Abstract
The protistan parasite Perkinsus marinus is a severe pathogen of the oyster Crassostrea virginica along the east coast of the United States. Very few data have been collected, however, on the abundance of the parasite in environmental waters, limiting our understanding of P. marinus transmission dynamics. Real-time PCR assays with SybrGreen I as a label for detection were developed in this study for quantification of P. marinus in environmental waters with P. marinus species-specific primers and of Perkinsus spp. with Perkinsus genus-specific primers. Detection of DNA concentrations as low as the equivalent of 3.3 × 10−2 cell per 10-μl reaction mixture was obtained by targeting the multicopy internal transcribed spacer region of the genome. To obtain reliable target quantification from environmental water samples, removal of PCR inhibitors and efficient DNA recovery were two major concerns. A DNA extraction kit designed for tissues and another designed for stool samples were tested on environmental and artificial seawater (ASW) samples spiked with P. marinus cultured cells. The stool kit was significantly more efficient than the tissue kit at removing inhibitors from environmental water samples. With the stool kit, no significant difference in the quantified target concentrations was observed between the environmental and ASW samples. However, with the spiked ASW samples, the tissue kit demonstrated more efficient DNA recovery. Finally, by performing three elutions of DNA from the spin columns, which were combined prior to target quantification, variability of DNA recovery from different samples was minimized and more reliable real-time PCR quantification was accomplished.
Perkinsus species are parasites of marine molluscs that can have a severe pathogenic effect on their hosts and cause significant economic losses. One of the most detrimental species in this group is Perkinsus marinus (27), a parasite of the eastern oyster, Crassostrea virginica (Gmelin). Since the 1950s, P. marinus has been responsible for severe mortalities among C. virginica populations along the mid-Atlantic region of the United States (1, 7).
P. marinus is waterborne and directly transmitted from oyster to oyster (26). Experimentally, transmission of P. marinus is dose dependent and all three known parasite life stages, trophozoite, prezoosporangia, and zoospore, have been shown to induce infection in oysters (1, 26, 31, 41). However, studies of transmission dynamics in the environment have been hindered by the inability to detect free-living stages of the parasite in water. Traditionally, detection of P. marinus in oysters has involved histology or culture of oyster tissues in fluid thioglycolate medium (FTM) (33, 34), and the latter is still the most commonly used diagnostic test to determine infection prevalence and intensity in oysters. The major drawbacks of the FTM assay are its lack of species specificity and its inability to detect low-intensity infections corresponding to fewer than 1,000 P. marinus cells per g of wet oyster tissue (9). In addition, the FTM assay has not been adapted for detection of Perkinsus spp. in the environment.
A technique was previously developed that allows detection of Perkinsus spp. in the environment, but its specificity has been problematic. With polyclonal antibodies to P. marinus (15) and flow cytometry immunodetection, Ragone Calvo et al. (32) were able to estimate the environmental parasite abundance and examine its functional relationship with local oyster mortality and oyster infection acquisition. However, cross-reactivity of the polyclonal antibody with several free-living, phototrophic, and parasitic dinoflagellate species has been reported (8), potentially leading to overestimation of the parasite's abundance and demonstrating the limitation of the polyclonal antibody assay.
Specificity and sensitivity limitations associated with the FTM assay and immunoassays have been overcome with nucleic acid-based tools. Specific PCR primers have been developed for P. marinus (28, 35, 37), and some have been shown to have greater sensitivity than the FTM assay (37). These tools have also been used in attempts to quantify P. marinus in oyster tissue by semiquantitative PCR (28) or by quantitative competitive PCR (46). However, these methods have not been adapted for quantification of the parasite in the environment.
Real-time PCR with P. marinus-specific primers may be a powerful method to quantify P. marinus not only in its host but also in environmental waters. Compared to non-real-time “quantitative” PCR methods developed earlier (28, 46), the real-time systems enable continuous monitoring of PCR products during the exponential phase of quantification (44). Quantification during the logarithmic phase is more reflective of the initial target concentration than is quantification during the plateau phase, which is analyzed by endpoint PCR quantification methods. Moreover, real-time systems are considerably less time consuming than the previously developed methods. Real-time quantitative PCR is currently considered a powerful tool for determining genome numbers and quantifying levels of gene expression in parasites (3). It has also been used for quantification in the environment of various organisms such as bacteria (2, 23, 38), yeasts (5), dinoflagellates (4, 18), or protistan parasites (19, 21).
The objective of this study was to develop a real-time PCR assay to detect and quantify P. marinus in environmental water samples. Methods of DNA extraction from environmental water samples were optimized with two major concerns—optimizing DNA recovery and limiting the presence or the effect of inhibitors that can be present in environmental samples (43).
MATERIALS AND METHODS
Cultured Perkinsus sp. cells.
DNAs from P. marinus, P. chesapeaki, P. andrewsi, and P. atlanticus, which was recently synonymized with P. olseni (29), were isolated from clonal cultures. P. marinus cells were maintained in culture at the Virginia Institute of Marine Science in accordance with the method of La Peyre et al. (25). Cultures of P. chesapeaki and P. andrewsi were obtained from C. Dungan at the Maryland Department of Natural Resources Oxford laboratory (14) and from the American Type Culture Collection (13). P. atlanticus cultures were obtained from J. La Peyre at Louisiana State University (11).
PCR primers.
PCR primers were designed to target the multicopy internal transcribed spacer (ITS) region of the rRNA gene unit (Table 1). Primers PerkITS-85 and PerkITS-750 (PerkITS) were described by Casas et al. (12) and were designed to target the ITS region of all Perkinsus species except P. qugwadi. In the present study, primers PmarITS-70F and PmarITS-600R (PmarITS) were designed to target P. marinus (Table 1). Primers were designed by aligning Perkinsus sp. ITS sequences with the CLUSTAL W (39) algorithm in the MacVector 7.0 DNA sequence analysis software package (Accelerys, San Diego, Calif.). The following ITS sequences from P. marinus, P. chesapeaki, P. andrewsi, P. atlanticus, and P. mediterraneus previously published and deposited in the GenBank database were used: accession no. PAU07697, PMU07700, POU07701, PSU07698, and PSU07699 (17); accession no. AF140295 (36); accession no. AF252288 and AF102171 (13); accession no. AF091541, AF091542, AF126022, AF150988, AF150989, and AF150990 (S. I. Kotob et al., unpublished data); accession no. AF149876, AF150985, AF150986, and AF150987 (G. D. Brown et al., unpublished data); AF150990, AY295199, AY295198, AY295188, AY295186, AY295190, AY295185, AY295189, AY295192, AY295177, AY295195, AF150989, AY295178, AY295187, AY295193, AF150985, AY295180, AY295191, AY295179, Y295181, AY295194, AY295196, AY295184, AY295183, AY295182, and AY295197 (6); accession no. AF369967 to AF369979 (12); accession no. AF440464 to AF440471 (14); accession no. AF472517 to AF472523 (11); accession no. AF441207 to AF441218 (11); and accession no. AY487834 to AY487843 (10). Additional sequences available in the laboratory of K. S. Reece (unpublished data) were also added to the alignment and used to identify regions of unique sequences in the P. marinus ITS region.
