Abstract
Epidemic methicillin-resistant Staphylococcus aureus 16 (EMRSA-16) and EMRSA-15 are the two most important and prevalent EMRSA strains found in the United Kingdom and have also been found in a number of European countries and the United States. We describe for the first time the spread of an EMRSA strain (EMRSA-16) from its point of origin in one hospital to the surrounding hospitals and regions over the following 2 years. In the first 18 months after its original appearance, 136 hospitals referred EMRSA-16 isolates for typing, and interhospital and intraregional spread were reported: it was more prevalent in males between 60 and 80 years old and was isolated from sputum and throat more often than EMRSA-15. Important characteristics, e.g., carriage of the enterotoxin A (sea) and toxic shock syndrome toxin (tst) genes and production of urease, are described. Phage-variant strains of EMRSA-16 which share some of the characteristics of the classical strain, including toxin carriage and urease production, emerged, but without genotypic investigations, their relationship could only be inferred. A total of 129 clinical isolates from 52 hospitals, collected between March 1998 and April 1999 and representing classical EMRSA-16 (49 isolates) or phage variants (80 isolates), were compared by phage typing, pulsed-field gel electrophoresis (PFGE) following SmaI macrorestriction, antimicrobial susceptibility testing, urease production, and PCR detection of toxin gene carriage. PFGE analysis revealed 29 profiles, A1 to A29, with A1 representing the prototypic strain, NCTC 13143. All other profiles differed from A1 by 1 to 6 bands, but some differed from each other by up to 10 bands.
In the United Kingdom, guidelines for the control of methicillin-resistant Staphylococcus aureus (MRSA), which were revised in 1990 and 1998, recommend that clinical laboratories refer isolates of MRSA to reference laboratories when infection control teams suspect that the MRSA is behaving in an epidemic or virulent fashion (12). Referral of isolates to the laboratory was also informed by regular discussions with infection control teams and a survey in 1995 (see below). Advice was provided on selection of isolates, both verbally and in written guidance that was audited and adjusted accordingly. We have thus been able to plot the emergence and spread of epidemic MRSA (EMRSA) strains, which we have numbered consecutively. EMRSA-16 was first observed in the English town of Kettering in Northamptonshire in 1992 (1, 2) and rapidly became one of the two predominant supraregional epidemic strains of MRSA currently circulating in the United Kingdom. EMRSA-16 is the only EMRSA strain for which it has been possible to plot the emergence from a single hospital and spread to other regions in the United Kingdom.
Epidemiological data and background.
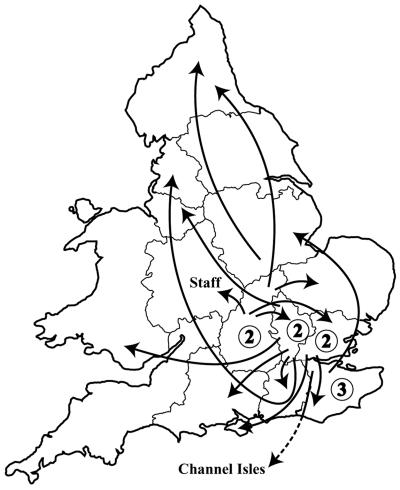
EMRSA-16 was not encountered prior to April 1992, when the initial outbreak occurred and affected 400 patients and 27 staff in three hospitals (one acute, one elderly care, and one psychogeriatric) in Kettering, a town located in Northamptonshire in the East Midlands of England (14). Between April 1992 and September 1994, 136 hospitals referred MRSA isolates to the S. aureus Reference Service (SaRS) at the Laboratory of Hospital Infection, now the Laboratory of Health Care-Associated Infection (LHCAI), that were found to be EMRSA-16 (previously unpublished data) (Fig. 1). Interhospital and intraregional spread of EMRSA-16 was reported by nine hospitals in four regions. Interregional hospital spread following patient transfer (14 hospitals, including one in the Channel Islands) or staff transfer (one hospital) was also evident. Initially, the outbreak spread from Northamptonshire to the three neighboring counties or regions (15 hospitals and 845 patients), which was followed by spread to London (21 hospitals and 309 patients) plus a further four hospitals and 18 patients in the more distant parts of the United Kingdom.
FIG. 1.
Reported route of acquisition by 24 of 136 hospitals submitting their first EMRSA-16 strains to the reference laboratory. Interhospital transfer of patients was reported in all instances but one, when the EMRSA-16 was introduced by a newly employed colonized member of the staff. Numbers in circles represent EMRSA-16 spread via patients transferred between hospitals in the same region.
