Abstract
Wild animals living close to cattle and pig farms (four each) were examined for verocytotoxin-producing Escherichia coli (VTEC; also known as Shiga toxin-producing E. coli). The prevalence of VTEC among the 260 samples from wild animals was generally low. However, VTEC isolates from a starling (Sturnus vulgaris) and a Norway rat (Rattus norvegicus) were identical to cattle isolates from the corresponding farms with respect to serotype, virulence profile, and pulsed-field gel electrophoresis type. This study shows that wild birds and rodents may become infected from farm animals or vice versa, suggesting a possible role in VTEC transmission.
The most important reservoir of verocytotoxin (VT)-producing Escherichia coli (VTEC) is considered to be ruminants, particularly cattle. However, VTEC (also known as Shiga toxin-producing E. coli [STEC]) strains have been isolated from a number of other domestic animals as well as wild-living animals and birds: e.g., goats, sheep, pigs, cats, dogs, deer, wild rabbits, birds, and rats (1-3, 5, 14, 15, 19). As most of these studies have only focused on one particular serotype of VTEC, O157:H7/H−, there is only limited knowledge of the occurrence of all VTEC serotypes in farm animals and wild animals.
The importance of wild-living animals for the transmission and/or persistence of VTEC within farms or between farms is unknown. The objective of the present study was to elucidate the possible role of wild animals in the transmission of VTEC. The study focused on wild birds, rodents, and insects in close contact with farm animals as carriers of VTEC. In addition, other animals kept on the farms were examined. Two types of farms, dairy cattle and pig farms, were included in the study. As cattle are known to be an important reservoir of VTEC, whereas pigs are rarely colonized with VTEC (except E. coli producing the subtype VT2e), the prevalence of VTEC is a priori expected to be lower in pigs than cattle. Likewise, if VTEC strains are transmitted from the farm animals to the wild fauna, VTEC would be expected less common in wild-living animals in and around pig farms than in animals in and around cattle farms.
Danish cattle and pig farms (four each) were sampled in August 2002 as part of a larger study investigating the presence of Salmonella (M. N. Skov, J. J. Madsen, C. Rahbek et al., unpublished data). All samples from a given location were obtained within 1 week. Wild-living animals were sampled: cloacal swab samples from wild birds (caught in mist-nets or by hand), fresh fecal samples from rodents (collected indoors in and around stables), and insects living in close contact with the production animals (pools of different species of flies collected by netting at or around production animals). Domestic animals were sampled: fecal samples from pigs or cattle (pooled fresh fecal samples from different places in the stables and representing different age groups), “pet animals ” (cats, dogs, and rabbits), and other animals on the farm (horses, hens, pigs, and cattle were classified as “other animals ” when only a few head were present on the farm). In total, 446 samples were analyzed, including 244 samples from wild birds, 10 samples from rats and mice, and 6 pools of insects (mostly Musca domestica and Stomoxys calcitrans) (Table 1). Twenty-four species of wild birds were represented: the four most common (24 to 63 birds each) were barn swallows (Hirundu rustica), tree sparrows (Passer montanus), house sparrows (Passer domesticus), and blackbirds (Turdus merula).
TABLE 1.
Numbers of samples positive in the vtx PCR screening and VTEC isolates obtained from these samples
| Farm | No. of PCR-positive samples/total [no. of isolates (no. of samples with isolates)]
|
|||||
|---|---|---|---|---|---|---|
| Production animals (cattle or pigs) | Birds | Rodents | Pet animals | Insects | Other animalsa | |
| Cattle | ||||||
| A | 4/20 [5 (1)] | 0/35 | 0/1 | |||
| B | 8/20 [12 (5)] | 1/32 [0] | 2/2 [1 (1)] | 0/2 | 0/1 | |
| C | 2/20 [6 (2)] | 1/28 [6 (1)] | 0/2 | |||
| D | 9/18 [0 (0)] | 0/30 | 0/1 | 0/11 | ||
| Total | 23/78c | 2/125 | 2/2 | 0/5 | 0/2 | 0/11 |
| Pigs | ||||||
| E | 1/20 [0] | 2/30 [0] | 0/2 | 0/2 | 0/1 | 1/3b [0] |
| F | 1/20 [0] | 0/30 | 0/4 | 0/2 | 0/1 | |
| G | 0/20 | 0/30 | 0/2 | 0/3 | 0/1 | 0/2 |
| H | 0/20 | 0/29 | 0/1 | |||
| Total | 2/80d | 2/119 | 0/8 | 0/7 | 0/4 | 1/5 |
Other animals included hens (2 samples), horses (2 samples), cattle (2 samples at a pig farm), and pigs (10 samples on a cattle farm).
The PCR-positive samples were from cows (categorized as “other animals” at a pig farm).
