Abstract
The cyanobacterium Microcystis aeruginosa is widely known for its production of the potent hepatotoxin microcystin. Microcystin is synthesized nonribosomally by the thiotemplate function of a large, modular enzyme complex encoded within the 55-kb microcystin synthetase (mcy) gene cluster. Also encoded within the mcy gene cluster is a putative ATP binding cassette (ABC) transporter, McyH. This study details the bioinformatic and mutational analyses of McyH and offers functional predictions for the hypothetical protein. The transporter is putatively comprised of two homodimers, each with an N-terminal hydrophobic domain and a C-terminal ATPase. Phylogenetically, McyH was found to cluster with members of the ABC-A1 subgroup of ABC ATPases, suggesting an export function for the protein. Two mcyH null mutant (ΔmcyH) strains were constructed by partial deletion of the mcyH gene. Microcystin production was completely absent in these strains. While the mcyH deletion had no apparent effect on the transcription of other mcy genes, the complete microcystin biosynthesis enzyme complex could not be detected in ΔmcyH mutant strains. Finally, expression levels of McyH in the wild type and in ΔmcyA, ΔmcyB, and ΔmcyH mutants were investigated by using immunoblotting with an anti-McyH antibody. Expression of McyH was found to be reduced in ΔmcyA and ΔmcyB mutants and completely absent in the ΔmcyH mutant. By virtue of its association with the mcy gene cluster and the bioinformatic and experimental data presented in this study, we predict that McyH functions as a microcystin exporter and is, in addition, intimately associated with the microcystin biosynthesis pathway.
The cyclic heptapeptide microcystin, first isolated from the freshwater cyanobacterium Microcystis aeruginosa, is a potent hepatotoxin (32). In addition to causing death and illness in animals, fish, and livestock, chronic exposure to sublethal doses of microcystin has been linked to the high incidence of certain types of liver cancer (hepatocellular carcinoma) in several communities in China (35).
Like many other biologically active secondary metabolites, microcystin is synthesized nonribosomally by the thiotemplate function of a large multifunctional enzyme complex (5). The gene cluster encoding this enzyme complex spans 55 kb and comprises 10 genes arranged in two gene clusters that are divergently transcribed, mcyA to -J, encoding three peptide synthetases (McyA to -C), a modular polyketide synthase (McyD), two hybrid enzymes comprising peptide synthetase and polyketide synthase modules (McyE and McyG), and enzymes putatively involved in the tailoring (McyJ, -F, and -I) and transport (McyH) of the toxin (33).
Release of microcystin into the extracellular environment has in the past been attributed to the death and lysis of cyanobacterial blooms (31). Recent cellular and genetic investigations, however, have since provided evidence in support of a microcystin export pathway. For example, Shi et al. (28) used in situ hybridization and electron microscopy to demonstrate the presence of microcystin in the wall and sheath area of intact M. aeruginosa cells. A later study by Kaebernick et al. suggested that microcystin is constitutively produced under low and medium light intensities and is exported when a certain higher threshold intensity is reached (15). The discovery of mcyH, situated within the mcy gene cluster, has thus led us to hypothesize that this putative ABC transporter may be responsible for microcystin export in M. aeruginosa.
Preliminary sequence analysis of the McyH protein suggested that it belonged to the ATP binding cassette (ABC) transporter superfamily (33). Members of this superfamily possess the characteristic A and B signature sequences described by Walker et al., (34) and are responsible for the ATP-dependent transport of a vast range of solutes (allocrites) across intracellular and cell surface biological membranes (for a review see reference 14). While to our knowledge, mcyH is the first ABC transporter-encoding gene to be described for M. aeruginosa, nearly 50% of all cellular transporters in the cyanobacterium Synechocystis belong to the ABC superfamily (22). Previously characterized cyanobacterial ABC transporters have been implicated in the transport of an extensive range of allocrites, including manganese (1), bicarbonate (21), osmoprotectants (10), and glycolipids (6). The association of ABC transporter genes with the gene clusters of nonribosomally synthesized peptides, such as microcystin, is not uncommon. While several such associations have been identified in cyanobacteria (e.g., with nostopeptolide [13] and microginin [D. Kramer, personal communication]), the vast majority have been discovered in gram-positive, sporulating bacteria. Some examples include putative transporters associated with the biosynthesis gene clusters of amphotericin B (2), rapamycin (26), bleomycin (3), frenolicin (accession number AF058302), bacitracin (16), yersiniabactin (9), and exochelin (36, 37). Such proteins have been proven or hypothesized to confer allocrite resistance to the producing organisms.
