Abstract
A shuttle vector designated pMAD was constructed for quickly generating gene inactivation mutants in naturally nontransformable gram-positive bacteria. This vector allows, on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates, a quick colorimetric blue-white discrimination of bacteria which have lost the plasmid, greatly facilitating clone identification during mutagenesis. The plasmid was used in Staphylococcus aureus, Listeria monocytogenes, and Bacillus cereus to efficiently construct mutants with or without an associated antibiotic resistance gene.
The study of chromosomal genes and the modification of bacterial strains require the development of new strategies and new genetic tools to facilitate the rapid screening of recombinant clones. Molecular genetic analyses require the exchange by homologous recombination of a chromosomal gene by a mutated allele or inactivated copy, and several strategies have been developed for generating gene replacements in bacterial chromosomes. Here, we describe a new plasmid for efficiently accomplishing such constructions in nontransformable bacteria. This plasmid, pMAD, overcomes the limitations of the available vectors, which were largely developed for highly transformable bacteria. For example, a one-step gene inactivation procedure has been developed for Bacillus subtilis by using a nonreplicative vector called pMutin (22). This vector carries a lacZ reporter gene and an inducible Pspac promoter which can be induced by IPTG (isopropyl-β-d-thiogalactopyranoside). Following chromosomal integration via a single crossover event of a pMutin recombinant vector containing an internal fragment of the target gene, the target gene is inactivated, lacZ becomes transcriptionally fused to the gene, and the Pspac promoter controls the transcription of the downstream gene(s) in an IPTG-dependent fashion.
To avoid the introduction of a complete plasmid and duplication of the target sequence in the chromosome, other strategies were described. These two-step strategies proceed by homologous recombination between a target gene and homologous sequences carried on a plasmid which is temperature sensitive for DNA replication. After transformation of the plasmid into the host, integration of the plasmid into the chromosome by a single crossover event is selected during growth at the nonpermissive temperature while maintaining selective pressure. Subsequent growth of the cointegrates at the permissive temperature (30°C) leads to a second recombination event, resulting in their resolution. Screening of clones where an efficient second recombination event has taken place (i.e., where the loss of the vector is concomitant with the deletion of the gene of interest) is rather difficult without positive selection or colorimetric screening of the candidate clones. A general system for generating gene replacement in bacterial chromosomes was developed by using the pORI vector (12). This conditionally replicating vector contains the Escherichia coli lacZ gene, which is expressed from a constitutive promoter recognized in gram-positive and gram-negative bacteria. The presence or the absence of the lacZ gene enables a simple blue-white screening of recombination events. However, these plasmids lack the repA gene encoding the replication initiation protein and are unable to replicate in any bacterial strains unless the RepA protein is provided in trans. In consequence, high transformation frequencies are required to obtain integrants and deletions or substitutions are produced after resolution of cointegrants, thus limiting the use of this plasmid vector to naturally competent bacteria with a high efficiency of transformation (12). A system for generating deletions in open reading frames of E. coli was developed by using plasmid pK03, which contains a temperature-sensitive origin of replication, a chloramphenicol resistance gene, and a counterselectable sacB gene to facilitate allelic exchange. Briefly, the in vitro altered sequences carried on the vector pK03 are integrated into the chromosome by using the sucrose-resistant phenotype, and the sucrose-resistant and chloramphenicol-sensitive colonies are screened for the desired gene replacement (15). In earlier work with the Tn917 transposon in B. subtilis (25), methods of recovering populations of bacteria containing chromosomal insertions of the transposon were based on the use of temperature-sensitive vectors derived from a small Staphylococcus aureus plasmid called pE194 (9). However, practical complications arose from the fact that the thermosensitivity of replication of the pE194-derived vectors was incomplete. Replication of the plasmid was impaired at high temperatures but not abolished. New vectors based on the use of pE194ts, a thermosensitive mutant derivative of pE194, were isolated by Alexandra Gruss in R. Novick's laboratory (A. Gruss, personal communication) (24). These vectors display an extremely tight replication block above 37°C yet maintain essentially wild-type copy numbers at temperatures below 32°C. A chimeric integrational vector called pKSV7, which contains temperature-sensitive replication functions derived from the pE194ts that functions in B. subtilis, was constructed (20). As shown by Ehrlich and Noirot (17), the recombination excision of integrated plasmids is greatly stimulated in the vicinity of an active replication origin. It was shown that excision rates of integrated pKSV7 were greater than with ordinary ColE1-derived vectors, facilitating the introduction of deletions into chromosomal genes (20). The thermosensitive replication origin of pE194ts was also used to construct delivery vectors for obtaining chromosomal insertions of Tn917 in B. subtilis (19) and in Listeria monocytogenes (2).
