85-kb uvrA gene from MR-1 was cloned into pJB321, creating pXQ03 (Table 1). pXQ03 was transferred from E. coli DH10B to P. aeruginosa UA11079 by triparental mating as described by Kim and Sundin (25). The survival after UVC exposure was assayed as described above.
RESULTS
UVC sensitivity in Shewanella strains.
The sensitivities to UVC of five Shewanella strains from different natural habitats were examined. E. coli strain K-12 was used as the control (Fig. 1). Both S. algae and S. oneidensis strain MR-4 were more tolerant to UVC radiation, whereas Shewanella putrefaciens 200 and S. oneidensis strains DLM-7 and MR-1 were more sensitive to UVC radiation. To compare the degrees of resistance of the five strains to UVC treatments, we calculated the D37 and D10 values from the regression line of the exponential slope of each survival curve (Table 2). S. algae showed the highest UVC resistance, with a D37 of 4.0 J m−2 and a D10 of 8.2 J m−2, which was about 74.1 and 79.6% of that for E. coli K-12. S. oneidensis strain MR-1 showed the highest UVC sensitivity, with a D37 of 0.3 J m−2 and a D10 of 1.7 J m−2, which was about 5.6 and 16.5% of E. coli K-12 values (Table 2). The UVC resistance and sensitivity within the Shewanella genus correlated well with the radiation exposure in the habitat from which the organisms were isolated. Both MR-1 and DLM-7, isolated from lake sediment, and S. putrefaciens 200, isolated from a crude oil pipeline, were from habitats with limited solar radiation exposure, whereas S. algae, isolated from the surface of a red alga, and S. oneidensis MR-4, isolated from the top 5 m of the Black Sea, were from habitats with more solar radiation exposure (Table 1).
FIG. 1.
Survival of E. coli K-12 (▪), S. algae (•), S. putrefaciens 200 (□), and S. oneidensis strain MR-4 (▴), strain DLM-7 (ο), and strain MR-1 (Δ) after UVC irradiation. Plates used for measuring CFU were LB for E. coli K-12, marine agar for S. algae, and TSA for others. Each datum represents the mean (± the standard error of the mean) from three replicates.
TABLE 2.
D valuesa (J m−2) and slopes of survival curves from various bacterial strains
| Strain | Slope | R2 | D37 (%) | D10 (%) |
|---|---|---|---|---|
| E. coli K-12 | −0.272 ± 0.037 | 0.964 ± 0.013 | 5.4 ± 0.5 (100) | 10.3 ± 1.1 (100) |
| S. algae | −0.311 ± 0.031 | 0.989 ± 0.005 | 4.0 ± 0.5 (74.1) | 8.2 ± 0.9 (79.6) |
| S. oneidensis MR-4 | −0.330 ± 0.020 | 0.990 ± 0.007 | 2.9 ± 0.8 (53.7) | 6.8 ± 0.9 (66.0) |
| S. putrefaciens 200 | −0.690 ± 0.014 | 0.991 ± 0.004 | 1.9 ± 0.1 (35.2) | 3.8 ± 0.1 (36.9) |
| S. oneidensis DLM7 | −0.879 ± 0.044 | 0.994 ± 0.005 | 1.0 ± 0.2 (18.5) | 2.5 ± 0.2 (24.3) |
| S. oneidensis MR-1 | −0.937 ± 0.028 | 0.962 ± 0.022 | 0.3 ± 0.2 (5.6) | 1.7 ± 0.2 (16.5) |
D values are the radiation dose that reduced a cell population to a specified percentage of the original number of the cells (33). D37 is the radiation does required to inactivate 63% of a bacterial population. D10 is the radiation dose which inactivated 90% of the bacterial population. The D values were calculated from the regression line of the exponential slope of the survival curve.
UVA sensitivity in S. oneidensis MR-1.
