Abstract
Thirty-six isolates of carbapenem-resistant Pseudomonas aeruginosa were studied. Pulsed-field gel electrophoresis revealed the presence of two clones. One clone carried a blaIMP-1 gene identical to that first described in Japan. The other clone carried a blaIMP-1 variant containing four silent mutations. One isolate with a unique pulsed-field gel electrophoresis pattern contained blaIMP-7.
Gram-negative bacilli which produce Ambler class B metallo-β-lactamases may be resistant to multiple antimicrobial agents, including carbapenems. The most common acquired class B enzymes belong to the IMP family, of which at least 13 have so far been described.
We have previously described blaIMP-1 in a single clinical isolate of Klebsiella pneumoniae in Singapore (T. H. Koh, L.-H. Sng, G. S. Babini, N. Woodford, D. M. Livermore, and L. M. C. Hall, Letter, Antimicrob. Agents Chemother. 45:1939-1940, 2001). Because blaIMP-1 has been found in Pseudomonas aeruginosa isolated in Japan, we undertook a study to see if metallo-β-lactamase genes could also be found in this species isolated in Singapore.
Between December 1999 and February 2001, we collected 96 nonduplicate isolates of carbapenem-resistant P. aeruginosa in our laboratory. Thirty-six isolates showed metallo-β-lactamase activity by the disk diffusion test described by Arakawa et al. (1). Twenty-one isolates were collected from patients in hospital A, which is a 1,400-bed tertiary care hospital. Fourteen isolates were collected from patients in hospital B, which is a 200-bed community hospital specializing in rehabilitation and geriatrics, and one isolate was from a patient in hospital C, which is another 200-bed community geriatric hospital. The isolates were identified on the basis of oxidase positivity, pigmentation, and API20E (bioMérieux, Marcy-l'Etoile, France) or Microbact 24E (Medvet Diagnostics, Thebarton, South Australia). Isoelectric focusing of crude cell extracts revealed that each isolate produced an enzyme with an isoelectric point of approximately 8 to 9 in keeping with an IMP-type metallo-β-lactamase.
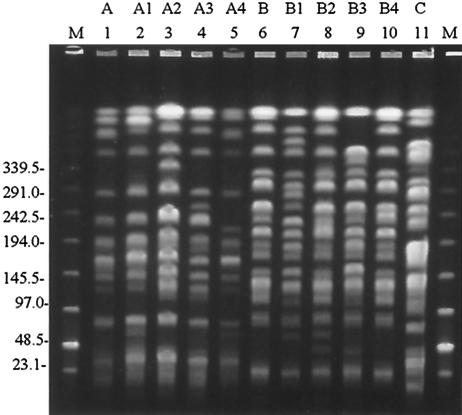
Pulsed-field gel electrophoresis (PFGE) was performed as previously described, using SpeI restriction endonuclease (6) Fig. 1. There were two major clones, designated A and B. Within each clone, there were a number of subclones (indicated by numbers) which differed from the main clone by 1 to 3 band positions.
FIG. 1.
Representative PFGE patterns of carbapenem-resistant P. aeruginosa. Lanes: M, low-range PFGE marker (size are given in kilobases); 1, strain DU20080/00; 2, strain DU16517/00; 3, strain DU31106/00; 4, strain DU34565/00; 5, strain DU8622/00; 6, strain DU9519/00; 7, strain DM10075/00; 8, strain DU40799/00; 9, strain DU6061/00; 10, strain DU14610/00; 11, strain DM727/00. The PFGE patterns are indicated by the letters above the lane numbers.
All isolates were positive for blaIMP by PCR using the primers described by Senda et al. (9) except DM727/00, which had a unique PFGE pattern. The entire blaIMP gene was amplified and sequenced from six strains belonging to clone A (DU8622/00, DM14158/00, DU16591/00, DU22812/00, DU11013/00, and DM11376/00) and five strains belonging to clone B (DU10114/00, DU32495/00, DU40799/00, DU45969/00, and DM10075/00), using the primers and conditions described by Yan et al. (11). In addition, PCR using the primers and conditions described by Lombardi et al. (7) was performed on strains DU8622/00, DM14158/00, DU10114/00, and DU32495/00 to determine if the blaIMP gene was sited on an integron.