In contrast to the situation observed with Streptococcus pneumoniae and B. subtilis, where the natural development of competence is concomitant with the increased expression of recombination genes, allelic replacement is a time-consuming and fastidious task in nonnaturally transformable bacteria, such as S. aureus, Bacillus thuringiensis, or L. monocytogenes. A shuttle vector (pE194ts::pBR322), which replicates in both E. coli and S. aureus, was constructed (A. Gruss, unpublished results). This vector has a thermosensitive pE194 replication origin (24) and confers erythromycin resistance to gram-positive hosts. This vector was used successfully for gene inactivation in B. thuringiensis (13, 14). However, the use of this vector or similar constructs, such as pKSV7, to obtain mutants remained labor intensive, due to the low efficiency of recombination and the lack of a simple method for screening recombinant clones where excision of the plasmid has taken place. In the postgenome era, there is an obvious need to develop vectors that facilitate the high-throughput mutagenesis required to analyze novel regulatory genes and potential virulence determinants in nonnaturally transformable gram-positive pathogens, such as S. aureus and L. monocytogenes. To this end, we have constructed a new vector called pMAD combining the different advantages of the various plasmids described above. This vector carries a constitutively expressed transcriptional fusion with the bgaB gene encoding a thermostable β-galactosidase from Bacillus stearothermophilus, allowing the easy screening of transformants on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates.
Construction of a novel vector, pMAD, for efficient gene replacement in nonnaturally transformable gram-positive bacteria.
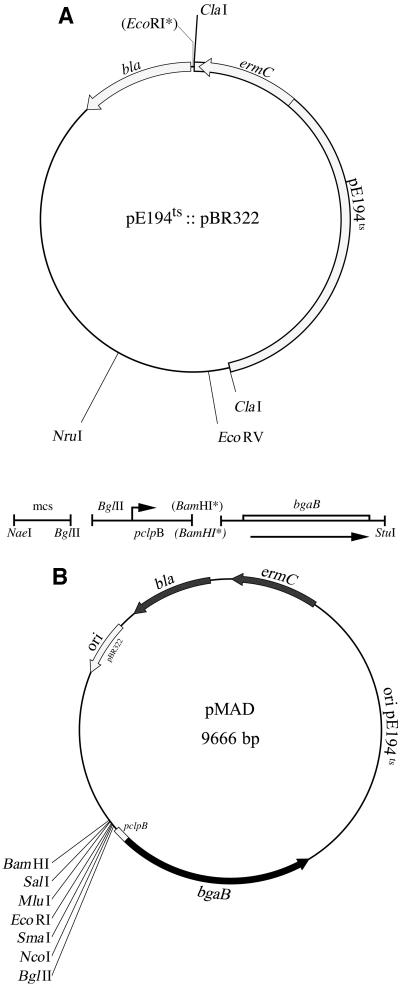
The pMAD vector was constructed by using a replication-thermosensitive mutant of pE194. This plasmid, conferring resistance to the macrolide, lincosamide, and streptogramin B antibiotics, was originally isolated from S. aureus and transferred to B. subtilis (7, 24). Plasmid pE194ts was linearized at the single ClaI restriction site and cloned into the corresponding site of pBR322 (1) to generate the shuttle vector called pE194ts::pBR322 (A. Gruss, personal communication).