TABLE 1.
Primers used in this study
| Forward primer | Sequence | Reverse primer | Sequence | Approximate size of amplification product (bp) | Reference |
|---|---|---|---|---|---|
| PerkITS-85 | CCGCTTTGTTTGGATCCC | PerkITS-750 | ACATCAGGCCTTCTAATGATG | 703 | 11 |
| PmarITS-70F | CTTTTGYTWGAGWGTTGCGAGATG | PmarITS-600R | CGAGTTTGCGAGTACCTCKAGAG | 509 | This study |
Enumeration of cultured P. marinus cells.
Media containing P. marinus cells were centrifuged for 10 min at 200 × g to pellet the cells without disrupting them. The supernatant was discarded, and cells were resuspended in 10 ml of new medium. Neutral red was added to a subsample of this culture, and stained parasite cells were counted under a microscope with a Hausser counting chamber. Eight cell counts were performed, and the mean cell concentration in the medium was calculated. On the basis of this value, subsamples were used to spike six replicates of ASW to obtain a final concentration of 100 cells/ml of water and a final volume of 100 ml (10,000 cells total per sample). The spiking of ASW with P. marinus cells allowed these DNAs to be treated (filtration, DNA extraction) as similarly as possible to an environmental water sample and to be used later to create standard curves. For each of the six replicates, the water was filtered onto a 47-mm-diameter, 3-μm-pore-size Nuclepore filter (Costar, Whatman, Clifton, N.J.) with a disposable apparatus (Nalgene Nunc International, Rochester, N.Y.) to minimize sample-to-sample contamination. The filter was placed in 180 μl of QIAGEN (Valencia, Calif.) lysis buffer with 20 μl (100 mg/ml) of proteinase K (QIAGEN) and incubated overnight at 55°C to lyse the cells for subsequent DNA extraction (see below).
DNA extraction methods.
During this study, two different extraction methods were tested on water samples from different origins. The first method was the QIAGEN DNeasy Tissue Kit (tissue kit) currently used in our laboratory to extract DNA from cell culture or tissues and for which DNA extraction was performed by following the manufacturer's protocol. The second method was the QIAGEN QIAamp DNA Stool Mini Kit (stool kit), involving InhibitEX tablets (QIAGEN) that adsorb inhibitory substances. Modifications of the manufacturer's protocol included a decrease by half of the volume of ASL buffer and use of half of an InhibitEX tablet. For this method, an overnight lysis step was added prior to following the manufacturer's stool kit protocol, which also included a lysis step.
For both protocols, DNA from the column was eluted by three to five buffer (AE; QIAGEN) loadings onto the column with 5 min of incubation before centrifugation. During this study, each eluate was kept in a separate tube and in some cases subsamples from the first three eluates were combined.
Standard PCR conditions.
Amplifications were done in 25-μl reaction mixtures with 10 to 50 ng of genomic DNA measured spectrophotometrically. Reagents were used as follows: 20 mM Tris-HCl (pH 8.4); 50 mM KCl; 1.5 mM MgCl2 (Invitrogen Corporation, Carlsbad, Calif.); 0.2 mM each dATP, dGTP, dCTP, and dTTP (Invitrogen); 25 pmol of each primer (Invitrogen); 0.625 U of Taq DNA polymerase (Invitrogen); and 0.2 mg of bovine serum albumin (Idaho Technology Inc., Salt Lake City, Utah) per ml. Bovine serum albumin was added because it has been shown to overcome inhibition of Taq DNA polymerase by substances commonly found in environmental samples (24). For PerkITS, the cycling parameters, a modified form of those of Casas et al. (12), were as follows: initial denaturation for 4 min at 94°C, followed by 40 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C and a final extension of 10 min at 72°C. For PmarITS, the cycling parameters were as follows: initial denaturation for 4 min at 94°C, followed by 40 cycles of 1 min at 94°C, 1 min at 57°C, and 3 min at 65°C and a final extension of 10 min at 65°C. Products were electrophoresed on 2% agarose (in 1× Tris-borate- EDTA) gels, stained with ethidium bromide, and then visualized with UV light.
Real-time PCR conditions.
Quantitative PCR was performed on the LightCycler (45) from Roche Diagnostics (Mannheim, Germany). Quantification of the amplified product was done on a cycle-by-cycle basis via the acquisition of a fluorescent signal generated by binding of the fluorophore SybrGreen I (Roche Diagnostics) to double-stranded DNA. The cycle number at which the fluorescence signal crosses a certain threshold (threshold cycle [CT] in correlation with the background fluorescence of the assay) was noted. This CT value is proportional to the logarithm of the target DNA concentration in the assay. From a dilution series of a DNA amount corresponding to a known concentration of cells, a standard curve was produced in which the CT was plotted versus the logarithm of the starting concentration of DNA corresponding to a known number of cells, with each cell containing multiple copies of the targeted ITS region (6, 14, 30). For determination of the signal corresponding to a particular number of cells, we assumed that the efficiency of extraction and recovery of DNA from a known (counted) number of cells was consistent and reproducible (see below).
The volume of the PCR mixture was 10 μl comprising 1× Fast Start Taq DNA polymerase mixture (Roche Diagnostics), 3 mM MgCl2, 0.5 μM concentrations of primers PerkITS-85 and PerkITS-750 (Invitrogen), and 1 μl of DNA extract. The same concentrations of MgCl2 and primers were found to be optimal for the primer pair PmarITS. For the real-time PCR assay, primers were high-performance liquid chromatography purified as recommended by Roche Diagnostics.
