Abstract
To characterize intracellular gram-negative bacteria associated with epitheliocystis in farmed Atlantic salmon (Salmo salar), gills with proliferative lesions were collected for histopathology, conventional transmission and immunoelectron microscopy, in situ hybridization, and DNA extraction during epitheliocystis outbreaks in Ireland and Norway in 1999 and 2000, respectively, and compared by ultrastructure and immunoreactivity to nonproliferative gills from Ireland archived in 1995. Genomic DNA from proliferative gills was used to amplify 16S ribosomal DNA (rDNA) for molecular phylogenetic analyses. Epitheliocystis inclusions from proliferative gills possessed variably elongate reticulate bodies, examples of binary fission, and vacuolated and nonvacuolated intermediate bodies, whereas inclusions in nonproliferative gills had typical chlamydial developmental stages plus distinctive head-and-tail cells. Immunogold processing using anti-chlamydial lipopolysaccharide antibody labeled reticulate bodies from proliferative and nonproliferative gills. 16S rDNA amplified directly from Irish (1999) and Norwegian (2000) gill samples demonstrated 99% nucleotide identity, and riboprobes transcribed from cloned near-full-length 16S rDNA amplicons from Norwegian gills hybridized with inclusions in proliferative lesions from Irish (1999) and Norwegian (2000) sections. A 1,487-bp consensus 16S rRNA gene sequence representing the chlamydia-like bacterium (CLB) from proliferative gills had the highest percent nucleotide identity with endosymbionts of Acanthamoeba spp. (order Chlamydiales). Molecular phylogenetic relationships inferred from 16S rRNA gene sequences using distance and parsimony indicated that the CLB from proliferative gills branched with members of the order Chlamydiales. “Candidatus Piscichlamydia salmonis” is proposed for the CLB associated with epitheliocystis from proliferative gills of Atlantic salmon, which exhibits developmental stages different from those identified in nonproliferative gills.
Epitheliocystis has been associated with heavy mortality and reduced growth of survivors in farmed Atlantic salmon (Salmo salar) (22). Ultrastructural studies of the epitheliocystis agent found in Atlantic salmon have revealed it to be an intracellular gram-negative coccoid bacterium with distinct developmental stages typical of bacteria of the order Chlamydiales (22). Epitheliocystis has been described in other salmonid hosts, e.g., juvenile steelhead trout (Oncorhynchus mykiss) (28) and cultured lake trout (Salvelinus namaycush) (3), as well as in a number of nonsalmonid species, including bluegill (Lepomis macrochirus) (16), striped bass (Morone saxatilis) (32), white perch (Morone americanus) (32), sea bream (Sparus aurata) (25), grey mullet (Liza ramada) (25), and cultured white sturgeon (Acipenser transmontanus) (14). Morphological studies of epitheliocystis agents in sea bream (S. aurata) have provided evidence for two distinct chlamydia-like developmental cycles associated with proliferative and nonproliferative host reactions (5). Although transmission electron microscopic examinations of intracellular inclusions have demonstrated that the agents of epitheliocystis in both salmonid and nonsalmonid hosts are gram-negative bacteria with developmental stages typical of members of the order Chlamydiales (5, 14, 22, 28, 32), the genetic relatedness of these bacteria has yet to be determined.
Sequence data from the rRNA operon have revised phylogenetic relationships between Chlamydia species and chlamydia-like bacteria (CLB) (7, 8, 9). Reclassifying chlamydial species on the basis of 16S rRNA gene sequence identity, 16S and 23S ribosomal DNA (rDNA) sequences, and phenotypic characterization is considered by some to be the best means of taxonomically categorizing chlamydiae (24). Based on this approach, species within the family Chlamydiaceae have 16S rRNA gene sequences that are >90% identical (26), whereas chlamydia-like bacteria, defined as obligate intracellular bacteria having reticulate (RBs) and elementary bodies (EBs) characteristic of chlamydia, have been shown to have >80% 16S rRNA gene sequence identity, e.g., Simkania negevensis strain Z (19) and “Candidatus Parachlamydia acanthamoebae” (1).
Unlike morphologically similar chlamydia-like bacteria, such as S. negevensis strain Z (19) and endosymbionts of Acanthamoeba spp. (11), the agents of epitheliocystis from fish have never been successfully cultured in vitro to facilitate genetic studies. Neither antigenic reactivity nor 16S rDNA sequence data have been obtained to further a molecular characterization of a chlamydia-like bacterium from a salmonid host. The objectives of this study were to compare the ultrastructures and immunoreactivities of developmental stages of inclusions from proliferative and nonproliferative gill lesions of farmed Atlantic salmon and to perform molecular phylogenetic analyses of 16S rDNA sequence data generated directly from proliferative gill lesions.
MATERIALS AND METHODS
Sample collection.
Samples of gill from farmed Atlantic salmon (S. salar) were collected at separate times by the staff of a multinational aquaculture company as part of its health surveillance program during periods of increased mortality, which were confirmed as outbreaks of epitheliocystis by histopathologic analysis of gill sections. Two sets of gill arches from 20 Atlantic salmon were submitted as pooled samples from one site in Ireland in 1999, and two sets of gill arches from 25 Atlantic salmon were submitted as individually identified samples from one site in Norway in 2000. One set of tissue samples from each location was submitted fixed by immersion in 10% formalin for histopathologic, electron microscopic, and in situ hybridization studies. A second set of tissues was submitted in 70% ethanol in the case of the Irish samples or immersed directly in tissue lysis buffer (ATL Buffer; QIAGEN Inc., Chatsworth, Calif.) in the case of the Norwegian samples for DNA extraction and PCR studies. In addition, paraffin- and resin-embedded gill samples from Atlantic salmon from Ireland accessioned in 1995 and processed for histopathology and transmission electron microscopy, respectively, were retrieved from departmental archives.
Histopathology.
Formalin-fixed gill samples were trimmed to fit plastic cassettes, processed routinely for paraffin embedding, sectioned at 4 μm, mounted on glass slides, and stained with hematoxylin and eosin (HE) according to standard histologic techniques (29). Tissue sections were examined by light microscopy to identify histopathologic lesions and inclusions of epitheliocystis based on previous descriptions (22).
Transmission electron microscopy and immunogold labeling.
For conventional transmission electron microscopy, trimmed gill samples were further fixed in a mixture of 1.25% formaldehyde, 2.5% glutaraldehyde, 0.03% CaCl2, and 0.03% trinitrophenol in 0.05 M cacodylate buffer, pH 7.3 (18), and then postfixed in 1% OsO4 in 0.1 M cacodylate buffer, stained en bloc with 1% uranyl acetate in 0.1 M maleate buffer, dehydrated in ethanol, and embedded in Poly/Bed 812 (Polysciences, Warrington, Pa.). For immunoelectron microscopy, trimmed samples were fixed in a mixture of 3% formaldehyde and 1% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2 (21); stained en bloc with 1% uranyl acetate in 0.1 M maleate buffer; dehydrated in ethanol; and embedded in LR White (SPI Supplies, West Chester, Pa.). Ultrathin sections were cut using a Reichert-Leica Ultracut S ultramicrotome and placed onto uncoated copper grids for conventional transmission electron microscopy or on nickel Formvar-carbon-coated grids for immunoelectron microscopy. All grids were examined with a Philips 201 or Philips CM-100 electron microscope at 60 kV after being stained with uranyl acetate and lead citrate.