This paper presents a bioinformatic investigation of the putative ABC transporter McyH in M. aeruginosa PCC 7806. Phylogenetic trees encompassing McyH and representative members from across the entire ABC superfamily have provided structural and functional predictions for the putative transporter in M. aeruginosa. We have, in addition, constructed two ΔmcyH mutant strains and tested their ability to produce microcystin. The transcription and translation of different mcy gene products in these mutant strains were investigated.
MATERIALS AND METHODS
Cyanobacterial strains and culturing.
The microcystin-producing strain M. aeruginosa PCC 7806 was kindly provided by J. Weckesser (Freiburg University, Freiburg, Germany) and R. Rippka (Pasteur Institute, Paris, France). Wild-type (WT) cells were cultured in BG11 medium (Fluka, St. Gallen, Switzerland), and ΔmcyA, ΔmcyB, and ΔmcyH mutants were cultured in BG11 medium supplemented with 3 μg of chloramphenicol · ml−1. Cells were grown under 16 μmol of photons of white light m−2 s−1. Light intensities were measured by using a LI-CORR LI-250 light meter (Walz, Effeltrich, Germany). The optical density at 750 nm (OD750) of cultures was measured with an Ultrospec II (LKB, Biochrom, Cambridge, United Kingdom).
Sequence analysis of mcyH.
Primary amino acid sequences were deduced and analyzed by using Translate (Genetics Computer Group) (ANGIS, Sydney, Australia). Scores of percent similarity and identity to other peptide sequences were determined by using the Blastp program (National Center for Biotechnology Information [NCBI], Bethesda, Md.). Sequence alignments were performed by using ClustalW (ANGIS) with default parameters. Determination of membrane spanning and motifs was achieved by using the programs DAS, HMMTOP, TMpred, and TOPpred2 (ExPASy) and BLAST CD-search (NCBI). Signal peptide probability was determined with SignalP (ExPASy). Codon usage was analyzed with the Kazusa Countcodon program (17).
Phylogenetic analysis was based on the most conserved region of the primary peptide sequences of the ABC proteins, including the Walker A and B consensus sequences and the linker peptide. The sequences used have a mean length of 170 amino acids (114 to 203 amino acids) (Table 1). Genetic distances were calculated by using an algorithm based on a protein weight matrix (PAM). Phylogenetic trees were generated by using the neighbor-joining method (23). Trees were displayed graphically by using Njplot and MacDraw II.
TABLE 1.
ABC modules used to create the phylogenetic tree
| Subgroup | Allocrite or class | Protein name | Organism | Positiona | Lengthb | Accession no.c |
|---|---|---|---|---|---|---|
| A1 | PgP | MsbA | Escherichia coli | 369-554 | 185 | AAG55399 |
| Proteins | PrtD | Erwinia chrysantemi | 357-541 | 184 | P23596 | |
| Yersiniabactin | YbtQ | Yersinia pestis | 375-555 | 180 | AAC69584 | |
| Amphotericin C | AmphG | Streptomyces nodosus | 387-570 | 183 | AAK73498 | |
| Amphotericin N | AmphH | Streptomyces nodosus | 386-568 | 182 | AAK73499 | |
| Bacterocins | ComA | Streptococcus pneumoniae | 510-692 | 182 | P59653 | |
| Bleomycin | Streptomyces verticillus | 386-524 | 138 | AAB00463 | ||
| CFTR (chloride ions) | CFTR(-N) | Homo sapiens | 451-622 | 171 | P13569 | |
| Exochelin | ExiT | Mycobacterium smegmatis | 930-1044 | 114 | AAC32046 | |
| Microcystin | McyH | Microcystis aeruginosa | 350-518 | 168 | AAF00956 | |
| Modified cyclic peptides | SyrD | Pseudomonas syringae | 373-551 | 178 | P33951 | |