The amplification programs were performed as follows: (i) heating at 95°C for 10 min to activate the FastStart Taq DNA polymerase; (ii) 50 cycles of increasing the temperature 20°C/s to 95°C, holding the temperature at 95°C for 10 s, decreasing the temperature 20°C/s to 62°C for the primer pair PerkITS or to 69°C for the primer pair PmarITS, holding this temperature for 5 s, increasing the temperature 20°C/s to 72°C, and holding this temperature for 30 s for the primer pair PerkITS or for 25 s for the primer pair PmarITS. The fluorescent signal was collected at 80°C for the primer pair PerkITS or at 81°C for the primer pair PmarITS at the last step of each cycle to minimize the signal from nonspecific products, particularly from primer dimers. A melting curve was acquired by heating the product at 20°C/s to 95°C, cooling it at 20°C/s to 60°C, and slowly heating it at 0.1°C/s to 95°C with fluorescence collection at 0.1°C intervals. The standards, as well as the samples tested, were run in duplicate on the LightCycler instrument.
Amplification of the product was visualized in the quantification curve analysis. The specificity of the amplified products was confirmed by a melting curve analysis in which positive samples showed a specific peak with a melting temperature of approximately 84°C with the PerkITS primers and 83°C with the PmarITS primers. In some instances, specificity was further confirmed by analyzing the PCR products on 2% agarose gels as described above.
Specificity of the PCR assays.
The specificity of the standard and real-time PCR assays was analyzed by testing DNA extracted with the tissue kit from cultured cells of P. marinus, P. chesapeaki, P. andrewsi, and P. atlanticus. DNA samples extracted from flat oysters, Ostrea edulis, infected with P. mediterraneus (10) and from several dinoflagellate species (Hematodinium perezi, Amyloodinium occelatum, Pfiesteria piscicida, Pfiesteria shumwayae, Amphidinium carterae, Crypthecodinium cohnii, Karlodinium micrum, Peridinium foliaceum, and Prorocentrum micans) were also tested.
Serial dilution of DNA.
The DNAs obtained from the combined eluates following extraction were serially diluted 10-fold with the elution buffer (AE; QIAGEN), leading to a serial dilution of cells corresponding to a range of 33 × 100 down to 3 × 10−3 cell per μl, with each genome equivalent containing multiple copies of the rRNA gene complex with the ITS region (6, 14, 30). The calculated cell numbers assume minimal variability associated with total cell counts and DNA extraction efficiency (see below).
Sensitivity of the PCR assays and real-time PCR standard curves.
Serial dilution of P. marinus DNA was used to test the sensitivity of the standard and real-time PCR assays with both sets of primers, PerkITS and PmarITS. For conventional PCR, 0.25 μl of DNA from each dilution in the series was used in a reaction mixture, which led to final concentrations ranging from 8 × 100 to 8 × 10−3 cell per reaction mixture. Products were analyzed on 2% agarose gels as described above. For real-time PCR, 1 μl of DNA was used per reaction mixture, leading to concentrations corresponding to 33 × 100 to 3 × 10−3 cell per reaction mixture. DNA amplification was visualized on the fluorescence graph displayed by the quantification analysis screen on the LightCycler system, and the specificity of the amplified product was verified by melting curve analysis.
For both primer pairs, real-time PCR standard curves were obtained in which the measured CT of each sample was plotted against the logarithm of the starting concentration of DNA corresponding to an estimated cell number. The largest amount of target DNA (or the corresponding number of cells) resulted in the lowest CT. Each dilution was analyzed by real-time PCR in duplicate with each of the primer pairs, PerkITS or PmarITS. The values used for the standard curves correspond to the mean of the three replicate ASW samples whose DNA was extracted with either the tissue kit or the stool kit.
Optimization of DNA extraction from environmental water samples.
Because environmental water samples can potentially contain inhibitors affecting DNA extraction or subsequent PCR amplification (43), the tissue kit and the stool kit were compared for DNA extraction efficiency, i.e., the quality and recovery of DNA. They were tested on ASW and environmental water samples spiked with equivalent numbers of P. marinus cells. ASW prepared in the laboratory was included in this study to test DNA extraction efficiency when the sample presumably contained no inhibitors. The two environmental water samples analyzed were collected in the James River, Virginia, at two different sites, Point of Shoals (PTS) and Deep Water Shoals (DWS), in October 2003. For each DNA extraction method, three replicates of 100 ml of each type of water (ASW, PTS, and DWS) were spiked with P. marinus cells to obtain a final concentration of 100 cells per ml or a total of 10,000 cells per sample. For the samples treated with the stool kit, a concentration of 20 cells per ml of water was also tested. An additional 500 ml of environmental water was sampled and tested by the real-time PCR technique developed in this study to ensure the absence of P. marinus DNA in the water samples to use for these optimization studies.
In order to test DNA recovery from the QIAGEN columns with the different water sample types, DNA was eluted by five consecutive buffer (AE) loadings onto the column with a 5-min incubation before centrifugation. Each eluate was kept in a separate tube. Samples were analyzed in duplicate by real-time PCR with PmarITS primers. In addition, for each replicate and each water type, DNA subsamples of equal volume from the first three elutions were mixed and analyzed in duplicate by real-time PCR with PmarITS primers. For those samples, four real-time PCR runs were performed for each sample to test the reproducibility of the real-time PCR technique.
For each type of water analyzed and each replicate, the CT values were measured with the LightCycler. To make the results more easily visualized, the corresponding cell concentration per reaction mixture was calculated. This calculation was done by using the standard curve regression formulas obtained with PmarITS primers on DNA extracted with the tissue or the stool kit (where y is the CT value and x is the logarithm of the cell concentration).
Effect of background DNA on P. marinus quantification.
Two DNA samples corresponding to two concentrations of P. marinus cells (52 cells and 5 cells per μl of elution buffer) were obtained from a serial dilution of P. marinus DNA extracted with the tissue kit as described above. These DNA samples were diluted 1:20 by using them to spike five different solutions: the elution buffer, two DNA samples isolated from the oyster C. virginica, and two DNA samples isolated from environmental water samples. After dilution, the final P. marinus DNA concentrations in the solutions corresponded to cell concentrations of 2.6 or 0.2 cell per μl.