In the first step, the EcoRI restriction site of pE194ts::pBR322 plasmid was filled in with the Klenow fragment of DNA polymerase I in the presence of the four deoxynucleoside triphosphates and religated with T4 DNA ligase. Three DNA fragments were cloned between the NruI and the EcoRV restriction sites of the shuttle vector (see Fig. 1A): (i) a NaeI-BglII DNA fragment containing the extended multiple cloning site from the pMTL22 plasmid (3); (ii) a chromosomal BglII-BamHI DNA fragment (128 bp) containing the constitutively expressed promoter region of the S. aureus clpB gene encoding a Clp-Hsp100 ATPase (5, 6) and amplified by PCR from chromosomal DNA of strain RN6390 of S. aureus (18) with the Pwo thermostable DNA polymerase and oligonucleotides O1 (5′-GGAAGATCTGTCTAGTTAATGTGTAACGTA-3′) and O2 (5′-CGCGGATCCGACCTTAATTATATTATAGTCCCAAT-3′); and (iii) a BamHI-StuI DNA fragment containing the promoterless bgaB gene from B. stearothermophilus encoding a thermostable β-galactosidase (8). A map of the resulting plasmid, pMAD, is shown in Fig. 1B. Expression of bgaB in pMAD is driven by the constitutive pclpB promoter.
FIG. 1.
Physical map of pE194ts::pBR322 and its derivative pMAD. (A) Construction of the pMAD plasmid. DNA fragments used to construct pMAD are indicated: mcs, multiple cloning site; pclpB, DNA fragment from S. aureus containing the clpB promoter region; bgaB, DNA fragment containing the promoterless bgaB gene encoding a thermostable β-galactosidase (8). Asterisks indicate restriction sites which have been lost during the construction steps. (B) Map of the pMAD plasmid. Unique restriction sites are indicated.
Utilization of the pMAD vector for gene replacement in S. aureus.
To assess the pMAD vector for the generation of stable chromosomal mutations in S. aureus, we constructed a derivative of pMAD carrying the upstream part of a target gene, an antibiotic resistance cassette (spectinomycin resistance), and the downstream part of the target gene. We chose to inactivate two contiguous genes called vraF and vraG (vra standing for vancomycin resistance associated). These genes are up-regulated in S. aureus strains Mu3 and Mu50. The encoded proteins constitute a likely ABC transporter. This ABC transporter gene was found to be transcribed in a greater amount in the Mu50 strain, and vraFG probably contributes to vancomycin resistance through the transport of the cell wall amino acid component (11). DNA fragments corresponding to the upstream region of vraF (extending from positions −665 to −33 with respect to the vraF translation initiation site) and the downstream region of vraG (extending from positions −57 to +805 with respect to the vraG stop codon) were amplified with the oligonucleotide pairs OMD189 and OMD190 (5′-GGAGGATCCTCGATGCTGGATACACAGCTG-3′ and 5′-CTCCTCGAGCCTTTAGGCTTTGGCACTTG-3′, respectively) and OMD191 and OMD192 (5′-AGAAGATCTGCTGTTTTTGCAGTGACGGC-3′ and 5′-GGAGGATCCCGAATTCAATAGCAGCACG-3′, respectively), the high-fidelity Triple Master polymerase (Eppendorf), and chromosomal DNA from strain RN6390 as a template. The PCR products were treated with XhoI and BglII restriction enzymes, respectively. A 1.3-kb DNA fragment containing the entire spectinomycin resistance gene spc (16) was amplified from plasmid pIC333 (21) with the oligonucleotides OMD156 (5′-AGAAGATCTCACCTAGATCCTTTTGACTC-3′) and OMD157 (5′-CTCCTCGAGAAAGTAAGCACCTGTTATTGC-3′) and was restricted with the XhoI and BglII restriction enzymes.
These purified PCR DNA fragments were mixed in equal amounts and ligated with DNA ligase. The ligation mixture was used as a template for the Triple Master polymerase to produce the complete tripartite DNA fragment by PCR with OMD189 and OMD192 oligonucleotides. The full-length 2.4-kb PCR fragment was then purified on an agarose electrophoresis gel, restricted with BamHI, and cloned in the corresponding restriction site of the pMAD vector. The recombinant pMADΔFG vector was extracted from E. coli, and a two-step procedure was then used for allele replacement in S. aureus, as detailed below.