| A2 | Antibiotics (class I) | VgA | Staphylococcus aureus | 298-456 | 158 | AAA26684 |
| Antibiotics (class II) | SrmB | Streptomyces ambofaciens | 380-500 | 120 | S25202 | |
| EF3 | EF3(-C) | Candida albicans | 857-973 | 116 | P25997 | |
| EF3 | EF3(-N) | Candida albicans | 459-600 | 141 | P25997 | |
| Heme | HelA | Rhodobacter capsulatas | 29-156 | 127 | P29959 | |
| B1 | Osmoprotectants | Synechocystis sp. | 29-210 | 181 | AAB41280 | |
| Phosphonates | Phnk | Escherichia coli | 31-222 | 191 | P16678 | |
| Polar amino acids | HisP | Escherichia coli | 32-228 | 196 | AAC75366 | |
| Rapamycin | Streptomyces hygroscopicus | 40-226 | 186 | T30218 | ||
| Bicarbonate | CmpC | Synechoccous sp. | 35-215 | 180 | BAA05388 | |
| Frenolicin | FrnD(-C) | Streptomyces roseofulvus | 308-477 | 169 | AAC18099 | |
| Frenolicin | FrnD(-N) | Streptomyces roseofulvus | 40-237 | 197 | AAC18099 | |
| Abc (multidrug resistance homolog) | MetN | Escherichia coli | 31-217 | 186 | P30750 | |
| Glycolipids | DevA | Anabaena sp. | 36-183 | 147 | A55541 | |
| Ions | SfuC | Serratia marcescens | 29-212 | 183 | P21410 | |
| Monosaccharides | AraG | Escherichia coli | 33-219 | 168 | P08531 | |
| Nitrate | NrtD | Synechococcus sp. | 45-226 | 181 | P38046 | |
| Oligopeptides | AmiE | Streptococcus pneumoniae | 49-236 | 187 | P18765 | |
| B2 | Yhgb | NtrA | Acidthiobacillus ferrooxidans | 30-213 | 183 | P24693 |
| Antibiotics (class III) | DrrA | Streptomyces peucetius | 34-215 | 181 | P32010 | |
| Branched-chain amino acids | BraF | Pseudomonas aeruginosa | 27-230 | 203 | P21629 | |
| Nodulation factors | NodI | Bradyrhizobium japonicum | 33-214 | 181 | 1914270A |
Amino acids encompassed in sequence used to create the phylogenetic tree.
Length of the amino acid sequence used.
References to the papers corresponding to database entries can be found in the corresponding database files.
Insertional inactivation of the mcyH gene.
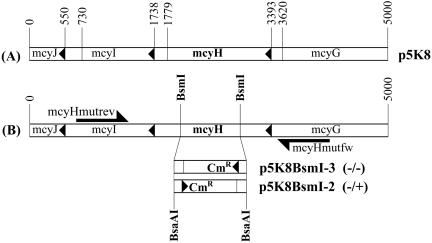
Oligonucleotide primers mcyH5KF (5′-ACTATTAACCCGCAGCCAGA-3′) and mcyH5KR (5′-GGGAGGTATGCTCGATCTTG-3′) were used to amplify by PCR a 5,000-bp fragment corresponding to nucleotides 1279 to 6279 of the of the mcy gene cluster (GenBank accession number AF183408). PCR amplifications were performed by using the Advantage genomic PCR kit (Clontech, Palo Alto, Calif.) according to the manufacturer's protocol. The resulting fragment was cloned into the pGEM-T vector (Promega, Madison, Wis.), giving rise to plasmid p5K8. A 1,123-bp internal fragment of the mcyH gene was excised from p5K8 with BsmI, and the remaining linear construct was subsequently treated with Klenow fragment (Fermentas, Vilnius, Latvia) to produce blunt ends. A 1,414-bp chloramphenicol resistance (Cmr) cassette, excised from plasmid pACYC184 (Fermentas) by using BsaAI, was then ligated into the blunt and linearized p5K8 vector. The resulting construct, p5K8BsmI, was transformed into Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.), which was subsequently grown under selection with chloramphenicol (34 μg · ml−1) and ampicillin (100 μg · ml−1). The resulting transformants were checked for p5K8BsmI via PCR and sequencing with primers Cmrfw (5′-TTTAGCTTCCTTAGCTCCTG-3′) and Cmrrev (5′-CCAACCGGTGATACCA-3′). The orientation of the inserted Cmr cassette was determined via PCR with primers mcyHmutfw (5′-CACGGGTTGTTTTAGGATAA-3′) and mcyHmutrev (5′-TTCAAACATTGCAAACTCCA-3′). Two constructs with differing Cmr cassette orientations, p5K8BsmI-2 and p5K8BsmI-3, were selected for subsequent transformation of M. aeruginosa PCC 7806 (see Fig. 3 for details).