Of the 7,500 MRSA strains submitted to the reference laboratory between July and September 1995, more than 50% of the total was made up of two strains, EMRSA-15 and EMRSA-16 (R. R. Marples, J. F. Richardson, G. White, and M. Ganner, Abstr. 8th Int. Symp. Staphylococci Staphylococcal Infect., abstr. 267, 1996). Chi-square analyses were used to explore the statistical significance of any relationships. An anatomical source was available for 6,383 isolates. EMRSA-15 was particularly prevalent in females older than 80 years and was found in pressure sores, urine, high-vaginal swabs, and eye swabs. EMRSA-16, however, was more prevalent in males between 60 and 80 years old and was more commonly isolated from sputum and throat swabs. The relationship to pneumonia was first noted in the initial outbreak (14), and clinical inquiries to the laboratory during these early years frequently commented on the propensity for the strain to cause pneumonia in ventilated patients in intensive care units (B. D. Cookson, unpublished).
In a survey of hospitals in the United Kingdom in 1995, it was found that 443 outbreaks of infection or colonization with EMRSA-16 that occurred over the previous 18 months affected surgical, medical, and acute elderly-care wards (19% for each of these), orthopedic surgical wards (14%), intensive care units (11%), long-stay elderly-care wards (9%), mixed medical and surgical wards (3%), and neonatal and dermatology wards (0.9% each) (B. Cookson and R. Marples, Abstr. 8th Int. Symp. Staphylococci Staphylococcal Infect., abstr. 313, 1996). In this same survey EMRSA-16 was found to affect 20% of the 150 nursing homes known to have patients colonized or infected with MRSA.
During the next 3 years, EMRSA-16, and indeed EMRSA-15, spread to most regions of the England and Wales (13). We analyzed the data for 2000 for this study, which showed that EMRSA-16 comprised 24% of all isolates of MRSA referred to SaRS from England and Wales (the corresponding value for EMRSA-15 was 48%). By region, EMRSA-16 accounted for 30% of all MRSA isolates in the south and east of England, 25% in the southwest, 23% in the Midlands, 12% in the north and northwest, and 30% in Wales (SaRS data). In addition, recent reports have indicated that EMRSA-16 comprises 26% of all MRSA isolates in Scotland (4), as well as 1 and 18% in Northern Ireland and the Republic of Ireland (3), respectively.
Initial characterization of EMRSA-16.
The original “classical” EMRSA-16 was typically identified by its phage pattern (29inh/52inh/75/77/83A/83C) and was characterized by carriage of staphylococcal enterotoxin A (sea) and toxic shock syndrome toxin-1 (tst) genes as well as production of urease. It was generally resistant to ciprofloxacin, erythromycin, gentamicin, kanamycin, and neomycin in addition to methicillin. Over the first 2 years, the gentamicin resistance was lost in the majority of isolates due to the loss of a labile gentamicin resistance plasmid of approximately 18 kDa (data not shown). Pulsed-field gel electrophoresis (PFGE) following SmaI macrorestriction of the chromosomal DNA resulted in a typical banding pattern that was distinct from those of the other EMRSA strains then recognized in the United Kingdom, EMRSA-1 to EMRSA-15.
MRSA populations are known to undergo rapid evolution (6). Over the years, a number of phage variants of EMRSA-16 have been detected and confirmed by PFGE analysis to be EMRSA-16 or a variant of EMRSA-16. These were designated 29 (29inh/47), 47 (29inh/47/75), D (29inh/52inh/42E/77/83A/81/83C ± 80inh/95), and T (29inh/52inh), plus a miscellaneous group of variants considered to be EMRSA-16-like phage variants. In contrast to the recent EMRSA-15 study in which the phage variants were associated with a broadening of the phage susceptibility pattern, some of the phage variants here were associated with a narrowing of their susceptibility (24). However, little is known of the relationship between these EMRSA-16 phage and PFGE variants, and so here we report on the additional characterization, including antimicrobial susceptibility testing, urease production, toxin carriage, and PFGE, performed on a collection of classical and variant phage patterns of EMRSA-16 to determine whether variation in phage pattern is related to variation in PFGE profile and whether any of the other characteristics vary with particular changes in phage pattern and/or PFGE profile. This “snapshot,” similar to the previous EMRSA-15 study (23), should provide the basis for future investigations.
MATERIALS AND METHODS
Bacterial strains.
One hundred twenty-nine isolates from 52 hospitals collected between March 1998 and April 1999 were selected from the S. aureus database of MRSA isolates submitted to SaRS at Colindale on the basis of the phage patterns obtained. These comprised 49 classical isolates chosen as control strains for all but five of the hospitals where variants were identified (three duplicates from three hospitals), 15 variant 29 isolates, 10 variant 47 isolates, 19 variant T isolates, 18 variant D isolates, and a group of 18 miscellaneous isolates. The variant isolates were single representatives from any hospital in which that variant had been identified. The prototypic strain of EMRSA-16, NCTC 13143, was used as a control. Isolates both were stored on nutrient agar (NA) slopes at room temperature and were recovered by subculture on NA plates followed by overnight incubation at 37°C. Identification of isolates was confirmed by coagulase testing with rabbit plasma broth.