All isolates showing the A pattern had sequences identical to that of the blaIMP-1 gene first reported in Japan and also found in K. pneumoniae (Koh et al., letter) and Pseudomonas putida in Singapore (4). All isolates showing the B pattern contained sequences for blaIMP-1 with four silent mutations at nucleotide positions 189 (C to T), 273 (C to T), 496 (T to C), and 702 (G to A), as described for Pseudomonas fluorescens from Singapore (4). The immediate flanking regions of blaIMP-1 in strains DU8622/00, DM14158/00, DU10114/00, and DU32495/00 were identical to that of an integron sequence containing blaIMP-1 in GenBank (accession number AB104852.1), showing that these genetic elements are probably important in the spread of blaIMP-1.
The blaIMP-7 allele, which codes for a metallo-β-lactamase with >86% homology with other IMP enzymes, has been found in neighboring Malaysia (3) and is not detected by the usual blaIMP primers. We therefore designed a custom primer, IMP-7ASF (5′-ATG AAA AAG TTA TCA GTA TTC-3′), which we used in combination with a 3′ integron primer (5) to amplify and sequence blaIMP-7 from isolate DM727/00. The 3′ region flanking this gene contained integron sequences (data not shown). We were unable to sequence the 5′ flanking region, however. The blaIMP-7 allele has also been found in a nosocomial outbreak of carbapenem-resistant P. aeruginosa in Canada (2).
The antimicrobial susceptibilities to piperacillin-tazobactam, imipenem, aztreonam, ceftazidime, and cefepime (BBL, Becton Dickinson and Company, Cockeysville, Md.) were determined by the disk diffusion method according to NCCLS guidelines (8). Since class B metallo-β-lactamases do not hydrolyze aztreonam, it was not surprising that a number of isolates appeared susceptible to this monobactam (Table 1). Interestingly, most isolates appeared susceptible to piperacillin-tazobactam even though IMP is known to hydrolyze piperacillin and tazobactam is not expected to inhibit metallo-β-lactamases. Susceptibility to piperacillin-tazobactam was also observed in blaIMP-7-positive P. aeruginosa from Canada (2). Therefore, apparent susceptibility to piperacillin-tazobactam does not exclude the possibility that an organism may produce IMP. All isolates were resistant to ceftazidime and cefepime.
TABLE 1.
Characteristics of carbapenem-resistant P. aeruginosa
| Strain no. | Source | Date isolatedb | Hospital/ward | Resistance phenotypea
|
PFGE pattern | ||
|---|---|---|---|---|---|---|---|
| TZP | IPM | ATM | |||||
| DU6061/00 | Urine | 2/21/2000 | B/1 | S | R | S | B3 |
| DU1900/00 | Fluid | 1/30/2000 | B/2 | S | R | I | A1 |
| DU8622/00 | Urine | 3/13/2000 | B/2 | S | R | I | A4 |
| DU9449/00 | Urine | 3/20/2000 | B/2 | S | R | S | A1 |
| DU15578/00 | Urine | 5/8/2000 | B/2 | S | R | I | A |
| DU16517/00 | Urine | 5/15/2000 | B/2 | R | R | I | A1 |
| DU16591/00 | Urine | 5/15/2000 | B/2 | S | R | I | A1 |
| DU20080/00 | Urine | 6/12/2000 | B/2 | S | R | I | A |
| DU22390/00 | Urine | 6/29/2000 | B/2 | S | R | I | A4 |
| DU22812/00 | Urine | 7/3/2000 | B/2 | S | R | I | A1 |
| DU29551/00 | Urine | 8/22/2000 | B/2 | S | R | I | A1 |
| DU32205/00 | Urine | 9/11/2000 | B/2 | S | R | S | A1 |
| DU35095/00 | Urine | 10/3/2000 | B/2 | S | R | S | A1 |
| DU36277/00 | Urine | 10/11/2000 | B/2 | S | R | I | A1 |
| DM10075/00 | Fluid | 6/3/2000 | A/42 | S | R | S | B1 |
| DU19510/00 | Urine | 6/6/2000 | A/42 | S | R | S | B |
| DU10114/00 | Urine | 3/24/2000 | A/45 | S | R | S | B |
| DM14158/00 | Catheter | 8/3/2000 | A/45 | S | R | R | A |
| DM14511/00 | Catheter | 8/8/2000 | A/45 | S | R | R | A |
| DM14668/00 | Wound | 8/11/2000 | A/45 | S | R | R | A |
| DM11013/00 | Wound | 6/19/2000 | A/46 | R | R | R | A |
| DM11376/00 | Wound | 6/25/2000 | A/46 | R | R | R | A3 |
| DU14610/00 | Urine | 4/29/2000 | A/47 | S | R | R | B4 |
| DM727/00 | Catheter | 1/12/2000 | A/48 | S | R | I | C |
| DU32495/00 | Urine | 9/13/2000 | A/48 | S | R | S | B2 |
| DU2658/00 | Urine | 1/23/2000 | A/54 | S | R | I | A |
| DU22185/00 | Urine | 6/28/2000 | A/63 | S | R | R | A3 |
| DU7670/00 | Urine | 3/4/2000 | A/73 | S | R | S | B |
| DU9519/00 | Urine | 3/20/2000 | A/73 | S | R | S | B |
| DU14720/00 | Urine | 4/29/2000 | A/73 | S | R | S | B |
| DU17318/00 | Urine | 5/22/2000 | A/73 | S | R | S | B4 |
| DU31106/00 | Urine | 9/2/2000 | A/74 | R | R | I | A2 |
| DU33961/00 | Urine | 9/25/2000 | A/74 | R | R | I | A |
| DM5599/00 | Wound | 3/25/2000 | A/75 | S | R | S | A1 |
| DU40799/00 | Urine | 11/17/2000 | A/76 | R | R | S | B2 |
| DU45969/00 | Urine | 12/30/2000 | C | S | R | S | B |
Abbreviations: TZP, piperacillin-tazobactam; IPM, imipenem; ATM, aztreonam; S, susceptible; I, intermediate; R, resistant.