29.5%.
2.5%.
Samples were enriched in buffered peptone water overnight (37°C) and examined for vtx1 and vtx2 genes (not including vtx2e) by real-time PCR as described previously (12). Isolation of VTEC from PCR-positive samples was carried out by colony replication using hydrophobic-grid membrane filters and detection by hybridization with vtx probes (12). VTEC colonies were further characterized by O:H serotyping by the use of the full set of E. coli antisera (Statens Serum Institut, Copenhagen, Denmark) and real-time PCR detection of a number of virulence factors: VT-encoding genes (vtx1 and vtx2), genes of the pathogenicity island locus of enterocyte effacement (primarily eae encoding intimin and, if positive for eae, variants of eae, tir, and espD), the adhesin Saa (saa), and plasmid-borne factors (ehxA, katP, espP, and etpD) (12).
The genotypes of VTEC isolates from wild-living animals were compared to isolates from production animals from the same farm by pulsed-field gel electrophoresis (PFGE). The restriction enzyme was XbaI, and the protocol recommended by PulseNet was used (7). Thiourea (50 μM) was added to the running buffer (16) to prevent DNA degradation.
The results of the PCR screening for vtx-positive samples are summarized in Table 1. Production animals were vtx positive by PCR on all cattle farms (29% of pooled samples) and on two pig farms (2.5% of pooled samples; P < 0.001 by chi-square test). More samples from calves (11 of 26 [42%]) were vtx positive by PCR than cow samples (11 of 52 [21%]; P = 0.09 by continuity-corrected two-tailed chi-square test). A relatively high prevalence of VTEC was expected in cattle (3, 4). However, as the analyzed samples were pools of feces likely to represent several individual animals, the prevalence could not be directly compared to the prevalence obtained in other studies. The finding that calves had a higher VTEC prevalence than cows was in agreement with the findings of Wilson et al. (21) and a number of studies showing that the prevalence of VTEC O157 gradually decreased in older animals (5, 13). Only a few pig samples were positive in the PCR screening, and no isolates could be obtained from any of these samples, indicating that the concentration of VTEC probably was low.
Two wild birds from two cattle farms and two wild birds from one pig farm were PCR positive (two tree sparrows, one barn swallow, and one starling (S. vulgaris)). Overall, the frequency of VTEC-positive wild birds was low: 1.6%, as judged by the PCR screening. Two Norway rat (R. norvegicus) samples from the same cattle farm were PCR positive, giving a relatively high prevalence among all 10 rodent samples (20%). VTEC strains were not detected in any of the insect or pet samples in this study. VTEC strains have only rarely been detected in wild birds in other studies: e.g., VTEC was not detected in any of 86 gulls in Finland, but one pigeon isolate carried vtx2f (9), a variant of vtx that has been associated with pigeons (17), but not with human disease. In Japan, one of 50 gulls carried VTEC strains (11). VTEC O157 strains were isolated from gull droppings at a landfill site and a seashore in the United Kingdom (19), but not in Canada geese or gulls sampled in Sweden (18). In a study of VTEC O157 occurrence in rodents and other wild animals on cattle farms in the United States, Hancock et al. (8) did not isolate any VTEC O157 among 300 samples of rats, but Cizek et al. (5) isolated VTEC O157 from 4 out of 10 rats in the barns of a cattle farm in the Czech Republic. The occurrence of non-O157 VTEC in rodents has not previously been investigated.
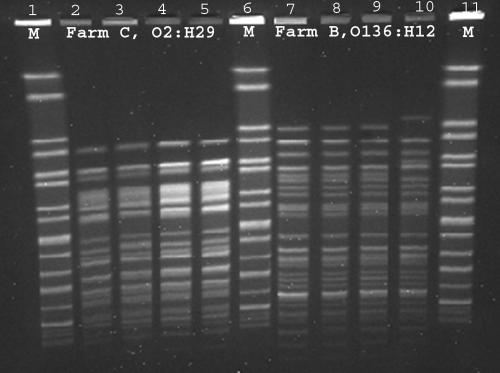
At least one VTEC isolate was obtained from 8 of the 14 PCR-positive cattle samples on three of the cattle farms, farms A, B, and C (1 to 5 isolates per sample). One VTEC isolate was obtained from one of the PCR-positive rat samples from farm B, and six VTEC isolates were obtained from the PCR-positive starling from farm C. No VTEC isolates were obtained from any samples taken on the pig farms (Table 1). The lack of success in isolating VTEC from all the PCR-positive samples can be explained by the high sensitivity of the PCR method as opposed to the general problem of isolating VTEC among the large and diverse E. coli flora of the intestine. The VTEC isolates were further characterized; the results are presented in Table 2. The rat isolate from farm B belonged to serotype O136:H12 and was positive for vtx1. This was also the case for seven of the isolates obtained from two of the pooled cattle fecal samples. PFGE typing of these isolates showed that the rat isolate had the same profile as six of the seven O136:H12 isolates from cattle (representative isolates shown in Fig. 1, lanes 7 to 9). The PFGE profile of the seventh O136:H12 cattle isolate differed only by two bands (Fig. 1; lane 10). Likewise, identical serotype (O2:H29) and virulence profiles (vtx2) were found for the six isolates from the starling from farm C as well as one of the cattle isolates from the same farm (Table 2). All O2:H29 isolates from farm C had identical PFGE profiles (Fig. 1, lanes 2 to 5). However, due to DNA degradation, the O2:H29 isolates did not result in a PFGE profile when the standard protocol was used. Addition of thiourea or HEPES to the running buffer has been recommended to prevent DNA degradation during electrophoresis (6, 10, 16). Here, addition of thiourea in the running buffer produced the profiles shown in Fig. 1.
