Abstract
The phylogenetic relationships of Enterobacter sakazakii strains were investigated using 16S ribosomal DNA (rDNA) and hsp60 sequencing. Each analysis distributed E. sakazakii strains among four clusters, indicating substantial taxonomic heterogeneity. The E. sakazakii type strain 16S rDNA sequence was 97.8% similar to that of Citrobacter koseri but 97.0% similar to that of Enterobacter cloacae.
In recent years, Enterobacter sakazakii has been associated with necrotizing enterocolitis, bacteremia, and infant meningitis through the ingestion of contaminated powdered infant formula milk (5). Since E. sakazakii has been reported to be 50% related to Enterobacter cloacae and Citrobacter koseri by DNA-DNA hybridization (3) and the latter organism produces brain abscesses similar to those caused by E. sakazakii (9), the 16S ribosomal DNA (rDNA) and hsp60 housekeeping gene sequences from these organisms were compared.
A total of 126 strains identified as E. sakazakii by biochemical test kits (API20E and ID32E; bioMérieux) were sequenced. These included the type strain, NCTC 11467, and strains NCIMB 5920, NCIMB 8272, ATCC 51329, and ATCC 29004. Seven E. sakazakii strains were from European hospitals; seven strains were supplied by Jeff Farber, Health Canada; six strains were supplied by Nestlé-CRN, Vers-chez-les-Blanc, Lausanne, Switzerland; six strains were from J. Hoogkamp-Korstanje, St Radboud, Nijmegen, The Netherlands; and a further five strains were from the Centers for Disease Control and Prevention, Atlanta, Ga. All other strains were food and environmental isolates from the culture collection at Nottingham Trent University, Nottingham, United Kingdom (6). Nine C. koseri strains were also sequenced, including the type strain, ATCC 25408, and strains ATCC 25409, ATCC 25410, and ATCC 27026. Four additional C. koseri strains were from S. Townsend, Children's Hospital, Los Angeles, Calif., and one strain was obtained from a European hospital.
Comparative 16S rDNA sequence analysis using the MicroSeq 500 16S rDNA bacterial sequencing kit (Applied Biosystems) was performed according to standard procedures (2) at Accugenix (Newark, Del.). Sequencing of the hsp60 gene was performed by AGOWA (Berlin, Germany) using the primer sequences GGT AGA AGA AGG CGT GGT TGC (forward) and ATG CAT TCG GTG GTG ATC ATC AG (reverse) (4) to generate a 341-nucleotide PCR product. Additional 16S and hsp60 DNA sequences from type strains of Enterobacter species and Citrobacter freundii were obtained from GenBank. Sequences were aligned, corrected for multiple base changes (Jukes-Cantor method), and compared by using Bionumerics version 3.0 (Applied Maths, Kortrijk, Belgium). Relationships between strains were depicted in dendrogram form using the neighbor-joining method. Gaps and unknown bases were not considered in the analysis. The statistical significance of the branch points was evaluated by use of bootstrap analysis using 1,000 replicates. The trees were rooted to Pantoea agglomerans, and the scale bars (see Fig. 1) represent percentage sequence divergence as measured by the length of horizontal lines connecting any two strains. Bootstrap values derived from 1,000 replicates indicating the statistical significance of the branch points are also shown.
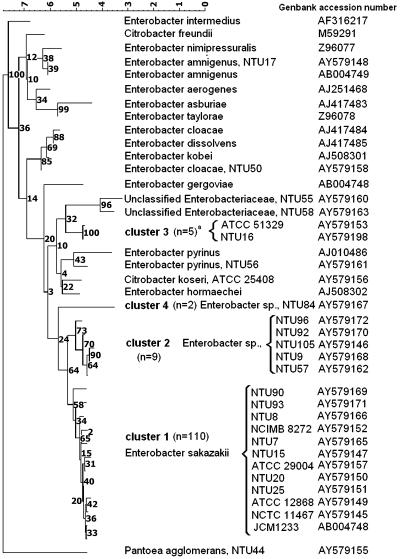
FIG. 1.
Neighbor-joining tree of E. sakazakii and related organisms based on partial 16S rDNA sequences. Strains presented are representative of the total strains sequenced. Numbers in parentheses indicate the number of strains in a given cluster.
According to 16S rDNA sequence comparisons, the type strain of E. sakazakii was closer to C. koseri (97.8% similar) than to any other Enterobacter species, although E. cloacae was 97.0% similar and C. freundii was 96.0% related by this method. Our findings are consistent with the polyphyletic nature of Enterobacter spp. revealed by both 16S rDNA and gyrB sequence comparisons (1). In the absence of defined phylogenetic criteria for delineating genera, the results observed with Citrobacter and Enterobacter species suggest that further studies are warranted to clarify their relationship. The 16S rDNA-based analysis divided the E. sakazakii strains into four clusters (Fig. 1). The majority (110 strains) were in cluster 1, with 0.1 to 1.2% difference from the type strain. These included 17 clinical strains and 3 nonpigmented strains. Nine strains (including one clinical strain) exhibited 1.6 to 1.9% sequence divergence from the type strain and formed a second cluster closely related to E. sakazakii (Fig. 1). The third cluster contained five strains (including one clinical strain), which despite being identified as E. sakazakii by the API20E and ID32E kits were more closely related (97.5 to 97.8% similar) to taxa including Enterobacter pyrinus, Enterobacter hormaechei, and C. koseri (Fig. 1). This cluster included the Preceptrol strain ATCC 51329, which was 3.0% divergent from the E. sakazakii type strain. The fourth cluster contained two strains, one of which was a clinical isolate. These strains were also identified as E. sakazakii by API20E and ID32E, but their 16S rDNA sequences were just 96.5% similar to the type strain of that species. Our 16S rDNA sequence analysis also confirmed that several strains identified as E. sakazakii by commercial biochemical kits belonged to distinct species, including Enterobacter amnigenus and E. cloacae (7), as well as potentially novel taxa (Fig. 1).