In immunoelectron microscopy procedures, grids were treated with blocking buffer (0.1% bovine serum albumin-0.01 M glycine in 0.05 M Tris-buffered saline) for 15 min at room temperature (RT), incubated in a wet chamber (floating on drops) with primary antibody for 1 h at RT and then overnight at 4°C, washed in blocking buffer, and incubated with secondary colloidal-gold-labeled antibody for 1 h at RT. The primary antibody was commercial mouse anti-chlamydial lipopolysaccharide (LPS) monoclonal antibody (Chemicon International, Inc., Temecula, Calif.) diluted 1:100 in diluting buffer (0.05 M Tris-buffered saline containing 1% bovine serum albumin). The secondary antibody was goat anti-mouse immunoglobulin G-immunoglobulin M (heavy plus light chains) labeled with 10-nm-diameter colloidal gold (AuroProbe EM GAMIgG+IgM G10, RPN 431; Amersham Biosciences, Piscataway, N.J.) diluted 1:20 in diluting buffer. Control grids were incubated in the same way as test grids, except that the primary antibody was omitted and replaced with diluting buffer.
DNA extraction, amplification, and cloning.
Genomic DNA from ethanol-fixed or fresh gill tissue was extracted using the DNeasy extraction system (QIAGEN Inc.) according to the mouse tail protocol of the manufacturer, with the following modification: during lysis, gill tissues in tissue lysis buffer were heated to 97 to 100°C for 15 min to assist in releasing bacterial DNA, lysis solutions were cooled to 55°C, proteinase K was added, and lysis was allowed to proceed overnight. Eluates were assessed by either spectrophotometric or fluorometric methods (VersaFluor Fluorometer; Bio-Rad, Inc., Hercules, Calif.). The DNA was aliquoted and stored at 4°C for immediate use or frozen at −80°C for future experiments. Negative control genomic Atlantic salmon DNA was extracted from commercially purchased skeletal muscle or from whole-blood samples from clinically unaffected Atlantic salmon (kindly provided by John Coll, Fish Health Center, Northeast Fishery Center, U.S. Fish and Wildlife Service, Lamar, Pa.). Muscle and blood were selected as sources of genomic Atlantic salmon DNA because the risk of encountering contaminating prokaryotic DNA from chlamydia-like or other bacteria in these tissues is minimal. Positive control genomic chlamydial DNA was extracted from a culture of Chlamydia trachomatis (ATCC VR1477; American Type Culture Collection, Manassas, Va.). Nucleic acid extractions were tested for amplifiable DNA by PCR amplification of host 18S rDNA using primers 18e and 18i or 18e and 18g (15).
Oligonucleotide primers used to amplify 16S rDNA of C. trachomatis and the CLB associated with epitheliocystis from proliferative gills were chosen to amplify three successively longer overlapping regions of the 16S rRNA gene and included those for detection of the 16S rRNA gene signature sequence of Chlamydiales (9) or were combinations of Chlamydiales 16S rRNA gene primers (9) and eubacterial primers (12, 27) (Table 1). The primers were synthesized by Invitrogen Corp. (Carlsbad, Calif.).
TABLE 1.
Primers, positions, and target regions of 16S rRNA gene-based PCR protocols used to amplify 16S rDNA from the CLB associated with epitheliocystis in farmed Atlantic salmon
| Protocol | Primer | Sequence (reference) | Position | Product size (bp) |
|---|---|---|---|---|
| Signature sequence | 16SIGF | 5′-CGGCGTGGATGAGGCAT-3′ (9) | 40-57 | 300 |
| 16SIGR | 5′-TCAGTCCCAGTGTTGGC-3′ (9) | 323-340 | ||
| 806R | 16SIGF | 5′-CGGCGTGGATGAGGCAT-3′ (9) | 40-57 | 766 |
| 806R | 5′-GGACTACCAGGGTATCTAAT-3′ (27) | 787-806 | ||
| Near-full-length 16S | 16SIGF | 5′-CGGCGTGGATGAGGCAT-3′ (9) | 40-57 | 1,487 |
| 16SB1 | 5′-TACGGYTACCTTGTTACGACTT-3′ (12) | 1505-1527 |
All 16S rDNA was amplified in individual 50-μl reaction mixtures containing 100 to 200 ng of sample DNA, 5 μl of 10× QIAGEN buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 15 mM MgCl2, 0.01% gelatin), 200 μM deoxynucleoside triphosphates, 20 pmol of forward and reverse primers/μl, and 1 U of QIAGEN HotStar Taq polymerase using a Perkin-Elmer model 2400 thermal cycler (Applied Biosystems, Foster City, Calif.). Three different cycling protocols were employed to amplify the three overlapping 16S rDNA targets. For the 16S signature sequence, a touchdown PCR protocol (23) was used to limit secondary priming. Reactions were initiated by a 15-min incubation at 94°C, followed by 40 cycles, each consisting of denaturation at 94°C for 45 s, primer annealing for 45 s, and extension at 72°C for 45 s. Annealing temperatures began at 66°C and decreased by 1°C every third cycle until 61°C, at which temperature the final 25 cycles were performed. After 40 cycles, a 7-min extension step at 72°C was performed. For the 806R protocol, PCR was initiated by a 15-min incubation at 94°C, followed by 40 cycles, each consisting of denaturation at 94°C for 30 s, annealing at 55°C for 45 s, and extension at 72°C for 45 s, with a final extension of 7 min. For near-full-length 16S rRNA amplicons, PCR was initiated by incubation at 94°C for 15 min, followed by 40 cycles, each consisting of denaturation at 94°C for 40 s, annealing at 58°C for 40 s, and extension at 72°C for 45 s, with a final 7-min extension step. The PCR products were separated by 10% polyacrylamide or 2% agarose gel electrophoresis, visualized by ethidium bromide staining and UV transillumination, and recorded digitally using a Stratagene (La Jolla, Calif.) Eagle Eye II-Still Video System. Reactions that yielded amplicons were repeated; samples that amplified twice were considered for DNA sequence analysis.
PCR products were excised and purified from agarose gels or purified directly from reactions (QIAGEN Mini-Elute Gel Extraction kit or PCR Purification kit). Purified products from 16S signature sequence PCRs were submitted directly for oligonucleotide-directed dideoxynucleotide chain termination sequencing reactions (HHMI Biopolymer/W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University, New Haven, Conn.), whereas those from other 16S rRNA gene PCRs were ligated into a T/A plasmid vector (TOPO TA Cloning Vector for Sequencing; Invitrogen Corp.), cloned, screened by PCR, and purified (QIAGEN Miniprep DNA Purification kit).
DNA sequencing and analysis.
PCR products and clones were selected based on the sample origin, i.e., Ireland or Norway, and the 16S rDNA target region, e.g., signature sequence, 806R, and near-full-length 16S. In each case, a minimum of four amplicons from several different samples and different PCR runs were designated for sequencing to account for intersample and intrasample variation. One clone from each separate cloning reaction was submitted for sequencing by primer walking in sense and antisense directions (HHMI Biopolymer/W. M. Keck Foundation Biotechnology Resource Laboratory). The nucleotide sequences of cloned PCR products were determined by assembling ABI sequence files for sense and antisense strands of each PCR product clone using Vector NTI version 7.0 (InforMax, Inc., Bethesda, Md.). Consensus sequences for 16S signature and 806R targets were constructed with respect to the sample source using multiple-sequence-alignment programs of DNAMAN (Lynnon Biosoft, Vaudreuil, Quebec, Canada) and Vector NTI version 7.0, in which base discrepancies were resolved by simple majority rulings. A 1,487-bp near-full-length 16S rDNA consensus sequence was constructed from amplicons from the year 2000 Norway samples and was aligned to all other consensus sequences to create an overall 16S rDNA consensus sequence of the CLB associated with epitheliocystis in proliferative gills, which was utilized in standard nucleotide-nucleotide BLAST searches of the National Center for Biotechnology Information databases (http://www.ncbi.nlm.nih.gov).