Phage typing.
All isolates were phage typed in duplicate by using the standard international set of phages (24) and local experimental phages 88A, 90, 83C, and 932 (20) at 100 times the routine test dilution (RTD) according to established protocols (8), and these results were compared with the results of the original phage typing to confirm their identity. Phage typing was performed at 100 times the RTD because many United Kingdom MRSA isolates are nontypeable at the RTD (19), and phage patterns were interpreted according to established criteria (8). The database at SaRS was examined to determine the prevalence of variant EMRSA-16 isolates across the United Kingdom and whether there were any significant differences in the geographical distribution.
PFGE.
Intact chromosomal DNA embedded in agarose blocks was prepared from overnight cultures grown at 37°C on NA (18), and plugs measuring 5 by 2 mm were digested with 20 U of SmaI (Boehringer, Mannheim, Germany) for 4 h at 25°C. Digested DNA was separated by using a DR-II contour-clamped homogeneous electric field apparatus (Bio-Rad Laboratories, Hercules, Calif.) in 1.2% agarose gels with 2 liters of 0.5× Tris-borate-EDTA recirculated at 12°C. A linear ramped pulse time of 1 to 80 s at 200 V was employed, for a total run time of 30 h. A lambda ladder (Bio-Rad Laboratories) was run every fifth or sixth lane to allow normalization of the gel, and the EMRSA-16 control strain NCTC 13143 was included as an internal control. Gel images were analyzed visually and with the aid of BioNumerics software version 1.5 (Applied Maths, Kortrijk, Belgium).
Antimicrobial susceptibility testing and urease production.
Antimicrobial susceptibility testing was performed by the agar dilution method according to the British Society of Antimicrobial Chemotherapy guidelines (10, 11) on Isosensitest agar (Oxoid, Basingstoke, United Kingdom) with 5% horse blood to determine the MICs of a panel of 14 antibiotics. Isolates that were found to be sensitive to methicillin were examined for carriage of the mecA gene by PCR as previously described (7). Isolates were tested for urease production by incubating inoculated Christensen's urea slopes for up to 5 days.
Enterotoxin detection.
Chromosomal DNA was prepared by using the Wizard genomic DNA preparation kit (Promega, Southampton, United Kingdom) according to the manufacturer's instructions. All isolates were screened in three multiplex PCR assays for the presence of enterotoxin genes A to E (sea to see) and G to J (seg to sei), exfoliative toxin genes (eta and etb), and the toxic shock syndrome toxin-1 gene (tst) (16). The PCR incorporated additional primers to detect the presence of the 16S ribosomal gene as an internal control.
RESULTS
Phage typing.
The most notable finding from repeat phage typing was that isolates belonging to phage variants 29 and 47 were indistinguishable, since the reaction with phage 75, which was originally thought to distinguish them, was highly variable. Thus, the characteristic phage pattern of this variant is 29/47 ± 52/75. One of the original variant 47 isolates was reclassified as variant T. Eighteen of the isolates with miscellaneous EMRSA-16-like phage patterns were also reclassified as either classical EMRSA-16 or one of the three variants (Table 1). Six isolates continued to give a miscellaneous or unclassifiable pattern that would usually be referred for PFGE analysis.
TABLE 1.
Comparison of original and repeat phage typing results
| Straina | No. of isolates
|
|
|---|---|---|
| Original phage result | Repeat phage result in duplicate | |
| Classical EMRSA-16 | 49 | 52 (includes 3 originally miscellaneous) |
| Variant 29 | 15 | |
| Variant 47 | 10 | |
| Variant 29/47b | 25 (includes 1 originally miscellaneous) | |
| Variant T | 19 | 23 (includes 3 originally miscellaneous and 1 originally variant 47) |
| Variant D | 18 | 23 (includes 5 originally miscellaneous) |
| Miscellaneous | 18 | 6 |
| Total | 129 | 129 |
Phage patterns are as follows: classical EMRSA-16, 29inh/52inh/75/77/83A/83C +; 29, 29inh/47; 47, 29inh/47/75; 29/47, 29inh/47 ± 52/75; T, 29inh ± 52inh; D, 29inh/52/42E/47/79/81/83C +; miscellaneous, combinations of the above, with or without additional reactions.
Variants 29 and 47 were determined to be the same on repeat phage typing and therefore renamed 29/47.