For the C. virginica DNA solutions, DNA was extracted from whole individuals (spat) with the tissue kit in accordance with the manufacturer's protocol. The DNA concentration was adjusted to 50 ng/μl. The environmental samples were obtained from two different sites (PTS and DWS) in the James River sampled in June 2003. Filtration of the water and DNA extraction with the stool kit were performed as described above. Spiked DNA solutions were analyzed in duplicate by real-time PCR with the PmarITS primers.
Statistical analysis.
CT values from the standard curves obtained as described in the section on the sensitivity of the PCR assays and real-time PCR standard curves were statistically analyzed by one-way analysis of variance (ANOVA) to determine if there was a significant difference (α = 0.05) between the CT values obtained by the two DNA extraction methods tested.
CT values, as well as calculated P. marinus cell concentrations obtained after combining the first three elutions as described in the section on optimization of DNA extraction from environmental water samples, were statistically analyzed to test the effect of the DNA extraction method and of the water origin on target DNA quantification. The whole data set was analyzed with a two-way ANOVA to test the significance (α = 0.05) of the differences in target DNA concentration depending on the type of DNA extraction method used (tissue or stool) or the type of water analyzed (ASW, PTS, or DWS).
CT values measured after using two concentrations of P. marinus DNA to spike different background DNA as described in the section on the effect of background DNA on P. marinus quantification were statistically analyzed by two-way ANOVA to test the significance (α = 0.05) of CT differences depending on the background DNA or the P. marinus DNA concentration.
RESULTS
Specificity of the PCR assays.
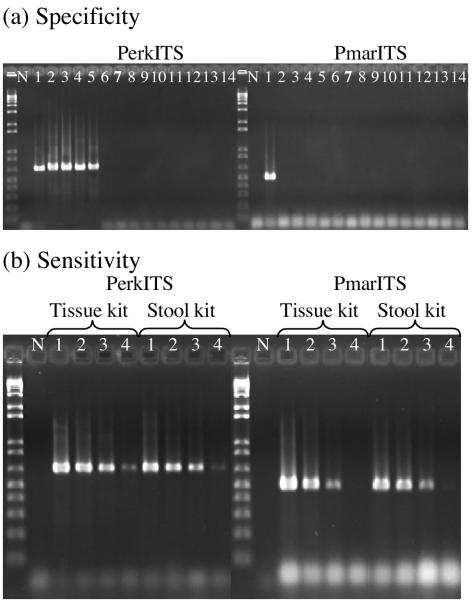
The standard (Fig. 1a) and real-time PCR assays showed equivalent specificities for the two primer pairs tested. The primer pair PerkITS amplified the DNA from P. marinus, P. chesapeaki, P. andrewsi, P. atlanticus, and P. mediterraneus, but no amplification was observed for the dinoflagellates H. perezi, A. occelatum, P. piscicida, P. shumwayae, A. carterae, C. cohnii, K. micrum, P. foliaceum, and P. micans. The primer pair PmarITS amplified only P. marinus DNA.
FIG. 1.
Standard PCR results. (a) Specificity of the PerkITS and PmarITS primers tested on no DNA (lane N) and P. marinus (lane 1), P. chesapeaki (lane 2), P. andrewsi (lane 3), P. atlanticus (lane 4), P. mediterraneus (lane 5), H. perezi (lane 6), A. occelatum (lane 7), P. piscicida (lane 8), P. schumwayae (lane 9), A. carterae (lane 10), C. cohnii (lane 11), K. micrum (lane 12), P. foliaceum (lane 13), and P. micans (lane 14) DNAs. (b) Sensitivity of PerkITS and PmarITS primers tested on DNA extracted with either the tissue or the stool kit and no DNA (N) and serial dilutions of P. marinus DNA corresponding to 8 × 100 (lane 1), 8 × 10−1 (lane 2), 8 × 10−2 (lane 3), and 8 × 10−3 cell per reaction mixture (lane 4).
Standard PCR sensitivity.
Strong amplification of P. marinus DNA was observed in a standard PCR assay with DNA concentrations as low as the equivalent of 8 × 10−2 cell per 25-μl reaction mixture with both sets of primers and with either DNA extraction method, the tissue kit or the stool kit (Fig. 1b). The ability to detect this very small amount of ITS region DNA, which corresponds to less than a single cell, can be attributed to the fact that there is evidence from many studies for multiple copies of the rRNA gene complex, including the ITS region, in the nuclear genome of each Perkinsus cell (6, 14, 30). Therefore, although the total amount of DNA in the reaction mixture is not equal to the amount of DNA in a complete Perkinsus sp. genome, there can still be several copies of ITS present in a subsample of the sheared genomic DNA. Weak amplification was observed for 8 × 10−3 cells per reaction mixture with the primer pair PerkITS, whereas at this concentration very light or no amplification was observed with the primer pair PmarITS.
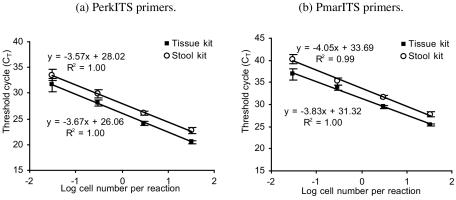
Real-time PCR sensitivity, standard curves.
Target DNA was detected in a real-time PCR assay with both primer sets and with either DNA extraction method with DNA concentrations as low as the equivalent of 3.3 × 10−2 cells per 10-μl reaction mixture for all of the replicates tested (Fig. 2). This indicated that even fewer copies of the ITS region DNA could be detected with the real-time assays compared to the standard PCR assays. A slight increase in the standard deviation was noted for both primer pairs and for concentrations of DNA corresponding to less than 3.3 × 10−1 cells per reaction mixture. This was due to a lack of consistency in the amplification and to an increase in primer dimers as observed on the melting peak analysis (data not shown). For both primer pairs, amplification of the target DNA concentration corresponding to 3.3 × 10−3 cells per reaction mixture was observed in one of the water sample replicates extracted with the tissue kit, while no amplification at this DNA concentration was observed for samples extracted with the stool kit. Because of the lack of reproducibility observed for the equivalent of 3.3 × 10−3 cells per reaction mixture, the corresponding CT values were not included in the standard curves presented in Fig. 2.