Sensitivity to UVA has been reported to depend greatly on the physiological conditions of the cell. Exponential cells are more sensitive to near-UV damage than stationary cells due to their active DNA replication (6), while the stationary phase triggers numerous protective pathways as well as enzymatic activities expected to confer some degree of UVA resistance to cells (6, 7, 32). Since UVA induces oxidative damage to cells, the survival rate is greatly dependent on the medium used for recovery after irradiation. MR-1 survived much better on tryptic soy agar (TSA) plates than LB plates for both exponential-phase and stationary-phase cells (Fig. 2). No significant difference in UVA sensitivity was observed between exponential cells and stationary cells at lower UVA doses. At high UVA dose, exponential cells were slightly more resistant to UVA. The difference in survival rate on LB plates was more than 10-fold at the dose of 30 kJ m−2 (Fig. 2).
FIG. 2.
Survival of S. oneidensis MR-1 after UVA irradiation. Both exponential- and stationary-phase MR-1 cells grown in Davis medium were irradiated with various UVA doses and plated on both LB and TSA plates to measure CFU. ▴, exponential phase, LB plates; Δ, exponential phase, TSA plates; ▪, stationary phase, LB plates; □, stationary phase, TSA plates. Each datum represents the mean (± the standard error of the mean) from at least three replicates.
Contribution of photoreactivation to survival of S. oneidensis strain MR-1 after UVR exposure.
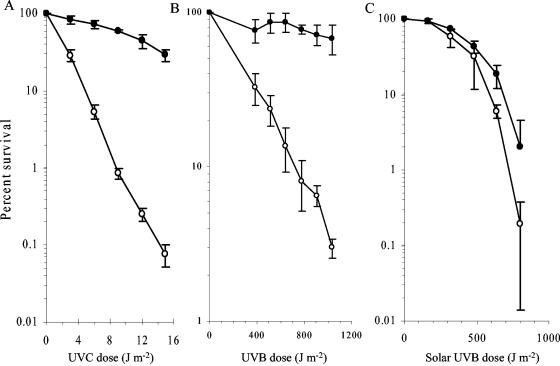
Annotated photolyase (512 amino acids) in S. oneidensis MR-1 shares 44% identity to that of E. coli K-12 (472 amino acids). The amino terminus contains the conserved domain for binding light harvesting cofactor, and the carboxyl terminus contains the conserved FAD binding domain of DNA photolyase. The tryptophans at enzyme active sites (W306 in E. coli K-12) and the one involved in substrate Pyr-Pyr-specific binding (W277 in E. coli K-12) are conserved in the photolyase of MR-1 (W342 and W312 in MR-1, respectively), which may indicate a similar catalyzing mechanism with that of E. coli K-12 (26). Photoreactivation conferred a significantly increased survival rate to S. oneidensis MR-1 in both UVB- and UVC-irradiated cells: as much as 177- and 365-fold after irradiation at UVC doses of 12 and 15 J m−2 (Fig. 3A) and 11- to 23-fold after irradiation at UVB doses of 774 to 1,032 J m−2 (Fig. 3B). For solar light-irradiated cells, further incubation under visible light for 1 h increased the survival rate 3- and 10-fold at solar UVB doses of 640 and 800 J m−2 (Fig. 3C) compared to those plated in the dark immediately after treatments.
FIG. 3.
Survial of S. oneidensis MR-1 following photoreactivation (•) or in the dark (ο) after UVC (A), UVB (B), or solar light (C) irradiation. LB plates were used for measuring CFU. Each datum represents the mean (± the standard error of the mean) from three replicates.
MDR activity in S. oneidensis MR-1.
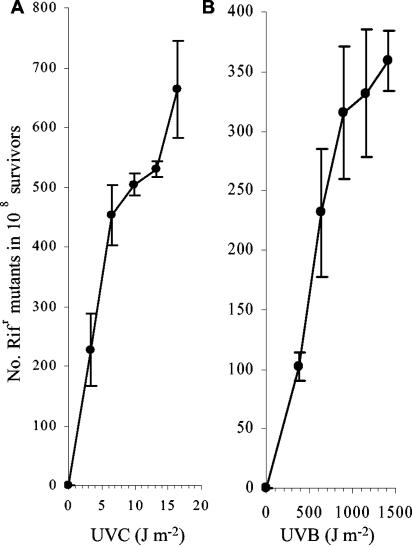
The umuDC operon in S. oneidensis MR-1 is located on the mega plasmid. The by-product of MDR, an increase in cellular mutation frequency, can be assayed by examining the increase in the occurrence of spontaneous mutants following irradiation. We examined the occurrence of Rifr mutants in both UVC- and UVB-treated samples (Fig. 4). The overall frequency was slightly higher in UVC-treated samples (Fig. 4A) than in UVB-treated samples (Fig. 4B) over the UV dose range used in this study. A mutation frequency as high as 6.6 × 10−6 was observed at 16.5 J of UVC m−2 (Fig. 4A). This result indicates that MDR-mediated translesion synthesis is functional in S. oneidensis MR-1.