Month/day/year.
In hospital A, 2,094 nonduplicate P. aeruginosa strains were isolated during the study period. Metallo-β-lactamase producers therefore represent 1.7% of all P. aeruginosa isolates in this hospital. This compares with 1.3% of P. aeruginosa in a study from Japan in 1996-1997 (H. Kurokawa, T. Yagi, N. Shibata, K. Shibayama, and Y. Arakawa, Letter, Lancet 354:955, 1999). In hospital B, the proportion of metallo-β-lactamase producers was 8%. However the data may not be representative, since only 174 nonduplicate P. aeruginosa strains were isolated from patients in this hospital during the study period.
In a large survey of carbapenem-resistant P. aeruginosa in 17 general hospitals in Japan, Senda et al. found different genetic backgrounds for 15 blaIMP-1-positive strains, although considerable similarity was observed with strains isolated from the same hospital (10). In our study, one subclone (A1) was prevalent in hospital B. In hospital A, two clones, each consisting of several subclones, coexisted, and these were distributed throughout the hospital and occurred in small, temporally and geographically related outbreaks. The difference in outbreak patterns may reflect differences in patient type, infection control practices, and antimicrobial pressure.
This study shows that the blaIMP determinant is not confined to the large tertiary hospital and can also be found in community hospitals. We were unable to confirm interhospital spread with this sample, but this is possible, since transfer of patients between different hospitals in Singapore is a common occurrence.
Nucleotide sequence accession numbers.
The following blaIMP sequences were submitted to GenBank: P. aeruginosa DU40799/00 (accession number AY168635), P. aeruginosa DM727/00 (accession number AY625685), P. aeruginosa DU10114/00 (accession number AY625686), P. aeruginosa DU32495/00 (accession number AY625687), P. aeruginosa DM14158/00 (accession number AY625688), and P. aeruginosa DU8622/00 (accession number AY625689).
Acknowledgments
We thank Ong Lan Huay and Tan Mee Lee, Department of Pathology, Singapore General Hospital, for their assistance.
This work was supported by a grant from the Department of Clinical Research, Singapore General Hospital.
REFERENCES
- 1.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-beta-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho, S. E., G. Subramaniam, S. Palasubramaniam, and P. Navaratnam. 2002. Carbapenem-resistant Pseudomonas aeruginosa in Malaysia producing IMP-7 beta-lactamase. Antimicrob. Agents Chemother. 46:3286-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koh, T. H., G. C. Y. Wang, and L.-H. Sng. 2004. IMP-1 and a novel metallo-β-lactamase, VIM-6, in fluorescent pseudomonads isolated in Singapore. Antimicrob. Agents Chemother. 48:2334-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling, M. L., and G. C. Wang. 2001. Epidemiological analysis of Salmonella enteritidis isolates in Singapore. J. Infect. 43:169-172. [DOI] [PubMed] [Google Scholar]
- 7.Lombardi, G., F. Luzzaro, J. D. Docquier, M. L. Riccio, M. Perilli, A. Coli, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2002. Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-beta-lactamase. J. Clin. Microbiol. 40:4051-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed. M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-beta-lactamase gene blaIMP in gram-negative rods resistant to broad-spectrum beta-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-beta-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum beta-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan, J. J., P. R. Hsueh, W. C. Ko, K. T. Luh, S. H. Tsai, H. M. Wu, and J. J. Wu. 2001. Metallo-beta-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]