FIG. 2.
Standard curves to quantify Perkinsus spp. and P. marinus in environmental samples by real-time PCR with PerkITS and PmarITS primers, respectively. The standard curves correspond to the CT versus the logarithm of the estimated cell concentration in the sample. The DNA concentrations tested were obtained by performing 10-fold serial dilution of DNA extracted with either the tissue or the stool kit after spiking ASW with cultured P. marinus cells. Each value corresponds to the mean of three replicate DNA samples for each dilution and the respective standard deviation. After the dilution of P. marinus DNA, the corresponding concentrations of cells per reaction mixture ranged from 33 × 100 to 3 × 10−2 cell per real-time PCR, with each cell containing multiple ITS copies.
For the same concentration of cells, the CT value obtained when the DNA was extracted with the tissue kit was significantly smaller (P < 0.0001) than that obtained when the DNA was extracted with the stool kit (Fig. 2). Because CT values are negatively correlated with the target concentration, this demonstrated that for the same cell concentration, more DNA is recovered when DNA is extracted with the tissue kit than when it is extracted with the stool kit.
Optimization of DNA extraction.
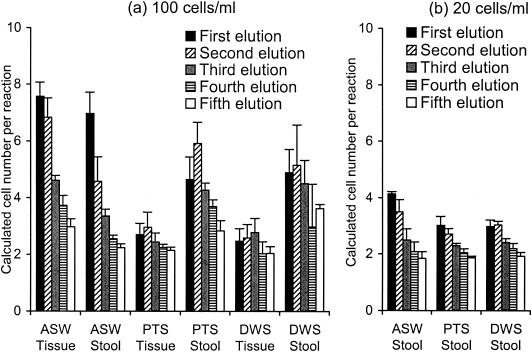
Recovery of DNA from the QIAGEN column after the DNA extraction step, with the tissue kit or the stool kit, was studied by performing five consecutive elutions on extracts from water samples spiked with P. marinus cells. The calculated concentrations of P. marinus cells per reaction mixture are shown in Fig. 3.
FIG. 3.
Recovery of DNA from the QIAGEN columns used with the tissue kit and the stool kit. Three replicates of each type of water sample (ASW, PTS, or DWS) were spiked with two P. marinus cell concentrations, 100 (a) and 20 (b) cells/ml of water. After each of the five elutions was performed, the mean cell concentration per reaction mixture and the standard deviation were calculated on the basis of the P. marinus DNA concentration, the CT values, and the regression formulas for the PmarITS primers.
DNA from P. marinus was still detectable by real-time PCR with PmarITS primers after the fifth elution independent of the type of water sample or the DNA extraction method (Fig. 3). For the ASW samples spiked with 10,000 cultured P. marinus cells, the amount of DNA recovered decreased with each elution (Fig. 3a). For the environmental samples, however, most of the time the concentration of DNA recovered from the second and third elutions usually was greater than or close to that obtained with the first elution. For the samples spiked with cultured P. marinus cells to a final concentration of 20 cells per ml and extracted with the stool kit (Fig. 3b), the ASW and PTS environmental water samples showed a decrease in the cell concentrations from the first elution to the third elution, while for the other environmental sample, DWS, the greatest recovery was obtained from the second elution.
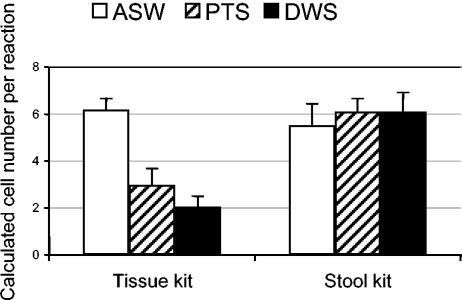
Considering the inconsistency in DNA recovery with subsequent elutions, the first three eluates were combined to minimize the variability in DNA recovery from sample to sample (Fig. 4). Statistical analysis of the data obtained on the basis of either the CT values or the corresponding calculated P. marinus cell concentration demonstrated the same level of significance. Differences in calculated P. marinus cell concentrations observed with the type of DNA extraction kit used or with the type of water spiked were significant (P < 0.0001). The interaction between the method of DNA extraction and the type of water was also significant (P < 0.0001). When analyzing each type of water separately, the calculated cell concentrations were significantly lower (P = 0.0018) when the spiked ASW samples were extracted with the stool kit than when they were extracted with the tissue kit. On the other hand, for PTS and DWS environmental samples, the calculated cell concentrations were significantly higher (P < 0.0001) when DNA was extracted with the stool kit than when it was extracted with the tissue kit (Fig. 4). With the stool kit, no significant difference (P = 0.1451) in calculated cell concentrations was observed between the ASW and the environmental seawater samples. The same result was observed when the stool kit was used on water samples spiked with a lower concentration of cells (20/ml; data not shown). With the tissue kit, however, cell concentrations in ASW samples were significantly higher (P < 0.0001) than in either PTS or DWS and the cell concentration in PTS was significantly higher (P = 0.0004) than that in DWS. Finally, cell concentrations in ASW extracted with the tissue kit were not significantly different from cell concentrations in the PTS (P = 0.0755) or DWS (P = 0.0828) samples extracted with the stool kit.
FIG. 4.
Quantification of P. marinus cell concentrations (cells per reaction mixture) after combining the first three elutions from DNA samples extracted with the tissue kit or the stool kit. Samples were obtained by spiking ASW or two environmental waters samples (PTS and DWS) with cultured P. marinus cells with a final concentration of 100 cells/ml of water. For each type of water and each cell concentration, three replicates were analyzed and the means and standard deviations are presented.
Effect of background DNA.
For the final concentration of P. marinus cells per reaction mixture equivalent to 2.6, the observed CT values obtained after the spiking of elution buffer, C. virginica DNA, or the environmental water DNA with P. marinus DNA were very similar (Table 2) and not significantly different (P = 0.2058) from each other. The CT values obtained by spiking with a lower concentration of P. marinus DNA (corresponding to 0.2 cell per reaction mixture) were also not significantly different (P = 0.6850).
TABLE 2.