Molecular phylogenetic analyses.
The consensus 16S rRNA gene sequence from the CLB was aligned with full-length 16S rRNA gene sequences selected from prior molecular systematic studies of members of the order Chlamydiales (9), Neochlamydia hartmannellae and endosymbionts of Acanthamoeba spp. (17), and members of the order Rickettsiales (6). Sequences downloaded for molecular phylogenetic analysis are listed with corresponding GenBank accession numbers in Table 2. Homology matrix analysis of the 16S rRNA gene sequence from the CLB with those of the organisms listed in Table 2 was performed using DNAMAN. Sequences for phylogenetic analyses were aligned, edited by visual inspection, and formatted using ClustalX version 1.81 (31; ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/), and distance- and parsimony-based analyses were conducted using PAUP* version 4.0b10 for Macintosh (30). Parsimony-based analyses were executed using heuristic search settings that included random sequence addition, i.e., independent of the order of the input file, with 10 replicates per sequence addition and random trees used as starting points, tree-bisection-reconnection branch swapping, and collapsing of branches to create polytomies when maximum branch lengths were zero. Distance-based, i.e., minimum evolution, analyses were executed using heuristic search settings employing neighbor-joining to obtain starting trees and tree-bisection-reconnection branch swapping. In heuristic parsimony searches, the number of parsimony-informative characters was 671 of a total of 1,596. Estimates of confidence at nodes were obtained through 1,000 bootstrap replicates of heuristic searches (10).
TABLE 2.
Percent nucleotide identities between the 1,487-bp 16S rRNA gene sequence of “Candidatus Piscichlamydia salmonis” and those of the four families of Chlamydia, along with representatives of Rickettsia and Ehrlichia
| No. | Species | % identity with species no.:
|
|||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 37 | 36 | 35 | 34 | 33 | 32 | 31 | 30 | 29 | 28 | 27 | 26 | 25 | 24 | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | 1 | |||
| 1 | Chlamydia muridarum MoPn [85718] | 78 | 68 | 68 | 68 | 69 | 67 | 69 | 67 | 70 | 68 | 66 | 67 | 66 | 66 | 66 | 79 | 85 | 86 | 86 | 82 | 84 | 92 | 95 | 98 | 98 | 98 | 91 | 93 | 91 | 98 | 98 | 99 | 94 | 93 | 97 | 100 | 100 | |
| 2 | Chlamydia muridarum SFPD [U68437] | 78 | 68 | 68 | 68 | 69 | 67 | 69 | 67 | 70 | 68 | 66 | 67 | 66 | 66 | 66 | 79 | 85 | 86 | 86 | 82 | 83 | 92 | 95 | 98 | 98 | 98 | 91 | 93 | 91 | 98 | 98 | 98 | 94 | 93 | 97 | 100 | ||
| 3 | Chlamydia suis S45 [U73110] | 78 | 68 | 68 | 68 | 69 | 67 | 68 | 66 | 69 | 68 | 66 | 67 | 65 | 67 | 66 | 80 | 84 | 86 | 86 | 82 | 83 | 91 | 94 | 97 | 97 | 97 | 90 | 92 | 90 | 97 | 97 | 97 | 93 | 94 | 100 | |||
| 4 | Chlamydia suis R22 [U68420] | 78 | 66 | 67 | 66 | 68 | 66 | 68 | 65 | 66 | 66 | 69 | 68 | 64 | 66 | 63 | 84 | 81 | 86 | 83 | 81 | 83 | 91 | 90 | 93 | 93 | 93 | 95 | 88 | 95 | 93 | 93 | 93 | 97 | 100 | ||||
| 5 | Chlamydia trachomatis L2/434/BU [U68443] | 78 | 66 | 68 | 67 | 68 | 67 | 68 | 66 | 66 | 66 | 69 | 68 | 65 | 66 | 64 | 84 | 81 | 82 | 83 | 82 | 82 | 92 | 91 | 95 | 91 | 91 | 99 | 93 | 95 | 95 | 95 | 95 | 100 | |||||
| 6 | Chlamydia trachomatis A/Har-13 [D89067] | 78 | 67 | 68 | 68 | 68 | 67 | 68 | 66 | 69 | 68 | 66 | 67 | 66 | 67 | 66 | 79 | 84 | 82 | 86 | 83 | 83 | 91 | 95 | 95 | 95 | 95 | 95 | 89 | 91 | 100 | 100 | 100 | ||||||
| 7 | Chlamydia trachomatis B/TW-5/OT [D85719] | 78 | 67 | 68 | 68 | 68 | 67 | 68 | 66 | 69 | 68 | 66 | 67 | 66 | 67 | 66 | 79 | 84 | 85 | 86 | 83 | 83 | 91 | 95 | 95 | 95 | 95 | 91 | 92 | 90 | 100 | 100 | |||||||
| 8 | Chlamydia trachomatis DUW-3/CX [D85721] | 78 | 67 | 68 | 68 | 68 | 67 | 68 | 66 | 69 | 68 | 66 | 67 | 66 | 67 | 66 | 79 | 84 | 85 | 86 | 83 | 83 | 91 | 95 | 95 | 95 | 95 | 91 | 92 | 91 | 100 | ||||||||
| 9 | Chlamydophila abortus EBA [U76710] | 78 | 66 | 68 | 67 | 68 | 67 | 68 | 66 | 66 | 66 | 69 | 68 | 64 | 66 | 63 | 84 | 80 | 85 | 83 | 81 | 83 | 94 | 92 | 92 | 94 | 94 | 91 | 92 | 100 | |||||||||
| 10 | Chlamydophila psittaci MN [AB001784] | 80 | 70 | 70 | 71 | 69 | 68 | 70 | 69 | 68 | 70 | 69 | 70 | 68 | 70 | 68 | 77 | 82 | 82 | 84 | 84 | 85 | 91 | 94 | 92 | 96 | 96 | 92 | 100 | ||||||||||
| 11 | Chlamydophila psittaci NJI [U68419] | 78 | 66 | 70 | 67 | 68 | 67 | 68 | 66 | 66 | 66 | 69 | 68 | 64 | 66 | 66 | 84 | 80 | 84 | 83 | 81 | 83 | 94 | 92 | 94 | 94 | 94 | 100 | |||||||||||
| 12 | Chlamydophila caviae GPIC [D85708] | 78 | 67 | 68 | 68 | 69 | 67 | 69 | 67 | 70 | 68 | 66 | 67 | 66 | 67 | 66 | 79 | 84 | 82 | 78 | 82 | 84 | 93 | 96 | 96 | 98 | 100 | ||||||||||||
| 13 | Chlamydophila felis FP Cello [D85706] | 78 | 67 | 68 | 68 | 69 | 68 | 69 | 67 | 70 | 68 | 66 | 67 | 66 | 67 | 66 | 80 | 84 | 86 | 78 | 82 | 84 | 93 | 96 | 96 | 100 | |||||||||||||
| 14 | Chlamydophila peccrum E58 [D88317] | 78 | 66 | 68 | 68 | 69 | 68 | 69 | 67 | 70 | 68 | 66 | 67 | 66 | 67 | 66 | 79 | 84 | 86 | 78 | 82 | 83 | 93 | 100 | 100 | ||||||||||||||
| 15 | Chlamydophila peccrum IPA [D85716] | 78 | 66 | 67 | 68 | 69 | 68 | 69 | 67 | 70 | 68 | 66 | 67 | 66 | 67 | 66 | 79 | 84 | 85 | 78 | 82 | 83 | 93 | 100 | |||||||||||||||
| 16 | Chlamydophila pneumoniae N16 [U68426] | 80 | 68 | 67 | 68 | 70 | 68 | 70 | 66 | 67 | 67 | 68 | 69 | 65 | 67 | 65 | 82 | 82 | 85 | 80 | 84 | 85 | 100 | ||||||||||||||||
| 17 | Endosymbiont UWC22 [AF083616] | 81 | 70 | 69 | 70 | 70 | 69 | 70 | 69 | 66 | 70 | 70 | 70 | 69 | 70 | 66 | 82 | 84 | 88 | 81 | 92 | 100 | |||||||||||||||||