FIG. 3.
(A) Construction of plasmid p5K8. A diagram illustrating the region of the mcy cluster amplified by primers mcyHSKF and mcyHSKR and cloned into pGEM-T to give rise to plasmid p5K8 is shown. (B) Construction of plasmids p5K8BsmI-3 and p5K8BsmI-2. A schematic diagram depicting the partial deletion of mcyH in p5K8, resulting in the knockout constructs p5K8BsmI-3 and p5K8BsmI-2, is shown. The directions of transcription of the mcyH and Cmr genes are indicated by (−/−) and (−/+), respectively.
The plasmids p5K8BsmI-2 and -3 were used to perform the partial deletion of mcyH in M. aeruginosa PCC 7806. Recombinant plasmid DNA (10 μg) was transformed via electroporation as previously described (33). Transformants were grown and selected from a chloramphenicol gradient (5). The microcystin contents of mutant and WT whole-cell extracts were measured via matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry as described previously (5). Mutant and WT cell extracts were also analyzed via immunoblotting with anti-McyH and anti-McyF anisera to check for the expression of McyH and McyF, respectively (see “Immunodetection of McyH and McyF” below).
RPA.
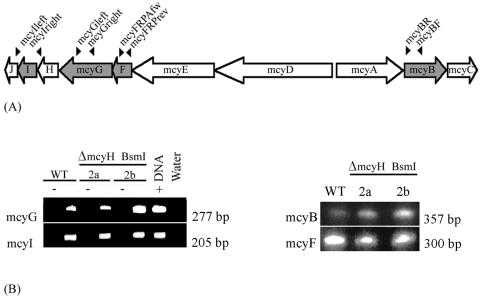
For RNA isolations, 50-ml cultures (mid-exponential phase; OD750 of 0.7 to 1.0) were incubated on ice for 10 min, centrifuged at 4,000 × g for 10 min at 4°C, and stored at −20°C. RNA extractions were performed as described previously (15), using Trizol reagent (Gibco BRL, Life Technologies, Gaithersburg, Md.). Oligonucleotides mcyBF (5′-AGGAACAAGTTGCACAGAATCCGCA-3′) and mcyBR (5′-ACTAATCCCTATCTAAACACAGTAACTCA-3′) were used to amplify a 357-bp DNA fragment from the mcyB gene (33), primers mcyFRPAfw (5′-GGTTACCGCCGAATTTCTTA-3′) and mcyFRPArev (5′-TGGAGTTCTTGGTCCGCTAT-3′) were used to amplify 300 bp of the mcyF gene, primers mcyGright (5′-GGGAATCAATCCCCATTTC-3′) and mcyGleft (5′-AACACAGGTTTTAATCGCCG-3′) were used to amplify 277 bp of the mcyG gene, and primers mcyIright (5′-GTTGCTCCTACTGTCTCCGC-3′) and mcyIleft (5′-AGCCTTGCTAAAAAGCTCC-3′) were used to amplify 205 bp of the mcyI gene (see Fig. 6A). PCR fragments were subsequently ligated into the pGEM-T vector in the antisense direction to the T7 promoter. After linearization of the vector with BcuI, the RNA probe was prepared by using the Maxiscript kit (Ambion) according to the manufacturer's instructions. RNase protection assays (RPAs) were carried out according to the supplied protocol (Roche, Mannheim, Germany).
FIG. 6.