TABLE 2.
Characterization of VTEC isolates with respect to O:H serotype, virulence factors, and PFGE type
| Farm | Animal | No. of isolates | Serotype | Virulence profilea | PFGE type |
|---|---|---|---|---|---|
| A | Cattle | 1 | O136:H12 | vtx1 | NDb |
| 3 | O116:H− | vtx2 | ND | ||
| 1 | Rough:H19 | vtx2 saa ehxA | ND | ||
| B | Cattle | 1 | O15:H16 | vtx2 espP | 4 |
| 2 | O116:H− | vtx2 | 2 | ||
| 1 | O116:H− | vtx2 | 6 | ||
| 6 | O136:H12 | vtx1 | 5 | ||
| 1 | O136:H12 | vtx1 | 3 | ||
| 1 | O172:H28 | vtx1 ehxA espP | 1 | ||
| Norway rat | 1 | O136:H12 | vtx1 | 5 | |
| C | Cattle | 1 | O2:H29 | vtx2 | 7c |
| 4 | O113:H− | vtx2 saa ehxA | 8 | ||
| Starling | 6 | O2:H29 | vtx2 | 7c |
Examined for the following factors: vtx1, vtx2, eae, saa, ehxA, katP, espD, and etpD.
ND, not determined.
A profile was only obtained when thiourea was added to the running buffer.
FIG. 1.
XbaI PFGE profiles of representative isolates from farm C (VTEC O2:H29 isolates) and from farm B (VTEC O136:H12 isolates). Lanes 1, 6, and 11, Salmonella enterica serovar Braenderup marker (M). Lane 2, VTEC O2:H29 isolate from cattle on farm C. Lanes 3 to 5, VTEC O2:H29 isolates from a wild bird sampled on farm C. Lane 7, VTEC O136:H12 isolate from a rat sampled on farm B. Lanes 8 to 10, VTEC O136:H12 isolates from cattle on farm B.
Despite the diversity of VTEC strains found in cattle, the VTEC isolates from the rat (farm B) and the starling (farm C) had serotype, virulence characteristics, and PFGE profiles indistinguishable from those of isolates obtained from cattle on the same farms. Likewise, Rice et al. (13) found indistinguishable PFGE types of VTEC O157 in cattle and deer sharing the same grazing area. Most examples of wild animals carrying VTEC have been animals living close to domestic animals (15, 20). It is therefore not likely that wild animals are important reservoirs of VTEC. However, it is possible that wild animals and birds act as vehicles for VTEC in transmission between farms or supporting the persistence of VTEC infections in domestic animals.
In conclusion, this study shows a low prevalence of VTEC in wild animals living in close proximity to cattle farms, but the positive findings of VTEC show that wild birds and rodents may become infected from farm animals or vice versa, suggesting a possible role in VTEC transmission.
Acknowledgments
We thank the farmers and their families, the team behind the project “Salmonella in the Wild Fauna,” the people collecting samples on location, and the laboratory technicians at the Danish Institute for Food and Veterinary Research and at Statens Serum Institut.
The project was financially supported by the Danish Ministry of Food, Fisheries and Agriculture (grant no. FØSI00-1).