Effect of background DNA on quantification of P. marinus DNA with PmarITSa
| Final P. marinus concn (cells/reaction mixture) | CT measured after spiking of background solution with P. marinus DNA
|
||||
|---|---|---|---|---|---|
| Elution buffer | C. virginica 1 | C. virginica 2 | PTS | DWS | |
| 2.6 | 33.21 ± 0.25 | 33.37 ± 0.31 | 33.29 ± 0.88 | 33.95 ± 0.63 | 33.94 ± 0.47 |
| 0.26 | 36.83 ± 0.79 | 37.13 ± 1.4 | 37.77 ± 1.12 | 37.55 ± 0.69 | 37.71 ± 1.14 |
Mean CT values and standard deviations were measured by real-time PCR after using two DNA samples corresponding to two P. marinus cells/μl of elution buffer (QIAGEN) to spike five different background solutions: elution buffer, two DNAs from C. virginica individuals, and two DNAs from environmental water samples (PTS, DWS).
DISCUSSION
Reaction conditions (primer and MgCl2 concentrations, annealing temperature) for the PerkITS and PmarITS primers were optimized on the LightCycler for real-time PCR to obtain specificity for the genus Perkinsus and for P. marinus, respectively. With both sets of primers, the sensitivity obtained with the real-time PCR assay was comparable to the sensitivity obtained with the standard PCR assay. The target DNA could be reliably detected and quantified for DNA concentrations as low as the equivalent of 3.3 × 10−2 cell per 10-μl reaction mixture, suggesting that there are hundreds to thousands of copies of the rRNA gene complex region (i.e., the ITS target) in the P. marinus genome. In practice, in order to reduce the cost of a study, this result suggests that when unknown samples have to be quantified it is important to first test them with a standard PCR assay to determine the presence or absence of P. marinus and then to run a real-time PCR on positive samples only.
For the real-time PCR assay, the double-stranded DNA intercalator dye SybrGreen I was used as a detection system. The other common real-time PCR detection system currently in use involves a sequence-specific probe(s) designed to specifically hybridize within the amplified fragment (22, 44). The major advantage of the SybrGreen I system resides in the application of primers designed for a standard PCR to the real-time PCR system, precluding the need to design new primers and probes. The sensitivities of the SybrGreen I and probe systems have been shown to be similar, and the main differences between the two systems reside in their levels of specificity (20, 23, 44). SybrGreen I is more tolerant of polymorphic targets than is the probe system, which can be advantageous with environmental samples in which the targeted organism may present slight genetic strain variation. With the SybrGreen I system, the melting curve analysis allows confirmation that the targeted region is amplified. One of the major limitations of the SybrGreen I system resides in its binding to any double-stranded DNA, potentially leading to overestimation of the quantity of target DNA. One source of nonspecific double-stranded product fluorescence is the primer dimers. In this study, primer dimers could be visualized in the melting curve analysis and showed a melting temperature (76°C with PerkITS and 78°C with PmarITS) lower than that of the targeted PCR product (83°C for PerkITS and 84°C for PmarITS). To minimize the increase in fluorescence due to primer dimers, fluorescence measurements were performed after an additional step that was added to the typical PCR cycles (denaturation, annealing of primers, and DNA elongation). This extra step consisted of a few seconds at a temperature higher (80°C for PerkITS and 81°C for PmarITS) than the melting temperature of primer dimers but lower than that of the targeted PCR product so that primer dimers melted while the specific PCR product remained double stranded. If nonspecific products with a melting temperature higher than that of the fourth step were produced, overestimation of the amount of target DNA in the sample could occur because of measurement of the fluorescence from the nonspecific product along with the fluorescence from the target. However, these types of nonspecific products were very rarely observed during this study.
Efficient and consistent recovery of DNA during the DNA extraction procedure is necessary in order to obtain reliable relative quantification of target DNA in environmental samples. In the present study, as in most of the published studies involving quantitative PCR, the DNA extraction procedure involves a column to which the DNA is bound while washes to eliminate contaminants are performed. DNA is subsequently eluted from the column with an elution buffer. As the quality and quantity of background compounds in environmental samples can vary, one of the objectives of this study was to determine if the differences among environmental samples could affect the efficiency of the extraction procedure, particularly the elution step. We found that for samples spiked with the same number of cells, differences were observed in the DNA recovery trends between the ASW and environmental samples after consecutive elutions. With both of the DNA extraction kits tested, spiked ASW samples showed a decrease in the calculated cell number per reaction mixture from the first elution to the fifth, as we had expected. On the other hand, the second and third elutions of some of the spiked environmental samples showed greater DNA recovery than the first elution. This was particularly evident when the water samples were spiked with a large number of cells to obtain a final concentration of 100 P. marinus cells per ml or a total amount of 10,000 cells per 100-ml sample. In this study, because variability in DNA recovery was primarily observed among the first three elutions, these eluates were subsequently combined for the quantification assays.
Differences in recovery efficiency observed between ASW and environmental water samples can be explained by the presence of various environmental compounds and other organisms in the environmental water samples. Thus, even if the amount of DNA from the P. marinus cells per sample remained in the range recommended by the manufacturer, overloading of the QIAGEN column might have occurred even with only 100 ml of filtered environmental water. Compared to the stool kit, when DNA was extracted with the tissue kit, significant differences in the quantification of P. marinus were observed between different water types. Environmental samples showed significantly lower cell numbers per reaction mixture than did ASW samples. This can be explained by the presence of PCR-inhibitory compounds in the environmental samples that may not have been eliminated during extraction with the tissue kit. Inhibitory compounds can interfere with several steps in the isolation and amplification protocols, including cell lysis, nucleic acid capture onto the column, and polymerase activity during the amplification of target DNA, or these compounds may even degrade the DNA if not eliminated during extraction (43). These inhibitors can cause the PCR to fail completely or result in an overall reduced sensitivity, depending on the type and quantity of the inhibiting compound. The identities of those compounds and their mode of action remain poorly understood. Although they have not been extensively studied, environmental samples have been found to contain phenolic compounds, clay particles, humic and fulvic acids, and heavy metals, all of which have been identified as PCR inhibitors (16, 24, 40, 42, 43). The impact of inhibitors on results obtained by real-time PCR, where quantification is desired, is potentially more detrimental than with a standard PCR, in which only the presence or absence of target DNA is assessed. With a standard PCR, as long as the inhibition does not induce complete failure of the reaction, the effect on the result is limited. With a quantitative PCR, however, reliable quantification of the target and sample-to-sample comparisons can be affected by the relative quality and quantity of inhibitory compounds in each sample.