In 2000, classical EMRSA-16 accounted for 80% of all EMRSA-16 isolates submitted to SaRS from England and Wales. Variants 29/47, T, and D comprised 2, 7, and 9%, respectively, of isolates, with the miscellaneous group accounting for the remaining 2%. Variant 29/47 was found primarily in eastern England but also in London and the northwest, while both T and D variants were more widespread. Together these variants accounted for one-third of all EMRSA-16 isolates in the northwest, while the D variant alone accounted for almost a third of isolates in the West Midlands.
PFGE profiles.
All isolates in the study produced a PFGE profile identical or similar to that of the EMRSA-16 type strain, NCTC 13143, according to established criteria (28). A similarity correlation coefficient of 100% was obtained with intergel pattern alignment of the internal control strain NCTC 13143 when analyzed by using BioNumerics. Among the 129 EMRSA-16 isolates, there were 29 PFGE profiles, designated A1 to -29 (Table 2). The EMRSA-16 type strain was designated A1, as were 35 other classical EMRSA-16 isolates (67% of all classical isolates). The remaining 17 classical EMRSA-16 isolates produced 12 PFGE profiles with one to three band differences from the progenitor pattern, A1. Variant EMRSA-16 isolates showed 23 different PFGE profiles, including A1, which accounted for 22 of these isolates (29%). Six profiles, A2, A3, A5, A14, A15, and A16, accounted for 35 isolates (45% of all phage variants), while the other 17 profiles were represented by just one or two isolates each, accounting for 20 isolates (26%). All but 6 of the 25 phage variant 29/47 isolates gave profile A1, compared with 3 of variant T and none of variant D isolates. A16 was the most common profile among the phage variant T isolates, accounting for 7 of the 23 isolates, and was otherwise found in only 2 isolates with the classical phage pattern. Profiles A3 and A15 were particularly associated with phage variant D, accounting for 13 and 5 isolates, respectively, with A3 occurring additionally in one classical isolate and A15 being unique to this phage variant. All of the miscellaneous group isolates had different PFGE profiles, two of which were seen in three other isolates belonging to one of the classical or variant phage patterns and the remainder of which were unique. All eight of the mecA-negative isolates showed PFGE profiles (A4, A18, A24, A26, and A29) not found among mecA-positive (methicillin-resistant) isolates and were found in all phage groups except variant D.
TABLE 2.
PFGE profiles obtained according to EMRSA-16 phage pattern
| PFGE profile | No. of isolates with EMRSA-16 phage pattern:
|
Total no. of isolates | ||||
|---|---|---|---|---|---|---|
| Classical | 29/47 | T | D | Miscellaneous | ||
| A1 | 35 | 19 | 3 | 0 | 0 | 57 |
| A2 | 3 | 2 | 2 | 0 | 0 | 7 |
| A3 | 1 | 0 | 0 | 13 | 0 | 14 |
| A4 | 0 | 0 | 2 | 0 | 0 | 2 |
| A5 | 1 | 2 | 1 | 0 | 0 | 4 |
| A6 | 1 | 0 | 0 | 0 | 0 | 1 |
| A7 | 0 | 0 | 0 | 1 | 0 | 1 |
| A8 | 0 | 0 | 1 | 0 | 0 | 1 |
| A9 | 0 | 0 | 0 | 0 | 1 | 1 |
| A10 | 0 | 0 | 1 | 0 | 0 | 1 |
| A11 | 1 | 0 | 0 | 0 | 0 | 1 |
| A12 | 0 | 0 | 0 | 1 | 1 | 2 |
| A13 | 0 | 0 | 0 | 2 | 0 | 2 |
| A14 | 3 | 2 | 1 | 0 | 0 | 6 |
| A15 | 0 | 0 | 0 | 5 | 0 | 5 |
| A16 | 2 | 0 | 7 | 0 | 0 | 9 |
| A17 | 1 | 0 | 0 | 0 | 0 | 1 |
| A18 | 0 | 0 | 1 | 0 | 0 | 1 |
| A19 | 0 | 0 | 0 | 1 | 0 | 1 |
| A20 | 0 | 0 | 0 | 0 | 1 | 1 |
| A21 | 0 | 0 | 1 | 0 | 0 | 1 |
| A22 | 1 | 0 | 0 | 0 | 0 | 1 |
| A23 | 0 | 0 | 1 | 0 | 0 | 1 |
| A24 | 1 | 0 | 1 | 0 | 1 | 3 |
| A25 | 0 | 0 | 0 | 0 | 1 | 1 |
| A26 | 0 | 0 | 0 | 0 | 1 | 1 |
| A27 | 1 | 0 | 0 | 0 | 0 | 1 |
| A28 | 1 | 0 | 0 | 0 | 0 | 1 |
| A29 | 0 | 0 | 1 | 0 | 0 | 1 |
| Total | 52 | 25 | 23 | 23 | 6 | 129 |
Visual analysis of PFGE patterns resulted in a band difference matrix (not shown) that showed that 65 isolates (of 72 non-A1) with 21 patterns fell within one to three band differences of the prototypic pattern, A1. The remaining seven patterns, each representing single isolates, were within four to six band differences of A1. The dendrogram of percent similarity (Fig. 2) showed that all 29 EMRSA-16 profiles clustered at >76% correlation. In addition, both classical and phage variants of EMRSA-16 were distinct from the other recognized EMRSA strains currently known to be circulating in the United Kingdom.
