Abstract
This study compared the sensitivity and viral load values of the AMPLICOR HIV-1 MONITOR microwell version 1.0, microwell version 1.5, and COBAS version 1.5 tests. Based on the percentage of positive replicates, the microwell version 1.5 and COBAS version 1.5 tests are more sensitive than the microwell version 1.0 test. Viral load values obtained with the COBAS version 1.5 test are lower than those obtained with either the microwell version 1.0 or microwell version 1.5 test.
The quantification of viral RNA is the standard of care in managing persons with human immunodeficiency virus type 1 (HIV-1) infection. Currently, there are three HIV-1 viral load tests that have been approved by the Food and Drug Administration: the AMPLICOR HIV-1 MONITOR test (Roche Diagnostics, Indianapolis, Ind.), the Quantiplex HIV-1 assay (bDNA; Bayer Corporation, Tarrytown, N.Y.), and the NucliSens HIV-QT assay (bioMerieux, Inc., Durham, N.C.). These tests are widely used in clinical practice, and each has its own strengths and weaknesses. One of the most significant limitations of the microwell plate AMPLICOR HIV-1 MONITOR version 1.0 (MWP v1.0) test is that it underquantifies non-B subtypes of HIV-1 (3). This problem has been rectified with the development of the MWP v1.5 test, which has changes to the primer sequences, allowing equivalent quantification of subtypes A through H (3, 5, 6).
For laboratories using the AMPLICOR test, version 1.5 has replaced version 1.0 in clinical practice. A recent study comparing versions 1.0 and 1.5 of the test showed that there was close agreement among the values obtained with the different test versions, and it was suggested that it was not necessary to reestablish a baseline viral load when changing versions of the test (2). However, since implementing the microwell plate version 1.5 test we have noted some patients with viral load values of <50 copies/ml of plasma with the version 1.0 test that were >50 copies/ml by the version 1.5 test. This observation led us to compare the sensitivity and viral load values between the MWP v1.0, the MWP v1.5, and the COBAS AMPLICOR HIV-1 MONITOR (COBAS v1.5) tests.
HIV-1 RNA (subtype B; AcroMetrix, Benicia, Calif.) was obtained at a concentration of 5,000 copies/ml and was diluted in human plasma to a concentration of 100 copies/ml of plasma. Quantification of the standard was confirmed by testing 10 replicates of the material in the MWP v1.5 test. After confirming the concentration, the control material was further diluted to 25 and 50 copies/ml in human plasma. Aliquots of the control material were frozen at −70°C until viral load testing was performed. Viral load testing was done using the ultrasensitive MWP v1.0 test, the MWP v1.5 test, and the COBAS v1.5 test. Testing was performed according to the manufacturer's protocols, using 500 μl of specimen. According to the manufacturer, the lower limit of quantification of the ultrasensitive tests is 50 copies/ml of plasma. One lot of reagents was used for each of the three tests. Forty replicates of the 25-copy/ml sample, 40 replicates of the 50-copy/ml sample, and 52 replicates of the 100-copy/ml sample were tested in each of the three assays. Two samples, a 25-copy/ml replicate in both the MWP v1.0 and COBAS v1.5 tests, gave invalid results due to failure to amplify the internal control and were eliminated from the data analysis. For this study, any sample with an optical density (OD) of >0.2 in the sample well was considered positive, even if the calculated viral load was <50 copies/ml of plasma. Data were analyzed using the χ2 or Fisher's exact test based on the percentage of positive samples for each concentration and test and combined across the three concentrations. Probit regression analysis of percent positive versus log10 copies/ml was performed with SAS Proc Probit software (version 8) to compare slope and intercept estimates between the three tests and to determine for each test the viral load value and the 95% confidence interval (CI) at which 95% of the results are expected to be positive (1). For replicates of the 100-copy/ml sample that had a detectable viral load, the geometric mean and 95% confidence interval were calculated for each of the three tests (4). The mean viral load was compared between the three tests with a one-way analysis of variance, and pairwise comparisons were made with t tests. Reported P values are two-sided. A Bonferroni adjustment (P < 0.0167; that is, 0.05/3) was used for the pairwise comparisons between the three tests.
The percentage of samples that were positive for each of the three tests is shown in Table 1. When the 25-copy/ml sample was tested, 30, 72, and 64% of the replicates were positive with the MWP v1.0, MWP v1.5, and COBAS v1.5 tests, respectively. The differences in sensitivity were statistically significant when both the COBAS v1.5 and MWP v1.0 tests (P = 0.003) and the MWP v1.5 and MWP v1.0 tests (P = 0.0002) were compared. The percentage of positive replicates increased for both the 50- and 100-copy/ml samples for all three tests and reached a high of 98% for the MWP v1.5 and COBAS v1.5 tests with the 100-copy/ml sample. The differences in the percentage of positive replicates for the 50- and 100-copy/ml samples were not statistically significant for the three tests. When the results for the 25-, 50-, and 100-copy/ml samples were combined, the MWP v1.5 and COBAS v1.5 tests were both more sensitive than the MWP v1.0 test (for COBAS v1.5 versus MWP v1.0, P = 0.0016; for MWP v1.5 versus MWP v1.0, P = 0.0002). A limitation of the study is that all testing was done using a single lot of reagents for each test, so the impact of lot-to-lot variation was not assessed.
TABLE 1.