| 18 | Endosymbiont UWE1 [AF083614] | 82 | 70 | 70 | 71 | 71 | 70 | 71 | 69 | 67 | 70 | 69 | 71 | 69 | 70 | 67 | 82 | 84 | 90 | 89 | 100 | ||||||||||||||||||
| 19 | Neochlamydia hartmannellae [AF177275] | 78 | 67 | 71 | 68 | 69 | 67 | 68 | 67 | 68 | 69 | 67 | 68 | 67 | 68 | 65 | 82 | 85 | 91 | 100 | |||||||||||||||||||
| 20 | Parachlamydia acanthamoebae [Y07556] | 79 | 67 | 68 | 68 | 69 | 68 | 69 | 67 | 70 | 68 | 66 | 67 | 66 | 67 | 66 | 84 | 87 | 100 | ||||||||||||||||||||
| 21 | Waddia chondrophila [AP042496] | 77 | 66 | 66 | 67 | 68 | 67 | 68 | 65 | 69 | 66 | 64 | 66 | 65 | 66 | 64 | 80 | 100 | |||||||||||||||||||||
| 22 | Simkania negevensis [L27666] | 77 | 65 | 66 | 65 | 66 | 65 | 67 | 64 | 65 | 65 | 68 | 67 | 64 | 65 | 62 | 100 | ||||||||||||||||||||||
| 23 | Agent of withering syndrome in abalone [AF069062] | 66 | 79 | 79 | 79 | 77 | 77 | 76 | 80 | 77 | 82 | 80 | 81 | 81 | 82 | 100 | |||||||||||||||||||||||
| 24 | Ehrlichia chaffeensis [M73222] | 69 | 83 | 83 | 83 | 81 | 83 | 82 | 85 | 80 | 92 | 91 | 92 | 98 | 100 | ||||||||||||||||||||||||
| 25 | Ehrlichia chaffeensis [U86664] | 68 | 82 | 81 | 81 | 79 | 82 | 80 | 84 | 78 | 91 | 89 | 90 | 100 | |||||||||||||||||||||||||
| 26 | Ehrlichia sp. [AB074460] | 71 | 83 | 84 | 83 | 81 | 82 | 82 | 83 | 79 | 91 | 92 | 100 | ||||||||||||||||||||||||||
| 27 | Ehrlichia sp. ‘HGE agent’ [AF093788] | 70 | 82 | 82 | 81 | 80 | 81 | 80 | 84 | 78 | 97 | 100 | |||||||||||||||||||||||||||
| 28 | Ehrlichia sp. ‘HGE agent’ [U02521] | 69 | 83 | 83 | 83 | 80 | 82 | 81 | 85 | 81 | 100 | ||||||||||||||||||||||||||||
| 29 | Ehrlichia risticii [M21290] | 66 | 77 | 78 | 78 | 79 | 92 | 79 | 94 | 100 | |||||||||||||||||||||||||||||
| 30 | Ehrlichia sennetsu [M73225] | 69 | 82 | 82 | 82 | 80 | 92 | 80 | 100 | ||||||||||||||||||||||||||||||
| 31 | Orientia tsutsugamushi [D38622] | 70 | 89 | 89 | 89 | 91 | 82 | 100 | |||||||||||||||||||||||||||||||
| 32 | Necrickettsia helminthoeca [U12457] | 70 | 80 | 81 | 81 | 82 | 100 | ||||||||||||||||||||||||||||||||
| 33 | Rickettsia typhi Wilmington [U12463] | 70 | 96 | 96 | 96 | 100 | |||||||||||||||||||||||||||||||||
| 34 | Rickettsia rickettsii strain R [L36217] | 70 | 98 | 98 | 100 | ||||||||||||||||||||||||||||||||||
| 35 | Rickettsia sp. [AY158006] | 70 | 98 | 100 | |||||||||||||||||||||||||||||||||||
| 36 | Rickettsia sp. (Ixodes symbiont) [D84558] | 69 | 100 | ||||||||||||||||||||||||||||||||||||
| 37 | “Candidatus Piscichlamydia salmonis” | 100 | |||||||||||||||||||||||||||||||||||||
In situ hybridization using riboprobes.
In situ hybridization was accomplished using riboprobes generated by in vitro transcription of the cloned ∼1.5-kb 16S RNA gene sequence amplified from gills infected with the CLB, based on procedures described by Brown (4) and using a dual promoter vector (pCRII TOPO Vector; Invitrogen Corp.) and Sp6/T7 DIG RNA labeling kit (Roche Applied Science, Indianapolis, Ind.). Plasmid inserts were confirmed and orientations were determined by DNA sequencing. To serve as a probe sequence control, the full-length 16S rRNA gene of Mycobacterium marinum was transcribed in vitro in identical fashion. Transcribed riboprobe concentrations were assessed by dot blot analysis.
Riboprobes were used in nonisotopic in situ hybridization experiments based on procedures described by Gan et al. (13) and Brown (4). Uninfected gill tissue from Atlantic salmon from a geographical location other than Ireland or Norway served as the negative tissue control. Gill sections stained with HE were examined by light microscopy for the number and distribution of inclusions, and successive serial tissue sections at 4 μm were mounted unstained on ProbeOn Plus glass slides (Fisher Scientific, Fairlawn, N.J.). Unstained sections were heated to 70°C for 10 min, deparaffinized in three 3-min washes with xylene (Sigma, St. Louis, Mo.), and rehydrated for 15 min at RT. Sections were digested with 20 ng of proteinase K/μl at 37°C for 15 min and then covered with prehybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5% blocking reagent, 48% formamide, 0.02% sodium dodecyl sulfate [SDS], 0.1% N-lauroylsarcosine) for 1 h at 37°C. The prehybridized slides were heated to 95°C for 10 min to denature target DNA, covered with ∼100 μl of hybridization solution (35 ng of digoxigenin-labeled riboprobe per 100 μl of prehybridization solution) per slide, covered with a glass coverslip, and allowed to hybridize overnight at 42°C in a humidified slide moat (Fisher Scientific). After hybridization, the slides were washed in 2× SSC with 1% SDS at 50°C for 20 min, followed by 1× SSC with 0.1% SDS at 50°C for 20 min and then 1× SSC for 10 min at RT and 0.1× SSC for 15 min at RT. Hybridization signals were developed by washing sections in buffer I (0.1 M Tris, 0.15 M NaCl, pH 7.5) for 5 min and then incubating the sections for 2 h at 37°C in a humidified chamber with 100 to 200 μl of buffer I containing 2% sheep serum and anti-digoxigenin Fab fragments conjugated to alkaline phosphatase at 1:275 dilution (Roche Applied Science). The slides received one 15-min wash in buffer I and then were washed twice in TBST (0.15 M NaCl, 2.7 mM KCl, 25 mM Tris, 0.05% Tween 20, pH 7.6) for 5 min. The signal was developed using the DAKO (Carpinteria, Calif.) Fuchsin-Substrate System with levamasole; the slides were counterstained and coverslipped according to the instructions of the manufacturer.