FIG. 4.
Analysis of MDR in S. oneidensis MR-1 following UVC (A) and UVB (B) exposure. The number of spontaneous mutations conferring Rifr in the absence of UVR treatment has been subtracted. LB plates were used for measuring CFU. Each datum represents the mean (± the standard error of the mean) from three replicates.
Expression of NER component genes after UVR exposure.
Expression of NER component genes (uvrA, uvrB, and uvrD) after UVC, UVB, and UVA irradiation were examined using a microarray that contained about 95% of MR-1 ORFs. In contrast to the NER system of E. coli, we did not observe any induction with any of the three UVR treatments at any of the incubation times. The ratio of irradiated sample to control (unirradiated sample) was in the range of 0.9 to 1.2 (Fig. 5). To confirm that the uvr genes were truly transcribed, we designed gene-specific primers that targeted both amino-terminal and carboxyl-terminal fragments of uvrA, uvrB, and uvrD (Table 1). Positive amplicons were detected in all UVR-treated cells as well as in the controls (unirradiated samples) by using reverse transcription-PCR (data not shown). This result indicated that, unlike in E. coli K-12, the uvrA, uvrB, and uvrD genes of MR-1 were not damage inducible. In agreement with the above observation, we were unable to identify any E. coli-like SOS box near the translation region (−200 to +40) (8, 27) for any of the three genes examined, whereas putative LexA binding sites were found for recA, lexA, umuDC, and dinP, a homolog of umuC, with a relatively low heterology index (HI) value (Table 3).
FIG. 5.
Relative gene expression of NER component genes uvrA, uvrB, and uvrD at 5 min (filled with dots), 20 min (filled with diagonal stripes), and 60 min (filled with lines) after UVC (A), UVB (B), and UVA (C) irradiation. Ratios for UVR-irradiated samples and unirradiated samples were taken at the same time points. Each datum is the mean (± the standard error of the mean) of 8 to 12 data points from three biological replicates and two technical replicates.
TABLE 3.
Examples of putative E. coli-like SOS boxes in S. oneidensis MR-1
| Protein (gene) | Putative SOS boxc | HIa | Relative positionb |
|---|---|---|---|
| LexA repressor (lexA) | TACTGTATATACTAACAGTA | 1.09 | −46 |
| LexA repressor (lexA) | AACTGTATAGAAAAACAGGA | 6.37 | −27 |
| RecA protein (recA) | TACTGTATGATTGTACAGTA | 4.05 | −127 |
| UmuD protein (umuD) | AACTGTATATTTATACAGTT | 5.20 | −32 |
| DNA damage-inducible protein P (dinP) | AACTGTTTTTATATACAGTA | 3.59 | −45 |
HI indicates the affinity of LexA to the SOS box (27). A low HI indicates a strong binding of the LexA to the SOS box.
Distance to a putative translation start condon.
Boldface indicates the conserved sites of the E. coli SOS box.
Functional analysis UvrA of S. oneidensis MR-1.
To confirm that the NER system of MR-1 is truly functional, we attempted to complement P. aeruginosa UA11079 (uvrA deficient) (Table 1) with uvrA of MR-1. Since we were not sure if the promoter of uvrA from MR-1 was functional in P. aeruginosa, we used the umuDC promoter from E. coli, which has been demonstrated to be functional in P. aeruginosa (24), for the expression of uvrA from MR-1. Complementation increased the UVC survival of the mutant by more than 3 orders of magnitude, but not to the level of the wild-type PAO1 strain (Fig. 6). The D37 (0.43 J m−2) and D10 (2.90 J m−2) values of the complemented strain were about 11.5 and 42.8% of that for PAO1 (D37, 3.73 J m−2; D10, 6.78 J m−2). Nonetheless, this result demonstrates that UvrA from MR-1 is functional in repairing UVC-induced damage, although the efficiency is not as high as with UvrA from PAO1.