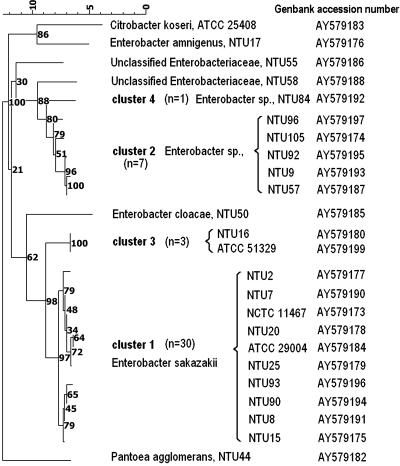
hsp60 sequences were obtained for 47 representative strains, including 41 strains of E. sakazakii that were representative of the different strain origins and of the four different 16S rDNA clusters (Fig. 2). Although the bootstrap values for the 16S rDNA-defined E. sakazakii-like clusters were somewhat low (Fig. 1), strains assigned to each of the 16S rDNA-based clusters also formed distinct clades in the hsp60 analysis (Fig. 2), although the tree topologies differed, reflecting the different similarity values obtained. Cluster 1 strains remained closely related (97.6 to 99.4% similar) to the E. sakazakii type strain, but cluster 2 (89.2 to 90.4% similar) and cluster 4 (88.8% similar) strains were more distinct. By contrast, cluster 3 isolates shared 95.2% hsp60 sequence homology with the E. sakazakii type strain, and their position in the hsp60-derived dendrogram supports our classification of these strains as E. sakazakii-like strains. In contrast, the distinct positions of strains NTU55 and NTU58 suggest that these strains should not be considered to be E. sakazakii despite their identification as this species by some biochemical test kits.
FIG. 2.
Neighbor-joining tree of E. sakazakii and related organisms based on partial hsp60 sequences. Strains presented are representative of the total strains sequenced. Numbers in parentheses indicate the number of strains in a given cluster.
This investigation has demonstrated that E. sakazakii is genetically diverse and potentially more taxonomically complex than hitherto realized. Clusters 2, 3, and 4 described here may represent novel species closely related to E. sakazakii; further study is necessary to confirm their taxonomic positions. This need is due to the nonlinear relationship often observed between 16S rDNA sequence similarities and the “gold standard” for species delineation, i.e., quantitative DNA-DNA hybridization data (8). Nonetheless, the implications of our study for accurate identification of E. sakazakii are self evident and relevant to clinicians, epidemiologists, and infant formula manufacturers.
Acknowledgments
We thank the Nottingham Trent University for the funding of this project and the technical support of Oxoid Ltd. (Basingstoke, United Kingdom) and Accugenix. We also thank the Danish Ministry of Agriculture for support.
REFERENCES
- 1.Dauga, C. 2002. Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studies. Int. J. Syst. Evol. Microbiol. 52:531-547. [DOI] [PubMed] [Google Scholar]
- 2.Drancourt, M., C. Bollet, A. Carlioz, R. Martelin, J.-P. Gayral, and D. Raoult. 2000. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 38:3623-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer, J. J., III, M. A. Asbury, F. W. Hickman, D. J. Brenner, and the Enterobacteriaceae Study Group. 1980. Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int. J. Syst. Bacteriol. 30:569-584. [Google Scholar]
- 4.Hoffman, H., and A. Roggenkamp. 2003. Population genetics of the nomenspecies Enterobacter cloacae. Appl. Environ. Microbiol. 69:5306-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iversen, C., and S. J. Forsythe. 2003. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci. Technol. 14:443-454. [Google Scholar]
- 6.Iversen, C., and S. J. Forsythe. 2004. Isolation of Enterobacter sakazakii and other Enterobacteriaceae from powdered infant formula milk and related products. Food Microbiol. 21:771-776. [Google Scholar]
- 7.Iversen, C., P. Druggan, and S. J. Forsythe. 2004. A selective differential medium for Enterobacter sakazakii, a preliminary study. Int. J. Food Microbiol. 96:133-139. [DOI] [PubMed] [Google Scholar]
- 8.Keswani, J., and W. B. Whitman. 2001. Relationship of 16S rRNA sequence similarity to DNA hybridization in prokaryotes. Int. J. Syst. Evol. Microbiol. 51:667-678. [DOI] [PubMed] [Google Scholar]
- 9.Kline, M. W. 1988. Pathogenesis of brain abscesses caused by Citrobacter diversus or Enterobacter sakazakii. Pediatr. Infect. Dis. J. 7:891-892. [PubMed] [Google Scholar]