REFERENCES
- 1.Asakura, H., S. Makino, T. Shirahata, T. Tsukamoto, H. Kurazono, T. Ikeda, and K. Takeshi. 1998. Detection and genetical characterization of Shiga toxin-producing Escherichia coli from wild deer. Microbiol. Immunol. 42:815-822. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, J. R., L. Warner, G. C. Pritchard, S. Williamson, T. Carson, G. Willshaw, T. Cheasty, and J. R. Bailey. 2002. Wild rabbits—a novel vector for Vero cytotoxigenic Escherichia coli (VTEC) O157. Commun. Dis. Public Health 5:74-75. [PubMed] [Google Scholar]
- 3.Beutin, L., D. Geier, H. Steinruck, S. Zimmermann, and F. Scheutz. 1993. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J. Clin. Microbiol. 31:2483-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco, M., J. E. Blanco, J. Blanco, A. Mora, C. Prado, M. P. Alonso, M. Mourino, C. Madrid, C. Balsalobre, and A. Juarez. 1997. Distribution and characterization of faecal verotoxin-producing Escherichia coli (VTEC) isolated from healthy cattle. Vet. Microbiol. 54:309-319. [DOI] [PubMed] [Google Scholar]
- 5.Cizek, A., P. Alexa, I. Literak, J. Hamrik, P. Novak, and J. Smola. 1999. Shiga toxin-producing Escherichia coli O157 in feedlot cattle and Norwegian rats from a large-scale farm. Lett. Appl. Microbiol. 28:435-439. [DOI] [PubMed] [Google Scholar]
- 6.Corkill, J. E., R. Graham, C. A. Hart, and S. Stubbs. 2000. Pulsed-field gel electrophoresis of degradation-sensitive DNAs from Clostridium difficile PCR ribotype 1 strains. J. Clin. Microbiol. 38:2791-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock, D. D., T. E. Besser, D. H. Rice, E. D. Ebel, D. E. Herriott, and L. V. Carpenter. 1998. Multiple sources of. Escherichia coli O157 in feedlots and dairy farms in the northwestern USA. Prev. Vet. Med. 35:11-19. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi, H., T. Pohjanvirta, and S. Pelkonen. 2002. Prevalence and characteristics of intimin- and Shiga toxin-producing Escherichia coli from gulls, pigeons and broilers in Finland. J. Vet. Med. Sci. 64:1071-1073. [DOI] [PubMed] [Google Scholar]
- 10.Koort, J. M. K., S. Lukinmaa, M. Rantala, E. Unkila, and A. Siitonen. 2002. Technical improvement to prevent DNA degradation of enteric pathogens in pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:3497-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino, S., H. Kobori, H. Asakura, M. Watarai, T. Shirahata, T. Ikeda, K. Takeshi, and T. Tsukamoto. 2000. Detection and characterization of Shiga toxin-producing Escherichia coli from seagulls. Epidemiol. Infect. 125:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen, E. M., and M. T. Andersen. 2003. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J. Clin. Microbiol. 41:2884-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen, E. M., C. Tegtmeier, H. J. Andersen, C. Gronbaek, and J. S. Andersen. 2002. Influence of age, sex and herd characteristics on the occurrence of Verocytotoxin-producing Escherichia coli O157 in Danish dairy farms. Vet. Microbiol. 88:245-257. [DOI] [PubMed] [Google Scholar]
- 14.Rice, D. H., D. D. Hancock, and T. E. Besser. 1995. Verotoxigenic E coli O157 colonisation of wild deer and range cattle. Vet. Rec. 137:524. [DOI] [PubMed] [Google Scholar]
- 15.Rice, D. H., D. D. Hancock, and T. E. Besser. 2003. Faecal culture of wild animals for Escherichia coli O157:H7. Vet. Rec. 152:82-83. [DOI] [PubMed] [Google Scholar]
- 16.Römling, U., and B. Tümler. 2000. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:464-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahlstrom, H., E. Tysen, E. E. Olsson, B. Brandstrom, E. Eriksson, T.Morner, and I. Vagsholm. 2003. Survey of Campylobacter species, VTEC O157 and Salmonella species in Swedish wildlife. Vet. Rec. 153:74-80. [DOI] [PubMed] [Google Scholar]
- 19.Wallace, J. S., T. Cheasty, and K. Jones. 1997. Isolation of vero cytotoxin-producing Escherichia coli O157 from wild birds. J. Appl. Microbiol. 82:399-404. [DOI] [PubMed] [Google Scholar]
- 20.Wasteson, Y., J. M. Arnemo, B. K. Johansen, L. Vold, S. D. Mathiesen, M. A. Olsen, O. Wiig, and A. E. Derocher. 1999. Analysis of faecal samples from wild animals for verocytotoxin producing Escherichia coli and E coli O157. Vet. Rec. 144:646-647. [DOI] [PubMed] [Google Scholar]
- 21.Wilson, J. B., S. A. McEwen, R. C. Clarke, K. E. Leslie, R. A. Wilson, D. Waltner-Toews, and C. L. Gyles. 1992. Distribution and characteristics of verocytotoxigenic Escherichia coli isolated from Ontario dairy cattle. Epidemiol. Infect. 108:423-439. [DOI] [PMC free article] [PubMed] [Google Scholar]