Percentage of replicates with a detectable viral load and probits for the three tests
| Log10 HIV copies/ml | No. of replicates tested | MWP v1.0
|
MWP v1.5
|
COBAS v1.5
|
|||
|---|---|---|---|---|---|---|---|
| % Detected | Probit | % Detected | Probit | % Detected | Probit | ||
| 2 | 52 | 88 | 6.18 | 98 | 7.05 | 98 | 7.05 |
| 1.7 | 40 | 75 | 5.71 | 85 | 6.04 | 85 | 6.04 |
| 1.4 | 40 | 30 | 4.48 | 72 | 5.58 | 64 | 5.36 |
For replicates of the 100-copy/ml sample that had a calculated viral load, the geometric mean viral load values obtained with the MWP v1.0, MWP v1.5, and COBAS v1.5 tests were 69 (95% CI, 59 to 83), 85 (95% CI, 72 to 100), and 39 (95% CI, 33 to 46) copies/ml, respectively (pooled geometric standard deviation = 1.79). The mean viral load value obtained with the COBAS v1.5 test was significantly lower than that seen for the MWP v1.0 and MWP v1.5 tests (P < 0.001). There were eight samples (one each in the COBAS v1.5 and MWP v1.5 assays and six in the MWP v1.0 assay) that had undetectable viral loads. A second analysis, was done; in this analysis including all specimens viral load values for these eight specimens were calculated based on the cutoff OD of the assay (an OD of 0.2 was used as the HIV-1 target value). The geometric mean viral load values obtained when all values were included in the analysis were 63, 81, and 37 copies/ml, respectively, for the MWP v1.0, MWP v1.5, and COBAS v1.5 tests. The mean viral load value obtained with the COBAS v1.5 test remained significantly lower than that seen for the MWP v1.0 and MWP v1.5 tests (P < 0.001).
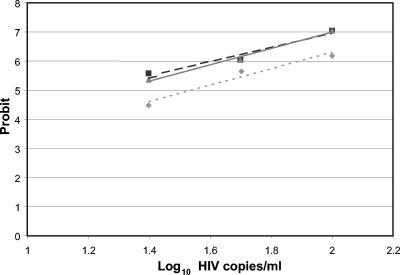
Probit analysis was used to estimate the limit of detection for the three tests (Fig. 1). The slopes of the three lines are not different (P = 0.74), but there are significant differences between the intercepts for the version 1.5 tests compared to the MWP v1.0 test (for COBAS v1.5 versus MWP v1.0, P < 0.001; for MWP v1.5 versus MWP v1.0, P < 0.001; for COBAS v1.5 versus MWP v1.5, P = 0.57). Viral load values that could be detected in 95% of the replicate tests were calculated to be 129 (95% CI, 94 to 238), 79 (95% CI, 56 to 220), and 77 (95% CI, 57 to 158) copies/ml for the MWP v1.0, MWP v1.5, and COBAS v1.5 tests, respectively.
FIG. 1.
Plot of probit (estimated probability) versus log10 copies/ml for the MWP v1.0 (⧫), MWP v1.5 (▪), and COBAS v1.5 (▴) tests.
Based on the percentage of positive specimens observed for the three sample concentrations (25, 50, and 100 copies/ml), the MWP v1.5 and COBAS v1.5 tests are both more sensitive than the MWP v1.0 test. Therefore, it is likely that when the v1.5 tests are used in clinical practice, some patients with undetectable viral load values in the MWP v1.0 test will have detectable viral load values in either the COBAS v1.5 or MWP v1.5 test. In addition, the viral load values obtained for the 100-copy/ml sample with the MWP v1.0 and MWP v1.5 tests are twofold higher than those seen with the COBAS v1.5 test. Though these differences in viral load are statistically significant, it is unlikely that they are clinically relevant given the variability of the assays. It is important to appreciate the differences in sensitivity and viral load values between the different versions and platforms for the AMPLICOR HIV-1 MONITOR tests as laboratories transition from the MWP v1.0 test to either the MWP v1.5 or COBAS v1.5 test.
REFERENCES
- 1.Finney, D. J. 1971. Probit analysis, 3rd ed. Cambridge University Press, London, England.
- 2.Hill, C. E., A. M. Green, J. Ingersoll, K. A. Easley, F. S. Nolte, and A. M. Caliendo. 2004. Assessment of agreement between the AMPLICOR HIV-1 MONITOR test versions 1.0 and 1.5. J. Clin. Microbiol. 42:286-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jagodzinski, L. L., D. L. Wiggins, J. L. McManis, S. Emery, J. Overbaugh, M. Robb, S. Bodrug, and N. L. Michael. 2000. Use of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J. Clin. Microbiol. 38:1247-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkwood, T. B. L. 1979. Geometric means and measures of dispersion. Biometrics 35:908-909. [Google Scholar]
- 5.Parekh, B., S. Phillips, T. C. Granade, J. Baggs, D. J. Hu, and R. Respess. 1999. Impact of HIV type 1 subtype variation on viral RNA quantitation. AIDS Res. Hum. Retrovir. 15:133-142. [DOI] [PubMed] [Google Scholar]
- 6.Pasquier, C., K. Sandres, G. Salama, J. Puel, and J. Izopet. 1999. Using RT-PCR and bDNA assays to measure non-clade B HIV-1 subtype RNA. J. Virol. Methods 81:123-129. [DOI] [PubMed] [Google Scholar]