FIG. 2.
BioNumerics-generated dendrogram of percent relatedness of PFGE profiles from putative EMRSA-16 isolates and United Kingdom EMRSA-15 and EMRSA-16 type strains (NCTC 13142 and NCTC 13143, respectively), calculated by using the Dice coefficient and represented by the unweighted pair group method with arithmetic mean. Band tolerances were set at 1.0%, and optimization was set at 0.5%.
Antimicrobial susceptibility patterns and mecA detection.
Nine isolates were sensitive to methicillin, and one of these was found to still carry the mecA gene. These isolates were included in the study despite being methicillin susceptible, since their phage patterns were as expected and loss of the mecA gene during storage is known to occur (A. Deplano, P. T. Tassios, Y. Glupczynski, E. Godfroid, and M. J. Struelens, Abstr. 9th Int. Symp. Staphylococci Staphylococcal Infect., abstr. 128, 2000). The most frequently observed profile was resistance to ciprofloxacin, erythromycin, kanamycin, neomycin, and methicillin, which was found in 63 isolates (49% of the total). The next most common profile was as described above with additional low-level resistance to mupirocin and was found in 26 isolates (a further 20% of the total). Both of these profiles were represented in the classical and all of the variant phage groups. Twelve isolates fit the classic picture of MRSA being multiresistant and exhibited additional resistance to gentamicin (nine classical isolates, two T isolates, and one miscellaneous isolate). Resistance to mupirocin was quite common, with low-level resistance (MIC, 16 to 256 mg/liter) in 33 isolates (26%) and high-level resistance (MIC, ≥512 mg/liter) in five isolates (4%). Occasional resistance to other antibiotics (streptomycin, fusidic acid, rifampin, and tetracycline) was observed in single isolates. All isolates were sensitive to vancomycin, teicoplanin, and pristinamycin.
Urease and enterotoxin gene carriage.
One hundred twenty-seven of the 129 EMRSA-16 isolates (one classical and one D variant) were found to be urease positive. All 129 isolates were positive for seg and sei and negative for seb, sec, sed, see, seh (one exception), sej, and also eta and etb. One hundred twenty isolates were also positive for sea and tst, with seven isolates negative for sea but positive for tst (three with PFGE profile A1 and one each with profiles A8, A18, A22, and A24) and two isolates negative for tst but positive for sea (profiles A16 and A27).
DISCUSSION
The United Kingdom was the first country to characterize epidemic MRSA strains and number them sequentially. This approach has since been adopted in a number of other countries. A group in Europe, including LHCAI, has recently developed a harmonized protocol for PFGE typing of MRSA (22) and has been collaborating with other researchers to compare this system with sequence-based typing approaches (reference 15 and unpublished data). Three of the most common EMRSA-16 variants (A2, A3, and A16) have been submitted to the Harmony collection of European EMRSA isolates (HARMONY project website, http://www.harmony-microbe.net/microtyping.htm) and are representative of phage variants with both narrower and wider lytic spectra. Along with the classical or prototypic strain of EMRSA-16, these variants have been shown to belong to multilocus sequence type ST36 and SCCmec type II (M. C. Enright, personal communication). In this paper, we have described data plotting the emergence and spread of EMRSA-16 from a single hospital and have characterized the recently identified phage variants of this strain. Given that EMRSA-16 was first recognized in the United Kingdom in 1992 and has since become widespread, it is not surprising that the number of both phage and PFGE variants is considerable, indicating the evolutionary pressure that the strain has been under over the past decade.
Identification of EMRSA-16 phage variants.
Two of the previously identified phage variants of EMRSA-16 (variants 29 and 47) can no longer be considered to be distinct, as repeat typing has revealed that they often present with only a minor, variable difference in phage pattern. Isolates with miscellaneous patterns were largely shown to belong to one of the already-recognized variants on repeat phage typing, while one isolate originally identified as 47 variant was found to give a pattern identifiable as a T variant (29/47 to 29/52). This could indicate a closer relationship between these phage variants than previously thought, especially as these two variants, along with classical EMRSA-16, share a number of common PFGE profiles. Where there is any uncertainty, phage typing should be repeated and the isolate should be examined by PFGE for a more definitive result.