To summarize the results of this study, the stool kit was more efficient than the tissue kit at removing inhibitory substances, as demonstrated by the higher calculated cell concentrations found in spiked environmental water samples extracted with the stool kit compared to those samples with the same cell concentration extracted with the tissue kit. Apparently, environmental inhibitors and background DNA did not have a significant effect on quantification of P. marinus DNA when the sample was extracted with the stool kit, in that nearly identical cell concentration values were obtained with ASW or environmental water samples spiked with equivalent numbers of cells. However, when the water contained no inhibitory substances (ASW), the tissue kit allowed significantly greater overall DNA recovery than the stool kit did. In addition, our results suggest that the tissue kit is preferred for extracting DNA from infected oysters, as no inhibitory effect was observed when P. marinus DNA was used to spike C. virginica DNA extracted with the tissue kit.
Real-time PCR can be used for determining the relative abundance of P. marinus in the environment; however, its absolute abundance may not be accurately measured and interpretation of P. marinus environmental abundance data should take into account the following limitations of the technique. The standard curves were developed with cultured cells that are potentially different from the cells that are found in water samples in the field. The ploidy of the different stages of the Perkinsus spp. is not known and might vary with different life stages. Diploidy has been postulated for cultured P. marinus cells (36), but more studies are needed to explore the ploidy of zoospores that may be found in environmental waters. Finally, the real-time PCR system developed in this study allows the detection of P. marinus DNA associated with viable and nonviable cells, as well as infective and noninfective cells. The development of a real-time PCR assay, along with optimization of the DNA extraction method for environmental water samples, however, offers new opportunities for water monitoring and for new studies such as those examining transmission dynamics, which require quantification of P. marinus in environmental water samples. Studies to examine correlations between the abundance of the parasite in the water column and environmental parameters, as well as the prevalence and intensity of the parasite infection in the host C. virginica and the acquisition of infection by naive oysters, are ongoing.
Acknowledgments
We thank N. Stokes and L. Ragone Calvo for valuable comments during the study. We also thank C. Dungan and J. La Peyre for providing the P. chesapeaki and P. atlanticus cultures, W. Litaker and M. Vanderdsea for providing A. ocellatum DNA, and D. Evans for comments on statistical analysis.
This research was funded in part by the NOAA Sea Grant Oyster Disease Research Program (grant NA16RG2207).
Footnotes
This report is Virginia Institute of Marine Science contribution number 2625.
REFERENCES
- 1.Andrews, J. D. 1988. Epizootiology of the disease caused by the oyster pathogen Perkinsus marinus and its effect on the oyster industry. Am. Fish. Soc. Spec. Publ. 18:47-63. [Google Scholar]
- 2.Bach, H.-J., J. Tomanova, M. Scholter, and J. C. Munch. 2002. Enumeration of total bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J. Microbiol. Methods 49:235-245. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A. B., and L. C. Ranford-Cartwright. 2002. Real-time quantitative PCR in parasitology. Trends Parasitol. 18:337-342. [PubMed] [Google Scholar]
- 4.Bowers, H. A., T. Tengs, H. B. Glasgow, J. R., J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinkman, N. E., R. A. Haugland, L. J. Wymer, M. Byappanahalli, R. L. Whitman, and S. J. Vesper. 2003. Evaluation of a rapid, quantitative real-time PCR method for enumeration of pathogenic Candida cells in water. Appl. Environ. Microbiol. 69:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, G. D., K. L. Hudson, and K. S. Reece. 2004. Multiple polymorphic sites at the ITS and ATAN loci in cultured isolates of Perkinsus marinus. J. Eukaryot. Microbiol. 51:312-320. [DOI] [PubMed] [Google Scholar]
- 7.Burreson, E. M., and L. M. Ragone Calvo. 1996. Epizootiology of Perkinsus marinus disease of oysters in Chesapeake Bay, with emphasis on data since 1985. J. Shellfish Res. 15:17-34. [Google Scholar]
- 8.Bushek, D., C. F. Dungan, and A. J. Lewitus. 2002. Serological affinities of the oyster pathogen Perkinsus marinus (Apicomplexa) with some dinoflagellates (Dinophyceae). J. Eukaryot. Microbiol. 49:11-16. [DOI] [PubMed] [Google Scholar]
- 9.Bushek, D., S. E. Ford, and S. K. Allen, Jr. 1994. Evaluation of methods using Ray's fluid thioglycollate medium for diagnosis of Perkinsus marinus infection in the eastern oyster, Crassostrea virginica. Annu. Rev. Fish Dis. 4:201-217. [Google Scholar]
- 10.Casas, S. M., A. Grau, K. S. Reece, K. Apakupakul, C. Azevedo, and A. Villalba. 2004. Perkinsus mediterraneus n. sp., a protistan parasite of the European flat oyster Ostrea edulis from the Balearic Islands, Mediterranean Sea. Dis. Aquat. Org. 58:231-244. [DOI] [PubMed] [Google Scholar]
- 11.Casas, S. M., J. F. La Peyre, K S. Reece, C. Azevedo, and A. Villalba. 2002. Continuous in vitro culture of the carpet shell clam Tapes decussatus protozoan parasite Perkinsus atlanticus. Dis. Aquat. Org. 52:217-231. [DOI] [PubMed] [Google Scholar]
- 12.Casas, S. M., A. Villalba, and K. S. Reece. 2002. Study of perkinsiosis in the carpet shell clam Tapes decussatus in Galicia (NW Spain). I. Identification of the aetiological agent and in vitro modulation of zoosporulation by temperature and salinity. Dis. Aquat. Org. 50:51-65. [DOI] [PubMed] [Google Scholar]