Nucleotide sequence accession numbers.
The 16S rRNA genetic sequence of a representative clone from the Norwegian gill samples and the same from the Irish gill samples were submitted to GenBank under accession numbers AY462244 and AY462243, respectively.
RESULTS
Histopathology.
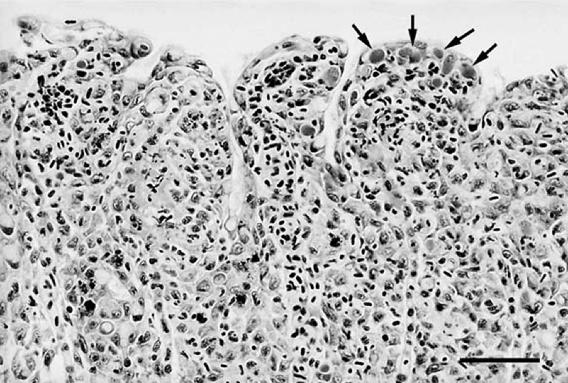
Gills collected from Ireland in 1999 and from Norway in 2000 had proliferative lesions and intracellular bacterial inclusions. Gills from both sources had multifocal to diffuse epithelial hyperplasia characterized by interlamellar filling, lamellar fusion, bridging across tips of adjacent lamellae, focal necrosis of epithelial cells, and infiltrates of tangible-body macrophages. Bacteria were located in granular basophilic inclusions in the cytoplasm of hypertrophied epithelial cells, often at the tips of lamellae (Fig. 1). However, as in previous descriptions (22), bacterial inclusions were not always associated with epithelial hyperplasia and were located on nonlesional lamellae as well. In subjective evaluations, lesions in gills from Ireland and Norway affected from 60 to 80% of lamellae, although bacterial inclusions were observed with slightly greater frequency in samples from Norway than in those from Ireland. In contrast, gills collected from Ireland in 1995 had no proliferative changes and few, widely separated bacterial inclusions, which were most often located at the distal tips of nonlesional lamellae.
FIG. 1.
“Candidatus Piscichlamydia salmonis” in proliferative gill lesion from year 2000 Norwegian farmed Atlantic salmon, with intracellular inclusions (arrows) at tips of lamellae accompanied by interlamellar filling by hyperplastic epithelium, macrophages, and pyknotic nuclei (HE). Bar = 50 μm.
Transmission electron microscopy and immunogold labeling.
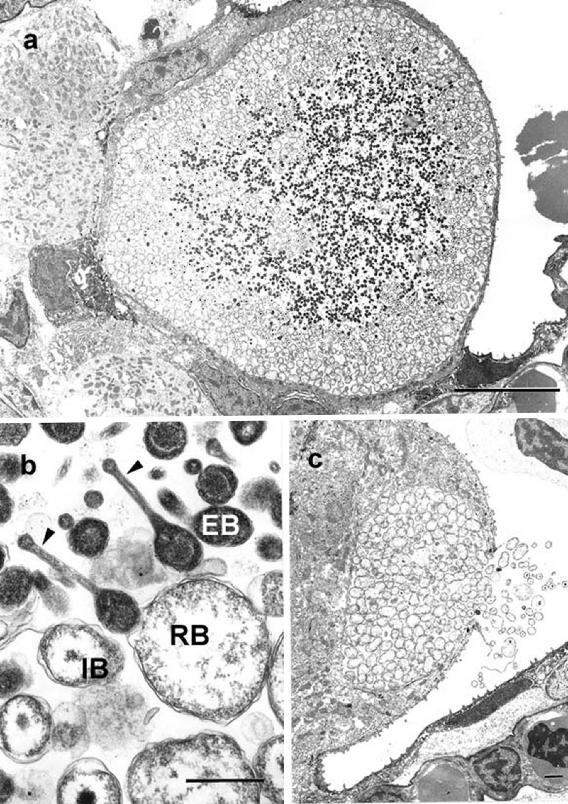
Ultrastructurally, in proliferative gill lesions from Norway, inclusions consisted of intracytoplasmic membrane-bound vacuoles containing mixtures of reticulate bodies (RBs) and intermediate bodies (IBs), with some smaller inclusions containing predominantly RBs. Reticulate bodies had different shapes, e.g., elongate, oblong, or spherical; ranged in length from 0.7 to 1.8 μm, with most at 1 μm; and had evenly dispersed granular osmiophilic material and ribosomes in the cytoplasm. Intermediate bodies were round to oval and 0.6 to 0.8 μm in length and had central electron-dense nucleoids. Both vacuolated IBs, i.e., those which contained single, sharply marginated, clear-space vacuoles at one pole, and nonvacuolated IBs were present, and there were elongate RBs with central narrowing and punctate condensations of nucleoids at opposite poles, suggestive of binary fission. Loosely dispersed, amorphous, osmiophilic, granular, and fibrillar material was present between RBs and IBs within vacuoles (Fig. 2). Morphological features of RBs and IBs from proliferative gills were consistent with descriptions of RBs and IBs of chlamydia (2) and the epitheliocystis agent from Atlantic salmon (22), although no distinct elementary bodies (EBs) were identified in this material. In nonproliferative gills from Ireland in 1995, vacuoles contained three morphologically distinct developmental stages characteristic of chlamydia, i.e., RBs, IBs, and EBs (2), as well as a fourth head-and-tail cell not observed in previous studies of farmed Atlantic salmon (22) or in proliferative gill lesions from Norway in 2000 (Fig. 3). RBs in inclusions from nonproliferative gills were round to oval with few, if any, elongate cells (Fig. 3), and IBs were round with central nucleoids and no vacuoles (Fig. 3b). Head-and-tail cells had central osmiophilic condensations and long unipolar cytoplasmic extensions with terminal knobs (Fig. 3b). Immunogold labeling experiments using mouse monoclonal anti-chlamydial LPS primary antibody and goat anti-mouse secondary antibody resulted in distinct labeling of membranes and the peripheral cytoplasm of RBs and IBs in the inclusions from both sources (Fig. 4). There was mild inconsistent cross-reactivity of the anti-chlamydial LPS primary antibody with organellar membranes of salmon epithelial cell cytoplasm, which was confirmed by negative control trials in which the goat anti-mouse secondary antibody was used alone and no reaction with salmon tissue was observed.
FIG. 2.
Transmission electron photomicrograph of typical “Candidatus Piscichlamydia salmonis” inclusions from year 2000 Norwegian specimen. (a) Inclusions are intracytoplasmic membrane-bound vacuoles containing round to oblong reticulate bodies and smaller, oval, intermediate bodies that have condensed osmiophilic nucleoids and vacuolated (arrows) or nonvacuolated cytoplasm. (b) Higher magnification of RBs and IBs, with some having constrictions in central regions (arrows) suggestive of binary fission. The space between RBs and IBs contains loose granular and fibrillar material (arrowheads). Bars = 1 μm.
FIG. 3.