Arbeit previously described the roles that phage insertions, deletions, and replacements within the chromosome play in determining the phage type of a bacterial strain (5). This study suggests that classical EMRSA-16 has given rise to two different population groups of phage variants: 29/47 and T with a narrower lytic spectrum and D with a much wider lytic spectrum than the prototypic strain. In the former group, the bacterial chromosome is much more likely to be heavily lysogenized, while in the latter, the degree of lysogeny is likely to be considerably less. The recent EMRSA-15 study showed that the broadening of the phage pattern for EMRSA-15, similar to that observed with the epidemic penicillinase-producing 80/81 strain of the 1950s that spread and persisted in several continents (24), could be associated with particular PFGE profiles. Replacement of a defective prophage, which was responsible for the resistance of the original 80/81 strain to phages 52 and 52A, by other phages resulted in additional lytic reactions with these phages (26). A similar mechanism could account for both the broadening and narrowing of the phage patterns seen with EMRSA-16, although, as with the EMRSA-15 study, the phages responsible have yet to be identified. Limited cross spotting of some isolates with phages was performed previously in LHCAI and showed that some band differences in PFGE patterns seen between isolates were probably due to the gain or loss of phage, which may or may not have a SmaI restriction site (17). Interestingly, the different phage patterns obtained over a 10-year period in a Belgian study were also investigated by PFGE, and a number of phage types were found to have the same PFGE profile, suggesting that there had been a drift in the epidemic phage patterns within a conserved PFGE profile (30). More-detailed studies are warranted and may inform these and other typing systems. Many of the isolates typed in this study were stored on agar slopes for several years. This could have introduced new banding patterns other than those related to the loss of mec-related genes. However, the majority of the profiles seen were also observed among fresh isolates seen recently in the laboratory, and we are confident that they are reliable indicators of the profiles seen in patients and relevant to providing the insights needed to inform investigation, prevention, and control strategies.
Discrimination between EMRSA-16 isolates by PFGE.
PFGE discriminated between the isolates with the classical phage pattern as well as among those with variant phage patterns. However, as the same PFGE profile was seen in isolates from different phage groups, associations were not always clear-cut. There was considerable overlap between the PFGE profiles detected in classical EMRSA-16 and those phage variants with a narrower lytic spectrum, 29/47 and T, suggesting a closer relationship between these. On the other hand, variant D, with a broader lytic spectrum, is associated with PFGE profiles not found among the classical and other EMRSA-16 phage variants, with just one exception (one A3 profile was found among classical EMRSA-16).
Criteria for interpretation of PFGE data.
Bacterial strains change over time, with the consequent accumulation of variant PFGE profiles, some of which will differ significantly from the prototypic strain but will still be closely related to one of the other profiles. Of the 28 PFGE profiles other than A1, 21 were found to be within one to three band differences and 7 were found to be within four to six band differences of the prototypic pattern, A1, which fits well with the criteria of Tenover et al. for determining strain relatedness for use within epidemiologically and temporally defined outbreaks (28). Referring back to the most common or prototypic profile will give a good indication that the profile belongs to the EMRSA-16 cluster. However, comparison with all recognized profiles may indicate another more closely related profile, while conversely, higher numbers of band differences will be seen among more divergent PFGE profiles indicating different strains if compared in isolation. Comparison with the other current United Kingdom strains, EMRSA-1, -3, -15, and -17 and Irish-01 and -02, shows that EMRSA-16 is unrelated to these, as they all differ by at least 10 bands (data not shown). In this study, the majority of PFGE profiles associated with EMRSA-16 clustered at ≥80% by the Dice coefficient, above which isolates are often considered to be related (27), but the correlation for all of the profiles was ≥76%. The eye can often distinguish very minor differences that may not be recognized by the computer software due to the tolerance settings chosen. For example, profiles A1 and A2 differ from each other by just a slight shift in the band at approximately 160 kb, which was not detected with a tolerance of 1.0% by using BioNumerics. By decreasing the tolerance to 0.8%, BioNumerics was able to distinguish between the two profiles; however, some isolates with the A1 profile appeared to be less closely related to other identical isolates, including the control strain NCTC 13143. It should be emphasized that applying recognized cutoffs for determining strain relatedness as used in outbreak situations should be done with caution with isolates such as these, which are neither geographically nor temporally linked.
Toxin carriage, urease production, and antibiograms.
Very little variation was observed in toxin gene carriage or urease production, nor was any association made between antimicrobial susceptibility patterns and phage or PFGE profiles. For the methicillin-susceptible isolates in this study, no obvious common band shift was noted to distinguish them from methicillin-resistant isolates, as occurred with EMRSA-15 (22).
Further geographic spread.