FIG. 6.
Survival of P. aeruginosa UA11079 (▴), P. aeruginosa UA11079 complemented with uvrA of MR-1(♦), and P. aeruginosa PAO1 (•) after UVC irradiation. LB plates were used for measuring CFU. Each datum represents the mean (± the standard error of the mean) from three replicates.
DISCUSSION
We evaluated the phenotypic responses important to all relevant wavelength groups of UVR and solar UVR in the environmentally relevant bacterium S. oneidensis MR-1. An analysis of the MR-1 genome (NC_004347 and NC_004349) indicated that this organism possesses genes that could encode major DNA repair systems, including NER and recombinational repair, and that MR-1 also encodes a photolyase enzyme and a plasmid-borne mutagenic DNA repair determinant. Regarding UVA survival, MR-1 contains several genes encoding proteins relevant to the removal of reactive oxygen species such as catalase (SO0725, SO4405, and SO1771.2), superoxide dismutase (SO2881), and proteins of the organic hydroperoxide resistance (Ohr) family (SO0976 and SO3409). The potential use in bioremediation, the availability of the genome sequence, and the phylogenetic relationship of S. oneidensis to other well-characterized organisms suggest that this strain is an effective model for physiological and genetic studies of UV and ionizing radiation effects on an environmental bacterium.
While the UVC resistance and sensitivity of isolates within the Shewanella genus correlated well with the radiation exposure in the habitat from which they were isolated, the uniform sensitivity of S. oneidensis MR-1 to UVA, UVB, UVC, and solar UVR may or may not be a result of lack of UVR exposure. For example, bacteria regarded as tolerant or resistant to UVR were recovered from solar radiation-exposed habitats, including aquatic habitats and the plant phyllosphere (16, 20, 47), but little or no correlation was observed between UVR resistance and the natural levels of solar radiation exposure (12). Great variability in sensitivity to UVR was observed among marine bacterial isolates (2, 20). UVR-tolerant organisms with active photoreactivation mechanisms were prevalent among deep subsurface bacteria which had been screened from solar radiation for more than one million years (1). Thus, the habitat of isolation is not always an indicator of the UVR sensitivity of an organism. The uniform UVR sensitivity of MR-1, however, could not be explained by gene content either. MR-1 possesses most of the important repair pathways and determinants compared to phylogenetically related E. coli and has even more DNA repair genes than Deinococcus radiodurans, a bacterium extremely resistant to radiation (http://www.usuhs.mil/pat/deinococcus/FrontPage_DR_Web_work/Pages/DNA_repair/dna_repair_pathways.htm). However, the resistance to UVC of D. radiodurans is more than 3 orders of magnitude higher than that of MR-1 (12, 33).
The sensitivity to DNA-damaging UVC and UVB wavelengths in MR-1 can be offset by photoreactivation. The contribution of photoreactivation to MR-1 survival is very similar to that observed in other bacteria (25, 56). Photoreactivation makes a larger contribution to survival following irradiation with UVB or UVC wavelengths in vitro compared to the increase in survival following exposure to solar UVR. This result is probably due to the additional lethal effects of UVA wavelengths present in solar UVR, but it also has implications for physiological studies aimed at determining the ecological importance of photoreactivation in microbial communities (19). The dramatic difference in survival rate between organisms on LB and TSA plates following UVA exposure indicates potential membrane damage caused by UVA (17, 53). We also observed an additional decrease in survival rate when the irradiated MR-1 cells were plated on old LB plates (relatively dry). Sensitivity to UVA radiation in MR-1 was also dose dependent. At lower doses, the survival of exponential-phase and stationary-phase cells was similar, whereas exponential-phase cells were more resistant to higher radiation doses. This result agrees with findings in studies using 4-thiouridine mutants, which showed that mutants possessing more DNA replication forks (similar to exponential growth cells) are more resistant to high UVA doses than are wild-type bacteria (18). This could explain the dramatic change in UVA-induced photodamage at lower and higher UVA doses.