(a) Transmission electron photomicrograph of an epitheliocystis inclusion in nonproliferative gill from year 1995 Irish specimen. The inclusion is located in the cytoplasm of a hypertrophied epithelial cell and consists of a membrane-bound vacuole containing multiple chlamydial developmental stages. Bar = 10 μm. (b) EBs, IBs, RBs, and head-and-tail forms. Head-and-tail forms have long unipolar extensions with terminal knobs (arrowheads) and central condensed nucleoids. Bar = 0.5 μm. (c) Rupture of vacuole from within a branchial epithelial cell with release of RBs, IBs, and EBs to the exterior. Bar = 1 μm.
FIG. 4.
Immunogold labeling with anti-chlamydial LPS antibody demonstrates distinct labeling mostly along membranes and at the periphery of cytoplasm of RBs in inclusions (a) of epitheliocystis from year 1995 Irish specimen and (c) of “Candidatus Piscichlamydia salmonis” from year 2000 Norwegian specimen. Note the absence of labeling of RBs or IBs in control sections from (b) year 1995 Irish and (d) year 2000 Norwegian specimens incubated with only secondary antibody. Bars = 0.5 μm.
DNA sequencing and analysis.
Amplicons were obtained from ethanol-fixed year 1999 gill samples from Ireland and freshly lysed year 2000 gill samples from Norway utilizing both the 16S signature sequence and the 806R protocols. Five 300-bp 16S signature sequence amplicons from five different gill samples from both Ireland and Norway were sequenced, whereas four cloned 806R PCR products from Ireland and six from Norway were sequenced. Only freshly lysed gills from Norway yielded amplicons utilizing the near-full-length 16S rRNA gene PCR protocol. Products of 1,487 bp were amplified from Norwegian gill samples, and a total of five clones from three salmon that generated amplicons in two repeat experiments were used to generate an ∼1.5-kb 16S rRNA gene sequence of the CLB.
Consensus 16S rRNA genetic sequences of the CLB associated with epitheliocystis from Irish and Norwegian gills, which were compiled separately, demonstrated 99% nucleotide sequence identity in pairwise alignments over those regions available for comparison, i.e., the 5′ half of the gene, and an overall consensus ∼1.5-kb 16S rRNA gene sequence of the CLB from proliferative Atlantic salmon gills was generated for further molecular analyses. Homology matrix analysis of the ∼1.5-kb consensus from the CLB identified members of the order Chlamydiales, e.g., endosymbionts of Acanthamoeba sp. strains UWE1 (82%) and UWC22 (81%), Chlamydophila psittaci MN (80%), and Chlamydophila pneumoniae N16 (80%), as having ≥80% nucleotide sequence identity. The percent nucleotide sequence identities between the consensus ∼1.5-kb 16S rRNA genetic sequence of the CLB and representative members of the orders Chlamydiales and Rickettsiales are shown in Table 2.
Molecular phylogenetic analyses.
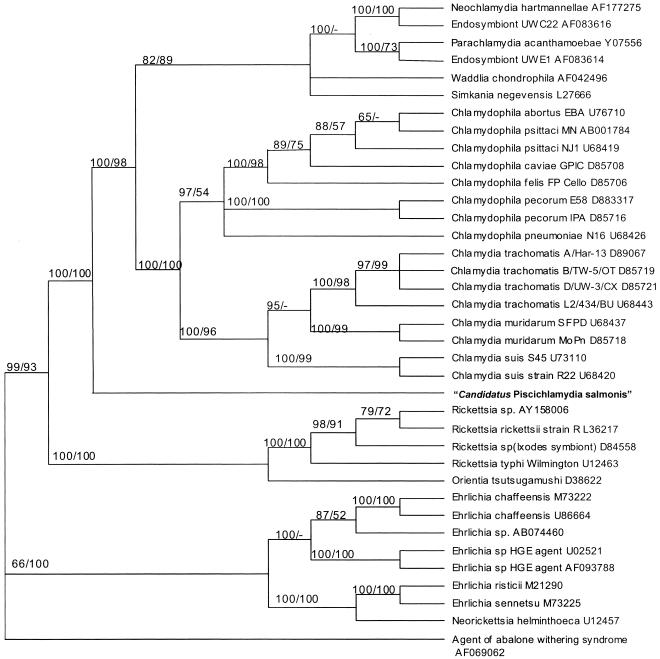
Analyses applying both distance and parsimony optimality criteria to alignments of 16S rDNA sequences, which included that of the CLB, yielded branching patterns for members of the Chlamydiales and Rickettsiales very similar to those previously described (6, 9, 17). The CLB associated with epitheliocystis from proliferative gills grouped in basal branches of trees with members of the order Chlamydiales and separately from members of the order Rickettsiales, e.g., Rickettsia spp., Ehrlichia spp., and Neorickettsia sp. Among members of the Chlamydiales, the CLB split into a separate basal branch and did not group with any members of the Chlamydiaceae or with the new families Parachlamydiaceae, Simkaniaceae, or Waddliaceae described by Everett et al. (9). Branching of the CLB with members of the Chlamydiales was supported statistically with a 100% bootstrap value in both distance and parsimony studies (Fig. 5).
FIG. 5.
Bootstrap consensus phylogenetic tree inferred from 16S rRNA gene sequences of select members of the order Chlamydiales, including N. hartmannellae and endosymbionts of Acanthamoebae spp.; members of the order Rickettsiales, including species of Rickettsia, Ehrlichia, and Neorickettsia; and near-full-length 16S rRNA gene sequence of “Candidatus Piscichlamydia salmonis” from farmed Atlantic salmon. Bootstrap values are included for clades observed using both distance and parsimony and are provided as percentages of 1,000 replicates. The sequence from the agent of abalone withering syndrome served as the designated outgroup.
In situ hybridization.
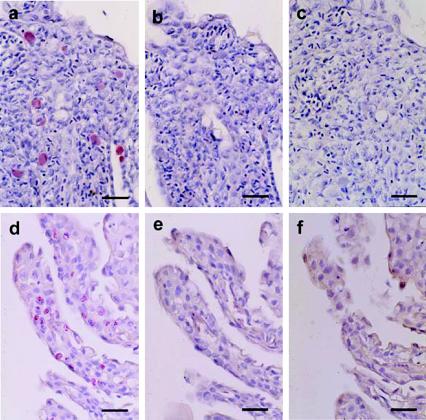
In the absence of a culture-positive control tissue, serial sections in threes were tested during each run using (i) a digoxigenin-labeled chlamydia-like 16S riboprobe transcribed from the ∼1.5-kb 16S rDNA amplicons from Norwegian gill samples with epitheliocystis, (ii) a digoxigenin-labeled 16S riboprobe transcribed from M. marinum, and (iii) no riboprobe. The ∼1.5-kb digoxigenin-labeled chlamydia-like 16S riboprobe demonstrated discrete and specific labeling localized to intracellular inclusions of epithelial cells in histologic sections of proliferative gill lesions from both Norway (2000) and Ireland (1999); concurrently processed slides that received digoxigenin-labeled M. marinum 16S riboprobe or no riboprobe had no signal (Fig. 6). No signal was obtained from negative control tissues processed with or without riboprobe.
FIG. 6.
In situ hybridization using digoxigenin-labeled 16S riboprobes performed on gill sections of year 2000 Norwegian (a, b, and c) and year 1999 Irish (d, e, and f) salmon. Distinct labeling limited to intracellular inclusions is visible in histologic sections of gills from Norway (a) and Ireland (d) using the chlamydia-like riboprobe transcribed from 16S rDNA amplified from Norwegian gill samples infected with “Candidatus Piscichlamydia salmonis”; no labeling is evident in sections incubated with riboprobe transcribed from 16S rDNA of M. marinum (b and e) and with no riboprobe (c and f) (Dako fuchsin substrate and hematoxylin). Bars = 20 μm.