Transcript analysis of WT and ΔmcyH mutants. (A) Schematic representation of the microcystin synthetase gene cluster, showing relative positions of primers used to amplify RPA probes within the mcyB, mcyF, mcyG, and mcyI genes. (B) mcyB, mcyF, mcyG, and mcyI transcripts from the WT and the ΔmcyH mutant, analyzed by the RPA. Positive (WT DNA) and negative (yeast tRNA) controls are indicated by + and −, respectively.
Expression and purification of polyhistidine-tagged McyH ATPase and McyF.
The DNA fragment encoding the putative ATPase domain of McyH (nucleotides 3056 to 3860; accession number AF183408) from M. aeruginosa PCC 7806 was amplified by PCR with primers AbcMidF (5′-GTTATGTTGAGCGTCTATCTG-3′) and AbcSR (5′-acgatgactactacttcacc-3′). The resulting 804-bp PCR product was cloned directly into the pGEM-T Easy vector and then excised with EcoRI and subcloned into pBluescript at the EcoRI site. After sequence verification, the putative ATPase-encoding fragment was excised from pBluescript with SacI and KpnI and ligated into the polyhistidine tag-incorporating pET30 expression vector (Novagen, Cambridge, Mass.) at the corresponding restriction sites. The resulting expression construct (pET-abc) was finally transformed into the Rosetta(DE3)(pLysS) E. coli strain (Novagen) for subsequent heterologous expression of the ATPase.
Expression of the recombinant peptide was performed on a 1-liter scale. Briefly, 1 liter of Luria-Bertani broth supplemented with 50 μg of kanamycin · ml−1 and 34 μg of chloramphenicol · ml−1 was inoculated with 1 ml of overnight culture and grown with shaking at 190 rpm and 37°C to an OD650 of 0.6. The incubation temperature was then reduced to 30°C, and the culture was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubated for a further 3.5 h. Cells were harvested via centrifugation at 6,000 × g for 10 min and stored at −80°C. The cell pellet was thawed, resuspended in 50 ml of ice-cold binding buffer (0.02 M Na2HPO4, 1 M NaCl [pH 7.2]), disrupted via three successive rounds of sonication (15 s at output 4; duty cycle, 40%) (Sonifier 250; Branson, Danbury, Conn.), and centrifuged for 30 min at 4°C and 20,000 × g. The pellet was resuspended in 25 ml of binding buffer with 8 M urea plus 20 mM imidazole and centrifuged as described above. The resulting supernatant was passed through a 0.2 μm-pore-size filter, and loaded onto an Ni2+-charged HiTrap column (Amersham Biosciences, Uppsala, Sweden), washed with 10 volumes of wash buffer (0.02 M Na2HPO4, 1 M NaCl, 8 M urea, 40 mM imidazole [pH 7.2]), and finally eluted in 2 volumes of elution buffer (0.02 M Na2HPO4, 1 M NaCl, 8 M urea, 300 mM imidazole [pH 7.2]). The eluate was collected in 0.5-ml fractions and analyzed via sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Coomassie brilliant blue staining.
The aspartate racemase (McyF) of the microcystin biosynthesis gene cluster was heterologously expressed in E. coli (by using the pET15b expression system [Novagen]) and purified as described previously (30).
Immunodetection of McyH and McyF.
Three hundred to 600 μg of the 90 to 95% pure recombinant McyH and McyF protein was used to raise anti-McyH and anti-McyF antibodies in rabbits (Pineda Antikörper Service, Berlin, Germany). Immunoglobulin G (IgG) antibodies from the final sera were precipitated by using 40% (NH4)2SO4, concentrated to 30% of the initial volume, and dialyzed 24 h in 1× phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]) at 4°C. The purified and concentrated IgG antibodies were used for subsequent immunodetection of native McyF.
Fifty milliliters of WT and mutant (ΔmcyH, ΔmcyA, and ΔmcyB) cultures were harvested via centrifugation (5,000 × g for 10 min at 4°C) and resuspended in buffer A (500 mM Tris-HCl [pH 7.5], 50 mM EDTA, 2 μM phenylmethylsulfonyl fluoride). The cells were then disrupted via freeze-thawing and vortexing (7). Total protein concentrations in samples were measured by using the Bio-Rad (Munich, Germany) protein assay.