The plasmid-encoded MDR determinant umuDCSo contributed to UVR-induced mutability in MR-1, but the contribution of this determinant to UVR survival is unclear. Although most MDR determinants transiently increase the mutation rate of cells following UVC irradiation, the contribution of these determinants to increased cell survival is only apparent in some cases. For example, the MDR determinant rulAB confers tolerance to UVC wavelengths in Pseudomonas syringae (48), but deletion of MDR determinants such as umuDC and samAB from E. coli and Salmonella enterica serovar Typhimurium, respectively, does not affect their cellular UVC sensitivity (36, 54).
Our investigation on the sensitivity to DNA-damaging UVC and UVB wavelengths centered on the NER system of MR-1. This system is probably functional, as organisms harboring mutations in NER component genes (e.g., uvrA, uvrB) are typically exquisitely sensitive to UVC (41). Indeed, we confirmed the functionality of UvrA through the ability of this protein to complement the UvrA defect in P. aeruginosa UA11079 (Fig. 6). Loss of the damage inducibility of the NER system in MR-1 may contribute to the UVR sensitivity of this organism. For example, in E. coli expression of the uvrA, uvrB, and uvrD genes is significantly induced following DNA damage. However, in P. aeruginosa, an organism that is more sensitive to UVC than E. coli, both uvrA and uvrB are not DNA damage inducible, although this bacterium possesses an SOS-like system (38, 39). In MR-1, we observed strong SOS induction following UVB or UVC exposure, which included increases in transcript levels of lexA and recA as well as the umuDC operon (unpublished data). The gene expression levels of uvrA, uvrB, and uvrD, however, remained constant following DNA damage.
We next examined the regulation of NER component genes among five organisms that are phylogenetically related to S. oneidensis, including E. coli, Haemophilus influenzae, Pasteurella multocida, P. aeruginosa, and Vibrio cholerae (15). Since LexA and RecA are highly conserved among these bacteria, it is reasonable to hypothesize that a similar mechanism is present in the regulation of the SOS response. Using the E. coli SOS box consensus sequence and three SOS box searching patterns (8), we searched for a putative SOS box near a putative translation start codon (−200 to +40) of either a uvrA, uvrB, or uvrD gene in five organisms. As expected, an SOS box was identified for all three genes in E. coli (Table 4). In V. cholerae, a strong putative SOS box was identified upstream of the uvrA gene, but no putative SOS box was identified upstream of both uvrB and uvrD (Table 4). Relatively strong putative SOS boxes were identified upstream of both uvrA and uvrD but not uvrB in both H. influenzae and P. multocida (Table 4). Similar to MR-1, no putative SOS box was identified upstream of uvrA, uvrB, or uvrD in P. aeruginosa PAO1 (Table 4). In agreement with their UVC sensitivity, both S. oneidensis MR-1 and P. aeruginosa PAO1 lost the damage inducibility of the NER system. Alternatively, the functional efficiency of the UvrABCD complex in NER may be diminished in both P. aeruginosa PAO1 and S. oneidensis MR-1. Further work is needed to understand the evolution and maintenance of NER in these organisms.
TABLE 4.
E. coli-like SOS boxes in species that are phylogenetically close to S. oneidensis MR-1
| Species |
E. coli-like SOS box sequence (HI value)f
|
||
|---|---|---|---|
| uvrA | uvrB | uvrD | |
| V. choleraea | AACTGTTTTTTTATCCAGTA (2.7) | ||
| E. colib | TACTGTATATTCATTCAGGT (9.6) | AACTGTTTTTTTATCCAGTA (2.7) | ATCTGTATATATACCCAGCT (9.4) |
| P. aeruginosac | |||
| P. multocidad | AACTGGATATTTGCACAGTT (7.8) | TACTGTATAAAAAAACAGTT (4.1) | |
| H. influenzaee | AACTGGATATTTGCACAGAT (10.7) | AACTGTAAATTTAAACAGAT (7.0) | |
V. cholerae O1 biovar eltor str. N16961 (genome accession number NC_002505.1). The SOS box of uvrA is from 420543 to 420562 (complement).