DISCUSSION
Epitheliocystis inclusions from proliferative gill lesions of year 2000 Norway salmon had histological and ultrastructural features identical to those described by Nylund et al. (22) in earlier morphological studies of the epitheliocystis agent from farmed Atlantic salmon. Inclusions from both studies were composed of mixtures of variably elongate RBs and vacuolated and nonvacuolated IBs. In contrast, inclusions from year 1995 Irish gills, which did not have proliferative lesions, contained mostly round RBs, IBs without vacuoles, and numerous EBs typical of chlamydial developmental stages (2), as well as distinctive head-and-tail cells. Head-and-tail cells were not identified in epitheliocystis inclusions of gills from Norway and were not seen in material studied by Nylund et al. (22). Head-and-tail cells have been reported in epitheliocystis inclusions of steelhead trout (28) and lake trout (3). Although it has been theorized that their occurrence may be evidence of abnormal EB development under certain environmental conditions (3), Nylund et al. (22) have proposed that morphological differences in RB (and IB) structure underscore the absence of head-and-tail cells in epitheliocystis inclusions from farmed Atlantic salmon and are evidence that the CLB from farmed Atlantic salmon in Norway is a different species from that of salmonids in freshwater in the United States. Similarly, morphological data from epitheliocystis inclusions investigated in this study identified ultrastructural differences between developmental stages of year 2000 Norwegian inclusions and year 1995 Irish inclusions that suggest that two different chlamydia-like developmental cycles representing two different species exist in farmed Atlantic salmon, one occurring in proliferative gill lesions and one occurring in nonproliferative gill samples.
Similarities in immunogold labeling of RBs in epitheliocystis inclusions of salmon gills from Norway in 2000 and Ireland in 1995 using a mouse monoclonal anti-chlamydial LPS antibody suggest the presence of related trisaccharide-like molecules. In light of the fact that no other macromolecules, except for the 16S rRNA gene from this study, have been characterized from this or other chlamydia-like bacteria in salmon, speculations as to the presence of biologically relevant molecules need to be made and interpreted cautiously. Immunogold labeling of RBs by an anti-chlamydial LPS antibody is an apparent contradiction of the 16S rRNA gene sequence results and the expected affinity of that antibody. Members of the family Chlamydiaceae are recognized by monoclonal antibodies that detect the LPS trisaccharide αKdo-(2→8)-αKdo-(2→4)-αKdo (9, 20). Cross-reactivity of this anti-chlamydial LPS antibody on the part of RBs of the CLB associated with epitheliocystis from Atlantic salmon may represent immunoreactivity to a related but yet uncharacterized trisaccharide-like molecule. At least one other CLB from a piscine source has been shown to be immunoreactive for this same genus-specific LPS target; antigenic and ultrastructural studies of epitheliocystis inclusions from cultured white sturgeon demonstrated positive immunohistochemical labeling of branchial inclusions in paraffin sections using the 11B5 murine monoclonal antibody from Diagnostic Products Corp. (Los Angeles, Calif.) (14). Additional nucleic acid-based studies are needed to define the genetic relatedness of the bacteria identified by the two morphologically different inclusions and to elucidate the degree of relatedness suggested by their similar immunoreactivities.
Gill samples having proliferative lesions and histopathologic evidence of epitheliocystis inclusions, i.e., proliferative epitheliocystis, from Ireland in 1999 and Norway in 2000 yielded 16S rDNA sequence data that demonstrated 99% nucleotide identity over the 5′ half of the 16S rRNA gene, i.e., the signature sequence and 806R target regions combined. Everett et al. (9) proposed that PCR amplification and direct sequencing of the 16S signature sequence would allow rapid identification of strains within the order Chlamydiales, and the nucleotide sequence identity of signature and 806R sequences amplified from gills from these two sources supports the assertion that the CLB associated with epitheliocystis from proliferative gill lesions from Ireland and Norway are most likely the same. This assertion is further substantiated by results of in situ hybridization, which demonstrated hybridization of the near-full-length ∼1.5-kb chlamydia-like 16S riboprobe with epitheliocystis inclusions in gills from both sources. The reasons why gill samples from Ireland in 1999 did not support near-full-length 16S rDNA amplification are more likely related to poor specimen preservation for maintenance of higher-molecular-weight DNA and differences in bacterial infection densities rather than actual differences in 16S rRNA gene sequences. Gill samples from 1995 were not archived in a manner that supported meaningful molecular testing like that performed on 1999 and 2000 samples.
The unique 16S sequence of the CLB associated with epitheliocystis from proliferative gill lesions of Atlantic salmon and its solo and basal grouping with members of the Chlamydiales in consensus trees having coherent groupings of bacteria in the orders Chlamydiales and Rickettsiales are evidence that this is a novel bacterium distinct from known chlamydia-like bacteria. To our knowledge, this is the first 16S rRNA gene sequence-based characterization of a CLB associated with epitheliocystis in any teleost. The fact that it was obtained from a commercially relevant species with morphological evidence of proliferative branchial lesions highlights the necessity for additional research to characterize other genetic loci, such as the major outer membrane protein gene, that would substantiate molecular phylogenetic studies and identify potential targets for nucleic-acid-based diagnostic tests. Additional sequence data from epitheliocystis agents of other piscine hosts are necessary in order to determine whether these organisms will form one or several coherent clusters in future molecular phylogenetic studies directed toward inferring relationships between chlamydia-like bacteria. Recent advances in our collective understanding of the ribosomal operons of many gram-negative intracellular bacteria have made ribosomal gene determination and analysis of these yet-unclassified bacterial agents, such as the CLB from Atlantic salmon, critical to their taxonomic classification and may also expand and potentially redefine the molecular systematics of other chlamydia-like or rickettsia-like bacteria.
Percent nucleotide sequence identities and pairwise sequence comparisons of the consensus ∼1.5-kb 16S rRNA gene sequence of the CLB to 16S rRNA gene sequences of select members of the orders Chlamydiales and Rickettsiales indicated that this CLB from Atlantic salmon has the highest nucleotide sequence identity with endosymbionts of Acanthamoeba sp. strains UWE1 (82%) and UWC22 (81%). Genetic relatedness to endosymbionts of Acanthamoeba spp. may provide insight into possible environmental sources of this CLB. Chlamydia-like endosymbionts have been identified in clinical and environmental isolates of acanthamoebae (1, 11), and the possibility exists that this CLB and others significant for farmed fish populations may passage through amoebae that are part of the microfauna of environments in which fish are cultured. The relatively high percent 16S rRNA gene sequence identity of the CLB from Atlantic salmon to those of these chlamydia-like endosymbionts supports the postulation that environmental carriage of this CLB may occur in amoebae. 16S rRNA gene sequence data from epitheliocystis-associated bacteria will make available to researchers templates from which genetic studies could be performed to detect distinctive DNA sequences in environmental samples or microbial isolates and to identify potential sources of agents of epitheliocystis.