McyH and McyF expression was analyzed via immunoblotting. Twenty micrograms of total cell protein was electrophoresed on SDS-12 to 15% polyacrylamide gels by using the Mini Protean apparatus (Bio-Rad). Proteins were then blotted to Hybond C-extra membranes (Amersham Biosciences) under semidry conditions (11). After blocking overnight in PBS-T (140 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 18 mM KH2PO4, 0.3% Tween) plus skim milk (5%, wt/vol), the membranes were incubated for 1 h with anti-McyH or anti-McyF primary antibodies (diluted 1/5,000 in blocking buffer). Membranes were washed three times (10 min each) in PBS-T and subsequently incubated for 45 min with anti-rabbit IgG (diluted 1/5,000 in PBS-T) coupled to horseradish peroxidase (Pierce, Rockford, Ill.). After being washed three times (10 min each) in PBS-T, blots were developed via chemiluminescence with the Supersignal West Pico chemiluminescent substrate according to the protocol of the manufacturer (Pierce).
Database entries.
The NCBI accession numbers for the M. aeruginosa microcystin biosynthesis (mcy) gene cluster and McyH protein are AF183408 and AAF00956, respectively (33). The accession numbers of proteins used in the sequence alignments in Fig. 1 are listed in Results. Those used to create the phylogenetic tree in Fig. 2 are listed in Table 1.
FIG. 1.
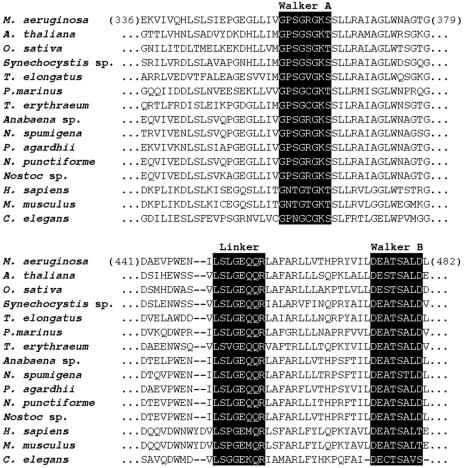
Alignment of McyH amino acid sequence (residues 336 to 379 and 441 to 482) with several homologous ABC transporters. The translated primary peptide sequence of McyH (NCBI accession number AAF00956) was aligned with peptide sequences from A. thaliana (NP175837), O. sativa (BAB16495), Synechocystis sp. (NP442354), T. elongatus (NP681014), P. marinus (NP895422), T. erythraeum (ZP00071302), Anabaena sp. (AAO62579), N. spumigena (AAO64410.), P. agardhii (CAD29796), N. punctiforme (ZP00110694), Nostoc sp. (AAO23332), H. sapiens (NP064719), M. musculus (NP033018), and C. elegans (NP495407). The signature sequences of the ABC superfamily (linker and Walker sites) are highlighted.
FIG. 2.
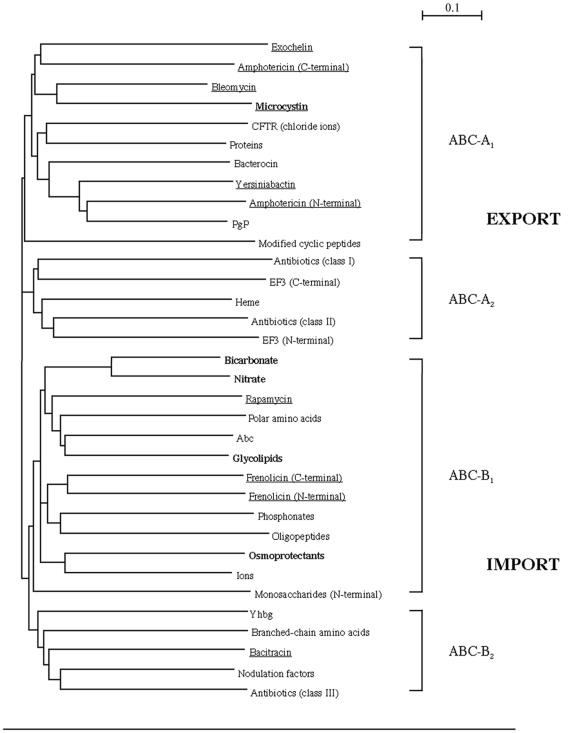
Global tree of collected ABC modules. The phylogenetic tree encompasses cyanobacterial ABC proteins (boldface), ABC proteins implicated in the transport of NRPs/PKs (underlined), and other representatives from across the ABC transporter superfamily. The major subdivisions of the tree are indicated according to the nomenclature given in the text (ABC-A, export systems; ABC-B, import systems). See Table 1 for a detailed list of modules present in each cluster. The scale bar corresponds to 10% divergence between sequences.