In the first step, the recombinant pMADΔFG vector was introduced by electroporation into S. aureus strain RN4220 (10). Transformants (106 transformants/μg of DNA) were selected after 2 days at 30°C on rich medium (tryptic soy broth; Difco) containing either spectinomycin (100 μg/ml) or erythromycin (5 μg/ml).
One blue colony was streaked onto the same medium and incubated at 30°C. In the second step, a single blue colony was inoculated in tryptic soy broth complete medium without antibiotic at an optical density at 600 nm of 0.01 and incubated with shaking for 2 h at 30°C followed by 6 h at 42°C. Serial dilutions of this culture were plated on tryptic soy agar (TSA) containing X-Gal and spectinomycin and incubated at 42°C overnight. Among a vast majority of light blue clones due to the integration of the vector by a single crossover event, 1 to 5% of white colonies were present and represent candidate clones resulting from a double crossover event and loss of the vector (Fig. 2). Several white colonies were isolated on the same medium at 37°C and verified for erythromycin sensitivity. To confirm the gene deletion, chromosomal DNA was extracted from five candidate clones and used in PCRs with oligonucleotides OMD202 (5′-GCGGAAACGCCATTTCAACG-3′) and OMD203 (5′-GCTGCCGTTTCTTCAATTGC-3′), hybridizing 926 bp upstream from the vraF translational start site and 1,006 bp downstream from the vraG stop codon, respectively. All of the spectinomycin-resistant, erythromycin-sensitive white colonies on X-Gal-containing plates had the expected deletion and replacement of the vraF/vraG locus. The pMAD vector was also used to create various deletions in the sarA and sigB genes in clinical isolates of S. aureus strain 15 981, producing biofilms. In addition to the well-characterized roles of SarA in the regulation of many virulence determinants, it has been shown that SarA plays a role in the regulation of biofilm development (23). The pMAD vector also replicates at 30°C in other gram-positive bacteria and was successfully used to create gene replacements in L. monocytogenes and Bacillus cereus as we describe here.
FIG. 2.
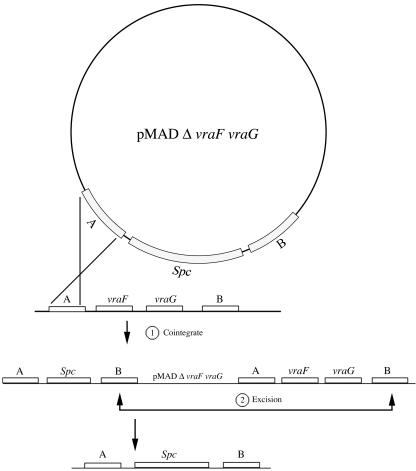
Schematic representation of a two-step procedure used to obtain gene replacement recombination. Areas labeled A and B represent DNA sequences located upstream and downstream from vraF and vraG genes. The crossed lines indicate crossover events. The integration of pMAD via homologous sequences can take place in area A or B. The cointegrate undergoes a second recombination event, regenerating the pMAD plasmid. Depending on whether the second recombination event occurs between the two homologous sequences in area A or B, the spectinomycin resistance marker will either remain in the chromosome (area B) or be excised along with the plasmid (area A). Gene replacement occurs only if the second recombination event occurs in area B, as shown.
Gene replacement in L. monocytogenes.