- 13.Coss, C. A., J. A. F. Robledo, G. M. Ruitz, and G. R. Vasta. 2001. Description of Perkinsus andrewsi n. sp. isolated from the Baltic clam (Macoma balthica) by characterization of the ribosomal RNA locus, and development of a species-specific PCR-based diagnostic assay. J. Eukaryot. Microbiol. 48:52-61. [DOI] [PubMed] [Google Scholar]
- 14.Dungan, C. F., R. M. Hamilton, K. L. Hudson, C. B. McCollough, and K. S. Reece. 2002. Two epizootic diseases in Chesapeake Bay commercial clams, Mya arenaria and Tagelus plebeius. Dis. Aquat. Org. 50:67-78. [DOI] [PubMed] [Google Scholar]
- 15.Dungan, C. F., and B. S. Roberson. 1993. Binding specificities of mono- and polyclonal antibodies to the protozoan oyster pathogen Perkinsus marinus. Dis. Aquat. Org. 15:9-22. [Google Scholar]
- 16.Frostegard, A., S. Courtois, V. Ramisse, S. Clerc, D. Bernillon, F. Le Gall, P. Jeannin, X. Nesme, and P. Simonet. 1999. Quantification of bias to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goggin, C. L. 1994. Variation in the two internal transcribed spacers and 5.8 ribosomal RNA from five isolates of the marine parasite Perkinsus (Protista, Apicomplexa). Mol. Biochem. Parasitol. 65:179-182. [DOI] [PubMed] [Google Scholar]
- 18.Gray, M., B. Warwick, J. Paul, and E. Casper. 2003. Molecular detection and quantitation of the red tide dinoflagellate Karenia brevis in the marine environment. Appl. Environ. Microbiol. 69:5726-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guy, R. A., P. Payment, U. J. Krull, and P. A. Horgen. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time PCR and application of this technique for examination of cheeses. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins, J. A., R Fayer, J. M. Trout, L. Xiao, A. A. Lal, S. Kerby, and M. C. Jenkins. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J. Microbiol. Methods 47:323-337. [DOI] [PubMed] [Google Scholar]
- 22.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreader, C. A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Peyre, J. F., M. Faisal, and E. M. Burreson. 1993. In vitro propagation of the protozoan Perkinsus marinus, a pathogen of the eastern oyster, Crassostrea virginica. J. Eukaryot. Microbiol. 40:304-310. [Google Scholar]
- 26.Mackin, J. G. 1962. Oyster diseases caused by Dermocystidium marinum and other microorganisms in Louisiana. Publ. Inst. Mar. Sci. Univ. Tex. 7:132-229. [Google Scholar]
- 27.Mackin, J. G., H. M. Owen, and A. Collier. 1950. Preliminary note on the occurrence of a new protistan parasite, Dermocystidium marinum n. sp. in Crassostrea virginica (Gmelin). Science 111:328-329. [DOI] [PubMed] [Google Scholar]
- 28.Marsh, A. G., J. D. Gauthier, and G. R. Vasta. 1995. A semiquantitative PCR assay for assessing Perkinsus marinus infections in the eastern oyster, Crassostrea vriginica. J. Parasitol. 81:577-583. [PubMed] [Google Scholar]
- 29.Murrell, A., S. N. Kleeman, S. C. Barker, and R. J. G. Lester. 2002. Synonymy of Perkinsus olseni Lester & Davis, 1981 and Perkinsus atlanticus Azevedo, 1989 and an update on the phylogenetic position of the genus Perkinsus. Bull. Eur. Assoc. Fish Pathol. 22:258-265. [Google Scholar]
- 30.Pecher, W. T., J. A. F. Robledo, and G. R. Vasta. 2004. Identification of a second rRNA gene unit in the Perkinsus andrewsi genome. J. Eukaryot. Microbiol. 51:234-245. [DOI] [PubMed] [Google Scholar]
- 31.Perkins, F. O. 1988. Parasite morphology, strategy, and evolution. Structure of protistan parasites found in bivalves molluscs. Am. Fish. Soc. Spec. Publ. 18:93-111. [Google Scholar]
- 32.Ragone Calvo, L. M., C. F. Dungan, B. S. Anderson, and E. M. Burreson. 2003. Systematic evaluation of factors controlling Perkinsus marinus transmission dynamics in lower Chesapeake Bay. Dis. Aquat. Org. 56:75-86. [DOI] [PubMed] [Google Scholar]
- 33.Ray, S. M. 1952. A culture technique for the diagnosis of infections with Dermocystidium marinum, Mackin, Owen and Collier, in oysters. Science 166:360-361. [DOI] [PubMed] [Google Scholar]
- 34.Ray, S. M. 1966. A review of the culture methods for detecting Dermocystidium marinum, with suggested modifications and precautions. Proc. Natl. Shellfish Assoc. 54:55-59. [Google Scholar]
- 35.Reece, K. S., D. Bushek, and J. E. Graves. 1997. Molecular markers for population genetic analysis of Perkinsus marinus. Mol. Mar. Biol. Biotechnol. 6:197-206. [Google Scholar]
- 36.Robledo, J. A. F., C. A. Coss, and G. R. Vasta. 2000. Characterization of the ribosomal RNA locus of Perkinsus atlanticus and development of a polymerase chain reaction-based diagnostic assay. J. Parasitol. 86:972-978. [DOI] [PubMed] [Google Scholar]
- 37.Robledo, J. A. F., J. D. Gauthier, C. A. Coss, A. C. Wright, and G. R. Vasta. 1998. Species specificity and sensitivity of a PCR-based assay for Perkinsus marinus in the eastern oyster, Crassostrea virginica: a comparison with the fluid thioglycolate assay. J. Parasitol. 84:1237-1244. [PubMed] [Google Scholar]
- 38.Stubner, S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreenTM detection. J. Microbiol. Methods 50:155-164. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, Y. L., and B. H. Olson. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volety, A. K., and F.-L. E. Chu. 1994. Comparison of infectivity and pathogenicity of meront (Trophozoite) and prezoosporangia stages of the oyster pathogen Perkinsus marinus in eastern oysters, Crassostrea virginica (Gmelin, 1791). J. Shellfish Res. 13:521-527. [Google Scholar]
- 42.Watson, R. J., and B. Blackwell. 2000. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 46:633-642. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-138. [DOI] [PubMed] [Google Scholar]
- 45.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCyclerTM: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]
- 46.Yarnall, H. A., K. S. Reece, N. A. Stokes, and E. M. Burreson. 2000. A quantitative competitive polymerase chain reaction assay for the oyster pathogen Perkinsus marinus. J. Parasitol. 86:827-837. [DOI] [PubMed] [Google Scholar]