E. coli K-12 (genome accession number NC_000913.1). The SOS box of uvrA, uvrB, and uvrD is from 4271534 to 4271553 (complement), 812655 to 812674, and 3995520 to 3995539, respectively.
P. aeruginosa PA01 (genome accession number NC_002516.1).
P. multocida (genome accession number NC_002663.1). The SOS box of uvrA and uvrD is from 2187327 to 2187346 and 480407 to 480426, respectively.
H. influenzae Rd (genome accession number NC_000907.1). The SOS box is from 282331 to 282350 for uvrA and from 1255918 to 1255937 for uvrD.
HI indicates the affinity of LexA to the SOS box (27). A low HI indicates a strong binding of the LexA to the SOS box.
It is very well known that UVR can induce prophage into the lytic cycle. Kidambi et al. reported that UVB can activate D3 prophage in P. aeruginosa in a RecA-dependent manner (23). The novel Shewanella phage lambdaSo shares syntenic regions with P. aeruginosa D3 and enterobacteria HK022 (15). Whether or not activation of prophage(s) on the MR-1 genome contributes to its high sensitivity to UVR needs to be investigated.
Despite possessing the relevant repertoire of oxidative damage repair genes, the results of our study indicate that S. oneidensis MR-1 is one of the most UVA-sensitive organisms known. Genome analysis showed that MR-1 has more c-type cytochromes than many organisms, including E. coli, V. cholerae, and P. aeruginosa (15). Cytochromes, along with flavins and quinones, are potential chromophores for UVA (5, 17, 53). Whether or not the high cytochrome content of MR-1 contributes to its high UVA sensitivity needs detailed investigation. As expected, MR-1 is also highly sensitive to ionizing radiation (M. Daly, personal communication). The radiation sensitivity of MR-1 may pose potential problems for environmental uses of this strain or its indigenous relatives in bioremediation of toxic metals or radionuclides, since a variety of DNA-damaging agents as well as ionizing radiation may be present at contaminated sites. Relatively little is known of the interrelationship of genetic systems and mechanisms involved in repairing cellular damage caused by UVR and ionizing radiation in organisms other than D. radiodurans. MR-1 is an excellent model to compare and understand the cellular function and regulation in response to various radiation stresses. This knowledge will contribute greatly to our fundamental understanding of the traits important in determining bacterial radiation resistance.
Acknowledgments
This work was supported by grant DE-FG-02-02ER63342 to Michigan State University from the Office of Biological and Environmental Research of the U.S. Department of Energy.
We also thank Jizhong Zhou for stimulating our work on strain MR-1, Liyou Wu for providing DNA microarrays, Alison Murray and Veronica Grüntzig for providing related bacterial strains, and members of the Shewanella Federation for helpful discussions on the biology and genomics of these organisms.
REFERENCES
- 1.Arrage, A. A., T. J. Phelps, R. E. Benoit, and D. C. White. 1993. Survival of subsurface microorganisms exposed to UV radiation and hydrogen peroxide. Appl. Environ. Microbiol. 59:3545-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrieta, J. M., M. G. Weinbauer, and G. J. Herndl. 2000. Interspecific variability in sensitivity to UV radiation and subsequent recovery in selected isolates of marine bacteria. Appl. Environ. Microbiol. 66:1468-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courcelle, J., A. Khodursky, B. Peter, P. O. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenstark, A. 1987. Mutagenic and lethal effects of near-ultraviolet radiation (290-400 nm) on bacteria and phage. Environ. Mol. Mutagen. 10:317-337. [DOI] [PubMed] [Google Scholar]
- 6.Eisenstark, A. 1989. Bacterial genes involved in response to near-ultraviolet radiation. Adv. Genet. 26:99-147. [DOI] [PubMed] [Google Scholar]
- 7.Favre, A., E. Hajnsdorf, K. Thiam, and A. Caldeira de Araujo. 1985. Mutagenesis and growth delay induced in Escherichia coli by near-ultraviolet radiation. Biochimie 67:335-342. [DOI] [PubMed] [Google Scholar]
- 8.Fernández de Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 79:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogliano, M., and P. F. Schendel. 1981. Evidence for the inducibility of the uvrB operon. Nature (London) 289:196-198. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 12.Gascon, J., A. Qubina, A. Perex-Lezaun, and J. Urmeneta. 1995. Sensitivity of selected bacterial species to UV radiation. Curr. Microbiol. 30:77-182. [DOI] [PubMed] [Google Scholar]
- 13.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. Earlehughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-562. [DOI] [PubMed] [Google Scholar]
- 15.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, R. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, J. L., and G. W. Sundin. 2001. Effect of solar UV-B radiation on a phyllosphere bacterial community. Appl. Environ. Microbiol. 67:5488-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagger, J. 1983. Physiological effects of near-ultraviolet radiation on bacteria. Photochem. Photobiol. Rev. 7:1-75. [Google Scholar]
- 18.Jagger, J. 1985. Solar UV actions on living cells. Praeger, New York, N.Y.