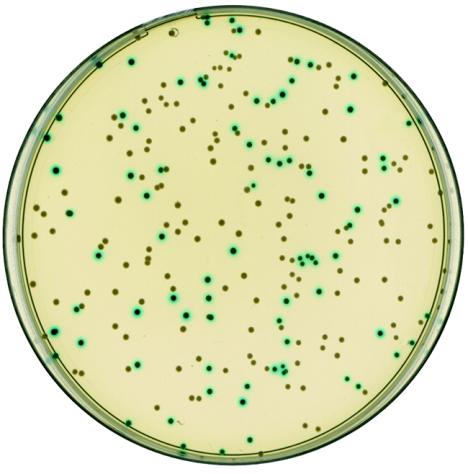
A similar strategy was used to inactivate the ctsR gene in L. monocytogenes strain LO28 (4). In this mutant, the ctsR gene was replaced through a double crossover event by the aphA3 kanamycin resistance gene deprived of its transcription initiation and termination sequences. However, the introduction of an antibiotic resistance marker can sometimes be undesirable for various reasons, such as the polar effect of the inserted DNA on the expression of the downstream genes in an operon or the multiple introductions of DNA fragments or different mutations within the same strain. To construct a deletion without insertion of foreign DNA, the clpB gene of L. monocytogenes was chosen as a target. A deletion eliminating the entire coding sequence of the clpB gene was constructed by generating through PCR two DNA fragments, of 829 and 761 bp, with oligonucleotides AC189 and AC190 (5′-AATGGATCCCACATCGGAGCGAGTAAACAC-3′ and 5′-TAAGTCGACTCATTCGTCCTCCTCCTTATAAAA-3′, respectively) and AC192 and AC193 (5′-CACGTCGACTGAAAGGGAAAACTTTGGTTG-3′ and 5′-TATCCATGGAATATTTATTTACTGGTTTTA-3′, respectively), corresponding to the chromosomal DNA regions located directly upstream and downstream, respectively, from the clpB gene. These two DNA fragments were cloned in tandem in pMAD, resulting in plasmid pMADΔclpB. A three-step procedure was used to obtain the L. monocytogenes mutant. The pMADΔclpB plasmid was introduced by electroporation into strain LO28 of L. monocytogenes, and transformants were selected at 30°C on brain heart infusion (BHI) plates containing erythromycin and X-Gal (50 μg/ml). One blue clone was reisolated on the same medium and incubated in BHI liquid medium at 39°C. To obtain integrants resulting from a single crossover event via homologous sequences, serial dilutions were plated on BHI erythromycin (5 μg/ml) plates containing X-Gal (50 μg/ml) and incubated at 39°C for 2 days. Clones resulting from the integration of the pMADΔclpB plasmid into the clpB gene were blue. Several blue colonies were cultivated at 30°C for 6 h, followed by incubation at 39°C for 3 h; dilutions were plated on TSA X-Gal plates in the absence of antibiotics. The appearance of white colonies (10 to 30%) on X-Gal plates indicates the excision and loss of the plasmid vector (Fig. 3). Five colonies were checked for erythromycin sensitivity, and PCR amplifications with oligonucleotides located upstream and downstream from the clpB gene were performed to confirm the gene deletion. One clone was shown to contain the expected deletion in clpB after the resolution process (4).
FIG. 3.
L. monocytogenes LO28 colonies following transformation with the pMADΔclpB plasmid. Cells were cultivated at 30°C for 6 h, followed by incubation at 39°C for 3 h; dilutions were plated on TSA X-Gal plates in the absence of antibiotics. White colonies have undergone the excision and loss of the plasmid vector, whereas blue colonies retain a copy of the plasmid integrated in the chromosome.
Gene replacement in B. cereus.
The pE194ts::pBR322 plasmid has been used previously to construct mutants of B. thuringiensis (13, 14). However, the low frequency of transformation and recombination and the lack of a clear phenotype allowing the screening of colonies where successful crossover events have taken place render the use of this plasmid tedious in this host. The pMAD vector was used successfully to delete the tetB gene (open reading frame RZC00709; Integrated Genomics, Inc.) from B. cereus strain ATCC 14579. Two DNA fragments of 877 bp and 1,013 bp corresponding to the regions located directly upstream and downstream from tetB were amplified by PCR with the following pairs of oligonucleotides: Tet1 and Tet2 (5′-CATGCCATGGCAGTAAGACCTCCATCTC-3′ and 5′-CCGGAATTGCGAAACACTTCATCTGTATG-3′, respectively) and Tet3 and Tet4 (5′-CCCAAGCTTCGTCATTTTAATTGCTGCTC-3′ and 5′-CGCGGATCCTGTTGTTAAGAAGAGATGATCGC-3′, respectively).