EMRSA-16 is not confined to the United Kingdom and Ireland. The recent HARMONY project, which has established a collection of epidemic strains of MRSA currently circulating in Europe and involves cooperation between 14 or more countries, has demonstrated the recent spread of EMRSA-16 in Scandinavia (HARMONY project website, http://www.harmony-microbe.net/microtyping.htm) (22). The Gothenburg strain from Sweden, originally identified as originating in Cyprus (hence also known as the Cyprus strain), has recently been documented as having spread to Denmark (C. S. Elsberg, J. Mondrup, N. Frimodt-Moller, L. Larsson, S. Murchan, and C. Welinder-Olsson. Abstr. 9th Int. Symp. Staphylococci Staphylococcal Infect., abstr. 232, 2000), while in Finland, the E5 strain is known to have originated in Turkey (S. Salmenlinna and J. Vuopio-Varkila [National Public Health Institute, Helsinki, Finland], personal communication). Both of these strains were shown to be closely related to the United Kingdom EMRSA-16 prototypic strain (data not shown), with both similar phage patterns when typed at SaRS and similar SmaI macrorestriction patterns. More recently, a Swiss strain (Geneva clone 2) related to EMRSA-16 was reported, with one isolate having originated in Greece (9). EMRSA-16 has also been identified in Norway (29), Spain (25), and the United States (21), where it is reported to be the second most common U.S. healthcare-associated pulsed-field type. Thus, the potential for further international spread of this strain is obvious. Already there appear to be four international foci of this strain of MRSA: the United Kingdom and Ireland, Scandinavia, the southeastern Mediterranean, and the United States. Further data on multilocus sequence type and SCCmec typing would be useful to validate these findings.
Conclusions.
We have described for the first time the origin and local and supraregional spread of a United Kingdom EMRSA strain. Phage typing distinguishes between isolates of EMRSA-16 with the classical pattern and three main variants, 29/47, T, and D. Investigation of these strains revealed that PFGE is more discriminatory than phage typing and has confirmed that classical isolates and the variants with a narrower phage pattern, 29/47 and T strains, are closely related. With one exception, the D variants, with a broader phage pattern, are associated with distinct PFGE profiles.
It is important to remember that no typing tool among those currently available is able to answer all epidemiological questions. Phage typing is currently the cheapest first-line approach for reference laboratories such as ours, which are typing approximately 10,000 isolates per year (in the mid 1990s we typed approximately 40,000 isolates per year), and PFGE is then used for validation of new types or additional characterization when needed on epidemiological grounds. When there are suspected examples of cross-infection with EMRSA-16 with the classical phage pattern and PFGE profile but epidemiological data are poor, it is impossible to provide more definitive reports to inform infection control teams, as the strain is now so widespread. We are currently exploring various sequence-based typing approaches to see if they can provide more discriminatory tools.
Acknowledgments
This study was supported by HARMONY, a European Union DGXII funded project (contract no. BMH4-CT96).
S.M. was the project typing leader, and B.D.C. was the overall project leader.
We thank Tyrone Pitt for critical comments on this manuscript, Polly Kaufmann and Jane Turton from the Epidemiological Typing Unit in the Laboratory of Healthcare-Associated Infection for assistance with the gel normalizations and analyses, and Marina Warner from the Antibiotic Resistance Monitoring and Reference Laboratory and Ana New from SaRS for technical assistance.
REFERENCES
- 1.Anonymous. 1993. EMRSA-16; a new epidemic strain of Staphylococcus aureus. Commun. Dis. Rep. 3:5 February. [PubMed]
- 2.Anonymous. 1993. The spread of EMRSA-16. Commun. Dis. Rep. 3:2 July. [PubMed]
- 3.Anonymous. 2000. North-South study of MRSA in Ireland in 1999. Brunswick Press Limited, Dublin, Ireland.
- 4.Anonymous. 2001. Methicillin-resistant Staphylococcus aureus (MRSA) reports to SCIEH. Scott. Ctr. Infect. Environ. Health Wkly. Rep. 35:147. [Google Scholar]
- 5.Arbeit, R. D. 1997. Laboratory procedures for epidemiologic analysis, p. 253-286. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 6.Ayliffe, G. A. J. 1997. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 24(Suppl. 1):S74-S79. [DOI] [PubMed] [Google Scholar]
- 7.Bignardi, G. E., N. Woodford, A. Chapman, A. P. Johnson, and D. C. E. Speller. 1996. Detection of the mecA gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance. J. Antimicrob. Chemother. 37:53-63. [DOI] [PubMed] [Google Scholar]
- 8.Blair, J. E., and R. E. O. Wilson. 1961. Phage typing of staphylococci. Bull. W. H. O. 24:771-784. [PMC free article] [PubMed] [Google Scholar]
- 9.Blanc, D., A. N. Banuls, P. M. Hauser, P. Moreillon, P. Francioli, M. Tibayrenc, et al. 2001. Methicillin-resistant Staphylococcus aureus: phylogenetic relatedness between European epidemic clones and Swiss sporadic clones. Microb. Drug Resist. 6:231-238. [DOI] [PubMed] [Google Scholar]
- 10.British Society for Antimicrobial Chemotherapy. 1991. A guide to sensitivity testing. J. Antimicrob. Chemother. Suppl. D 27:1-50. [PubMed] [Google Scholar]
- 11.British Society for Antimicrobial Chemotherapy. 1998. BSAC standardised disc sensitivity testing method. Newsl. Br. Soc. Antimicrob. Chemother. 1998:Summer.