In summary, morphologically different chlamydia-like developmental stages that were immunoreactive for chlamydial LPS were identified in proliferative and nonproliferative gills of farmed Atlantic salmon. Developmental stages from proliferative gills closely resembled those described in other morphological studies of epitheliocystis in farmed Atlantic salmon (22), and ultrastructural differences between epitheliocystis inclusions are taken as evidence of species differences in the CLB infecting proliferative and nonproliferative gills of Atlantic salmon. A unique 16S rRNA genetic sequence was generated directly from proliferative-gill lesions from salmon from both Ireland and Norway and localized to epitheliocystis inclusions in in situ hybridization experiments. Molecular phylogenetic analyses indicate that this CLB is novel and groups with members of the order Chlamydiales, although separately from the Chlamydiaceae and other chlamydia-like bacteria. The highest percent 16S rRNA gene sequence identity existed with endosymbionts of Acanthamoeba spp. Based on its novel sequence and solo molecular systematic assortment, the name “Candidatus Piscichlamydia salmonis” is proposed to identify this CLB in future references. Additional research is needed to identify other genetic loci capable of further defining the relationship of this bacterium to other chlamydia-like bacteria and to identify genetic loci that may be relevant to applied environmental studies of organism distribution.
Acknowledgments
We thank Oyvind Olauson and the technical managers of the salmon farms for specimen collection and for their patience and support of this project and its research team. We thank Julie Wen and Violet Han of the Electron Microscopy Laboratory of The University of Texas Medical Branch at Galveston for assistance with sample processing and sectioning for electron microscopy. We are grateful to Hugh W. Ferguson for his identification of sources representative of epitheliocystis disease, Julie Bebak-Williams for facilitating important collaborations, and Kathleen Nevis for her assistance with in situ hybridization.
This research was funded by grants from the University of Connecticut Research Foundation, the USDA Cooperative State Research, Education and Extension Service and Storrs Agricultural Experiment Station, and the Connecticut Sea Grant College Program.
REFERENCES
- 1.Amann, R. I., N. Springer, W. Schonhuber, W. Ludwig, E. N. Schmid, K. D. Muller, and R. Michel. 1997. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avakyan, A. A., and V. L. Popov. 1984. Rickettsiae and chlamydiae: comparative electron microscopic studies. Acta Virol. 28:159-173. [PubMed] [Google Scholar]
- 3.Bradley, T. M., C. E. Newcomer, and K. O. Maxwell. 1988. Epitheliocystis associated with massive mortalities of cultured lake trout Salvelinus namaycush. Dis. Aquat. Org. 4:9-17. [Google Scholar]
- 4.Brown, C. 1998. In situ hybridization with riboprobes: an overview for veterinary pathologists. Vet. Pathol. 35:159-167. [DOI] [PubMed] [Google Scholar]
- 5.Crespo, S., C. Zarza, F. Padrós, and M. Marín de Mateo. 1999. Epitheliocystis agents in sea bream Sparus aurata: morphological evidence for two distinct chlamydia-like developmental cycles. Dis. Aquat. Org. 37:61-72. [DOI] [PubMed] [Google Scholar]
- 6.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 7.Engel, J. N., and D. Ganem. 1987. Chlamydial rRNA operons: gene organization and identification of putative tandem promoters. J. Bacteriol. 169:5678-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett, K. D. E., and A. A. Andersen. 1997. The ribosomal intergenic spacer and domain I of the 23S rRNA gene are phylogenetic markers of Chlamydia spp. Int. J. Syst. Bacteriol. 47:461-473. [DOI] [PubMed] [Google Scholar]
- 9.Everett, K. D. E., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 11.Fritsche, T. R., M. Horn, M. Wagner, R. P. Herwig, K.-H. Schleifer, and R. K. Gautom. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 66:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukatsu, T., and N. Nikoh. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl. Environ. Microbiol. 64:3599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gan, F. Y., G. D. Luk, and M. S. Gesell. 1994. Nonradioactive in situ hybridization techniques for routinely prepared pathology specimens and cultured cells. J. Histotechnol. 17:313-319. [Google Scholar]
- 14.Groff, J. M., S. E. LaPatra, R. J. Munn, M. L. Anderson, and B. I. Osburn. 1996. Epitheliocystis infection in cultured white sturgeon (Acipenser transmontanus): antigenic and ultrastructural similarities of the causative agent to the chlamydiae. J. Vet. Diagn. Investig. 8:172-180. [DOI] [PubMed] [Google Scholar]
- 15.Hillis, D. M., and M. T. Dixon. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66:411-446. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman, G. L., C. E. Dunbar, K. Wolf, and L. O. Zwillenberg. 1969. Epitheliocystis, a new infectious disease of the bluegill (Lepomis macrochirus). Antonie Leeuwenhoek 35:146-158. [DOI] [PubMed] [Google Scholar]
- 17.Horn, M., M. Wagner, K.-D. Müller, E. N. Schmid, T. R. Fritsche, K.-H. Schleifer, and R. Michel. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231-1239. [DOI] [PubMed] [Google Scholar]
- 18.Ito, S., and Y. Rikihisa. 1981. Techniques for electron microscopy of rickettsiae, p. 213-227. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, N.Y.
- 19.Kahane, S., K. D. E. Everett, N. Kimmel, and M. G. Friedman. 1999. Simkania negevensis strain Z: growth, antigenic and genome characteristics. Int. J. Syst. Bacteriol. 49:815-820. [DOI] [PubMed] [Google Scholar]
- 20.Löbau, S., U. Mamat, W. Brabetz, and H. Brade. 1995. Molecular cloning, sequence analysis, and functional characterization of the lipopolysaccharide biosynthetic gene kdtA encoding 3-deoxy-alpha-d-manno-octulosonic acid transferase of Chlamydia pneumoniae strain TW-183. Mol. Microbiol. 18:391-399. [DOI] [PubMed] [Google Scholar]
- 21.McDowell, E. M., and B. F. Trump. 1976. Histologic fixatives suitable for diagnostic light and electron microscopy. Arch. Pathol. Lab. Med. 100:405-406. [PubMed] [Google Scholar]
- 22.Nylund, A., A. M. Kvenseth, and E. Isdal. 1998. A morphological study of the epitheliocystis agent in farmed Atlantic salmon. J. Aquat. Anim. Health 10:43-55. [Google Scholar]
- 23.Palumbi, S. R. 1996. Nucleic acids II: the polymerase chain reaction, p. 205-248. In D. M. Hillis, C. Moritz, and B. K. Mable (ed.), Molecular systematics, 2nd ed. Sinauer Associates, Inc., Sunderland, Mass.
- 24.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 25.Paperna, I. 1977. Epitheliocystis infection in wild and cultured sea bream (Sparus aurata) and grey mullets (Liza ramada). Aquaculture 10:169-176. [Google Scholar]
- 26.Pettersson, B., A. Andersson, T. Leitner, O. Olsvik, M. Uhlen, C. Storey, and C. M. Black. 1997. Evolutionary relationships among members of the genus Chlamydia based on 16S ribosomal DNA analysis. J. Bacteriol. 179:4195-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Relman, D. A. 1993. Universal bacterial 16S rDNA amplification and sequencing, p. 489-495. In D. H. Pershing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 28.Rourke, A. W., R. W. Davis, and T. M. Bradley. 1984. A light and electron microscopic study of epitheliocystis in juvenile steelhead trout, Salmo gairdneri Richardson. J. Fish Dis. 7:301-309. [Google Scholar]
- 29.Sheehan, D. Z., and B. B. Hrapchak. 1980. Theory and practice of histotechnology. Bettelle Press, Columbus, Ohio.
- 30.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Inc., Sunderland, Mass.
- 31.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolke, R. E., D. S. Wyand, and L. H. Khairallah. 1970. A light and electron microscopic study of epitheliocystis disease in the gills of Connecticut striped bass (Morone saxatilis) and white perch (Morone americanus). J. Comp. Pathol. 80:559-563. [DOI] [PubMed] [Google Scholar]