- 19.Jeffrey, W. H., P. Aas, M. M. Lyons, R. B. Coffin, R. J. Pledger, and D. L. Mitchell. 1996. Ambient solar radiation-induced photodamage in marine bacterioplankton. Photochem. Photobiol. 64:419-427. [Google Scholar]
- 20.Joux, F., W. H. Jeffrey, P. Lebaron, and D. L. Mitchell. 1999. Marine bacterial isolates display diverse responses to UV-B radiation. Appl. Environ. Microbiol. 65:3820-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenyon, C. J., and G. C. Walker. 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. USA 77:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenyon, C. J., and G. C. Walker. 1981. Expression of E. coli uvrA gene is inducible. Nature (London) 289:808-810. [DOI] [PubMed] [Google Scholar]
- 23.Kidambi, S. P., M. G. Booth, T. A. Kokjohn, and R. V. Miller. 1996. recA-dependence of the response of Pseudomonas aeruginosa to UVA and UVB irradiation. Microbiology 142:1033-1040. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J.-J., and G. W. Sundin. 2000. Regulation of the rulAB mutagenic DNA repair operon of Pseudomonas syringae by UVB (290 to 320 nanometers) radiation and analysis of rulAB-mediated mutability in vitro and in planta. J. Bacteriol. 182:6137-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J.-J., and G. W. Sundin. 2001. Construction and analysis of photolyase mutants of Pseudomonas aeruginosa and Pseudomonas syringae: contribution of photoreactivation, nucleotide excision repair, and mutagenic DNA repair to cell survival and mutability following exposure to UVB radiation. Appl. Environ. Microbiol. 67:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S.-T., and A. Sancar. 1993. Photochemistry, photophysics, and mechanism of pyrimidine dimmer repair by DNA photolyase. Photochem. Photobiol. 57:895-904. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, L. K., G. R. Harlow, L. A. Gregg-Jolly, and D. W. Mount. 1994. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 241:507-523. [DOI] [PubMed] [Google Scholar]
- 28.Lin, J.-R., and A. Sancar. 1992. (A)BC excinuclease: the Escherichia coli nucleotide excision repair enzyme. Mol. Microbiol. 6:2219-2224. [DOI] [PubMed] [Google Scholar]
- 29.Little, J. W., and D. W. Mount. 1982. The SOS regulatory system of Escherichia coli. Cell 29:11-22. [DOI] [PubMed] [Google Scholar]
- 30.Liu, C., Y. A. Gorby, J. M. Zachara, J. K. Fredrickson, and C. F. Brown. 2002. Reduction kinetics of Fe(III), Co(III), U(VI), Cr(VI), and Tc(VII) in cultures of dissimilatory metal-reducing bacteria. Biotechnol. Bioeng. 80:637-649. [DOI] [PubMed] [Google Scholar]
- 31.Middleton, S. S., R. B. Latmani, M. R. Mackey, M. H. Ellisman, B. M. Tebo, and C. S. Criddle. 2003. Cometabolism of Cr(VI) by Shewanella oneidensis MR-1 produces cell-associated reduced chromium and inhibits growth. Biotechnol. Bioeng. 83:627-637. [DOI] [PubMed] [Google Scholar]
- 32.Miller, C. D., W. S. Mortensen, G. U. L. Braga, and A. J. Anderson. 2001. The rpoS gene in Pseudomonas syringae is important in surviving exposure to the near-UV in sunlight. Curr. Microbiol. 43:374-377. [DOI] [PubMed] [Google Scholar]
- 33.Moseley, B. E. B. 1983. Photobiology and radiobiology of Micrococcus (Deinococcus) radiodurans. Photochem. Photobiol. Rev. 7:223-274. [Google Scholar]