These two DNA fragments were cloned in the pMAD vector on either side of a DNA fragment containing the spc spectinomycin resistance gene, producing the pMADΔtetB plasmid. This plasmid was introduced by electroporation into Bacillus cereus, and transformants were obtained after 2 days at 30°C on Luria-Bertani (LB) plates containing 250 μg of spectinomycin/ml, 3 μg of erythromycin/ml, and 50 μg of X-Gal/ml. A pool of individual blue clones was used to inoculate a culture of LB medium without antibiotic incubated at 40°C to stationary phase. Two more cycles of growth were carried out by diluting the stationary phase culture into fresh medium. Finally, single colonies were isolated by plating dilutions of the overnight culture onto LB plates containing X-Gal and 300 μg of spectinomycin/ml. Several white colonies were isolated and checked for the erythromycin-sensitive phenotype. Chromosomal DNA was purified from several clones and used to make PCRs with oligonucleotides hybridizing outside the cloned DNA fragments to verify the extent of the deletion and insertion in the tetB gene. All the candidate clones were shown to contain the deletion and insertion in the tetB gene (S. Fedhila, unpublished results).
We note that although this vector has proven to be a valuable tool and been used successfully in several gram-positive bacteria (S. aureus, L. monocytogenes, and B. cereus), the pE194 origin of replication is not functional in streptococci, enterococci, and lactococci, preventing the use of pMAD in these bacteria (P. Trieu-Cuot, personal communication).
Acknowledgments
We thank Tarek Msadek, Georges Rapoport, and Patrick Trieu-Cuot for many helpful discussions and critical reading of the manuscript. We thank Olivier Poupel for invaluable assistance with graphic representations, Fadhila Rayah for assistance with the early construction stages of pMAD, and Sinda Fedhila for sharing unpublished results.
This work was supported by research funds from the European Commission (grant QLRT-1999-01455), the Centre National de la Recherche Scientifique, and the Institut Pasteur. Arnaud Chastanet was the recipient of a Ph.D. thesis fellowship from the Ministère de la Recherche.
Footnotes
This paper is dedicated to the memory of Maryvonne Arnaud, who died on 1 February 2004.
REFERENCES
- 1.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 2.Camilli, A., D. A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. [DOI] [PubMed] [Google Scholar]
- 4.Chastanet, A., I. Derré, S. Nair, and T. Msadek. 2004. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 186:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat-shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 6.Frees, D., A. Chastanet, S. Qazi, K. Sørensen, P. Hill, T. Msadek, and H. Ingmer. Submitted for publication.
- 7.Gryczan, C. S., and D. Dubnau. 1978. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J. Bacteriol. 134:318-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirata, H., T. Fukazawa, S. Negoro, and H. Okada. 1986. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iordanescu, S. 1976. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch. Roum. Pathol. Exp. Microbiol. 35:111-118. [PubMed] [Google Scholar]
- 10.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Me50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 12.Leenhouts, K., G. Buist, A. Bolhuis, A. ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed] [Google Scholar]
- 13.Lereclus, D., H. Agaisse, M. Gominet, and J. Chaufaux. 1995. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Biotechnology 13:67-71. [DOI] [PubMed] [Google Scholar]
- 14.Lereclus, D., M. Vallade, J. Chaufaux, O. Arantes, and S. Rambaud. 1992. Expansion of insecticidal host range of Bacillus thuringiensis by in vivo genetic recombination. Biotechnology 10:418-421. [DOI] [PubMed] [Google Scholar]
- 15.Link, A. J., D. Philipps, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: applications to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy, E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3′′) (9). Mol. Gen. Genet. 200:33-39. [DOI] [PubMed] [Google Scholar]
- 17.Noirot, P. M. A., and D. Ehrlich. 1987. Plasmid replication stimulates DNA recombination in Bacillus subtilis. J. Mol. Biol. 196:39-48. [DOI] [PubMed] [Google Scholar]
- 18.Peng, H.-L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins, J. B., and P. Youngman. 1986. Construction and properties of Tn917-lac, a transposon derivative that mediates transcriptional gene fusions in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 83:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 21.Steinmetz, M., and R. Richter. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 23.Valle, J., A. Toledo-Arana, C. Berasain, J.-M. Ghigo, B. Amorena, J. R. Penadés, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 24.Villafane, R., D. H. Bechhofer, C. S. Narayanan, and D. Dubnau. 1987. Replication control genes of plasmid pE194. J. Bacteriol. 169:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youngman, P. J., J. B. Perkins, and R. Losick. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc. Natl. Acad. Sci. USA 80:2305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]