- 12.British Society for Antimicrobial Chemotherapy, Hospital Infection Society, and Infection Control Nurses Association. 1998. Revised guidelines for the control of methicillin-resistant Staphylococcus aureus infection in hospitals. J. Hosp. Infect. 39:253-290. [DOI] [PubMed] [Google Scholar]
- 13.Cookson, B. D. 1999. Nosocomial antimicrobial resistance surveillance. J. Hosp. Infect. 43(Suppl.):S97-S103. [DOI] [PubMed] [Google Scholar]
- 14.Cox, R. A., C. Conquest, C. Mallaghan, and R. R. Marples. 1995. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 29:87-106. [DOI] [PubMed] [Google Scholar]
- 15.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, W. M., S. D. Tyler, E. P. Ewan, F. E. Ashton, D. R. Pollard, and K. R. Rozee. 1991. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin-1 in Staphylococcus aureus by the polymerase chain reaction. J. Clin. Microbiol. 29:426-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorgensen, M., R. Givney, M. Pegler, A. Vickery, and G. Funnell. 1996. Typing multi-drug-resistant Staphylococcus aureus: conflicting epidemiological data produced by genotypic and phenotypic methods clarified by phlyogenetic analysis. J. Clin. Microbiol. 34:398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufmann, M. E. 1998. Pulsed-field gel electrophoresis, p. 33-50. In N. Woodford and A. P. Johnson (ed.), Methods in molecular medicine, vol 15. Molecular bacteriology: protocols and clinical applications. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 19.Kerr, S., G. E. Kerr, C. A. Mackintosh, and R. R. Marples. 1990. A survey of methicillin-resistant Staphylococcus aureus affecting patients in England and Wales. J. Hosp. Infect. 16:35-48. [DOI] [PubMed] [Google Scholar]
- 20.Marples, R. R., J. F. Richardson, and M. J. de Saxe. 1986. Bacteriological characters of strains of Staphylococcus aureus submitted to a reference laboratory related to methicillin resistance. J. Hyg. (Cambridge) 96:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. Elsberg Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A., A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis for epidemiological typing of methicillin-resistant Staphylococcus aureus by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Neill, G. L., S. Murchan, A. Gil-Setas, and H. M. Aucken. 2001. Identification and characterization of phage variants of a strain of epidemic methicillin-resistant Staphylococcus aureus (EMRSA-15). J. Clin. Microbiol. 39:1540-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker, M. T. 1983. The significance of phage typing patterns in Staphylococcus aureus, p. 33-62. In C. S. F. Easmon and C. Adlam (ed.), Staphylococci and staphylococcal infections. Academic Press Inc., London, England.
- 25.Perez-Roth, E., F. Lorenzo-Diaz, and S. Mendez-Alvarez. 2003. Establishment and clonal dissemination of the methicillin-resistant Staphylococcus aureus UK-16 epidemic strain in a Spanish hospital. J. Clin. Microbiol. 41:5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rountree, P. M., and E. H. Asheshov. 1961. Further observations on changes in the phage-typing pattern of phage type 80/81 staphylococci. J. Gen. Microbiol. 26:111-122. [DOI] [PubMed] [Google Scholar]
- 27.Struelens, M. J., A. Deplano, C. Godard, N. Maes, and E. Serruys. 1992. Epidemiological typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macro-restriction analysis of genomic DNA using pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tveten, Y., A. Jenkins, A. G. Allum, B.-E. Kristiansen, et al. 2003. Heterogeneity of methicillin-resistant Staphylococcus aureus isolated in Norway. Clin. Microbiol. Infect. 9:886-892. [DOI] [PubMed] [Google Scholar]
- 30.Wildenamauwe, C., C. Godard, G. Verschraegen, G. Claeys, M.-C. Duyck, H. De Beenhouwer, and R. Vanhoof. 2004. Ten years of phage typing of Belgian clinical methicillin-resistant Staphylococcus aureus isolates (1992-2001). J. Hosp. Infect. 56:16-21. [DOI] [PubMed] [Google Scholar]