- 34.Murray, A. E., D. Lies, G. Li, K. Nealson, J. Zhou, and J. M. Tiedje. 2001. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc. Natl. Acad. Sci. USA 98:9853-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 36.Nohmi, T., A. Hakura, Y. Nakai, M. Watanabe, S. Y. Murayama, and T. Sofuni. 1991. Salmonella typhimurium has two homologous but different umuDC operons: cloning of a new umuDC-like operon (samAB) present in a 60-megadalton cryptic plasmid of S. typhimurium. J. Bacteriol. 173:1051-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeifer, G. P. 1997. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem. Photobiol. 65:270-283. [DOI] [PubMed] [Google Scholar]
- 38.Rivera, E., L. Vila, and J. Barbe. 1996. The uvrB gene of Pseudomonas aeruginosa is not DNA damage inducible. J. Bacteriol. 178:5550-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera, E., L. Vila, and J. Barbe. 1997. Expression of the Pseudomonas aeruginosa uvrA gene is constitutive. Mutat. Res. 377:149-155. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sancar, A., and M. Tang. 1993. Nucleotide excision repair. Photochem. Photobiol. 57:905-921. [DOI] [PubMed] [Google Scholar]
- 42.Sancar, A. 1994. Mechanisms of DNA excision repair. Science 266:1954-1956. [DOI] [PubMed] [Google Scholar]
- 43.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 44.Schroeder, R. G., L. M. Peterson, and R. D. Fleischmann. 2002. Improved quantitation and reproducibility in Mycobacterium tuberculosis DNA microarrays. J. Mol. Microbiol. Biotechnol. 4:123-126. [PubMed] [Google Scholar]
- 45.Siegel, E. C. 1983. The Escherichia coli uvrD is inducible by DNA damage. Mol. Gen. Genet. 191:397-400. [DOI] [PubMed] [Google Scholar]
- 46.Smith, B. T., and G. C. Walker. 1998. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics 148:1599-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundin, G. W., and J. L. Jacobs. 1999. Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.). Microb. Ecol. 38:27-38. [DOI] [PubMed] [Google Scholar]
- 48.Sundin, G. W., S. P. Kidambi, M. S. Ullrich, and C. L. Bender. 1996. Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulAB genes. Gene 177:77-81. [DOI] [PubMed] [Google Scholar]
- 49.Sundin, G. W., and J. Murillo. 1999. Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290-320 nm) radiation and distribution of rulAB among P. syringae pathovars. Environ. Microbiol. 1:75-87. [DOI] [PubMed] [Google Scholar]
- 50.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insight into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 51.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. V. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 53.Webb, R. B. 1977. Lethal and mutagenic effects of near-ultraviolet radiation. Photochem. Photobiol. Rev. 2:169-261. [Google Scholar]
- 54.Woodgate, R. 1992. Construction of umuDC operon substitution mutation in Escherichia coli. Mutat. Res. 281:221-225. [DOI] [PubMed] [Google Scholar]
- 55.Woodgate, R., and S. G. Sedgwick. 1992. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol. Microbiol. 6:2213-2218. [DOI] [PubMed] [Google Scholar]
- 56.Yasui, A., and A. P. M. Eker. 1998. DNA photolyases, p. 9-32. In J. A. Nickoloff and M. F. Hoekstra (ed.), DNA damage and repair, vol. 2. DNA repair in higher eukaryotes. Humana Press, Inc., Totowa, N.J.