RESULTS
Sequence analysis of mcyH.
The approximately 1.8-kb mcyH open reading frame is predicted to encode a 61.48-kDa peptide with a pI of 6.02. Comparison of the inferred primary peptide (amino acid) sequence of mcyH with other sequences in the NCBI database revealed significant similarity (up to 80% identity and 87% similarity as determined by Blastp) to several members of the ABC transporter superfamily. Sequences of particularly high similarity included hypothetical ABC transporters from the microcystin biosynthesis gene clusters of Planktothrix agardhii (80% identity and 87% similarity), Nodularia spumigena (71% identity and 86% similarity), and Anabaena sp. (72% identity and 85% similarity) and the nostopeptolide biosynthesis cluster of Nostoc sp. (65% identity and 81% similarity). Other uncharacterized homologs were identified in species belonging to the cyanobacterial genera Nostoc, Thermosynechococcus, Trichodesmium, Synechocystis, Prochlorococcus, and Synechococcus (37 to 67% identity and 57 to 86% similarity) and the plant species Arabidopsis thaliana (35% identity and 55% similarity) and Oryza sativa (34% identity and 54% similarity). McyH also shared sequence similarity with the peroxisomal membrane proteins of various eukaryotic organisms, including Caenorhabditis elegans, Mus musculus, and Homo sapiens (up to 31% identity and 49% similarity). Figure 1 shows a section of the alignment of these peptide sequences with McyH.
By using several topology prediction programs, a large, hydrophobic N-terminal domain with three to five putative transmembrane regions was identified between residues 1 and 247 of the McyH peptide sequence. SignalP analysis gave a signal peptide probability of 0.975 with a maximum cleavage site probability of 0.697 between positions 20 and 21of the peptide sequence. The C-terminal domain of the sequence (residues 248 to 538) containing the diagnostic ABC ATPase Walker motifs (beginning at residues 356 and 469) was largely hydrophilic. A sequence corresponding to the linker region LSGGQQ/R/KQR, (25) was also identified at residues 341 to 347 (Fig. 1). The EAA motif, characteristic of ABC import systems (4), was not present.
Analysis of McyH codon usage with Countcodon (Kazusa) revealed a high proportion of rare E. coli codons. Present in particularly high frequencies were those triplets encoding leucine (UUA and CUA; 8.3%), isoleucine (AUU and AUA; 7%), arginine (CGA and AGA; 2.2%), and glycine (GGA; 1.9%). Subsequent heterologous expression of the McyH peptide was therefore performed in the Rosetta(DE3)(pLysS) expression strain containing the pLysSRARE plasmid carrying tRNA genes for “problematic” rarely used codons (20).
To gain an overview of the relative position of McyH in the ABC superfamily, a phylogenetic tree was constructed (Fig. 2). This tree was based on the most conserved part of the ABC proteins, the ATPase domains, which are known to partition phylogenetically according to function (24). The less conserved membrane domains were not analyzed. The global unrooted tree encompassed sequences from 33 proteins spanning the entire known ABC superfamily. Among these were several cyanobacterium-specific ABC proteins and ABC proteins implicated in the transport of nonribosomal peptides (NRPs) and polyketides (PKs). The ABC proteins used in this study are listed in Table 1. The global tree is largely in agreement with that published by Saurin et al. (24), with most members from the ABC-A1, -A2, -B1, and -B2 subgroups forming distinct branches. McyH was found to cluster within the ABC-A1 (export) subgroup on the same branch as the transporters associated with the nonribosomally produced peptides exochelin, amphotericin, and bleomycin. Other ABC proteins, putatively involved in transporting the NRPs rapamycin, frenolicin, and bacitracin, clustered in the ABC-B1 and -B2 subgroups.
Insertional inactivation of mcyH and mutant characterization.
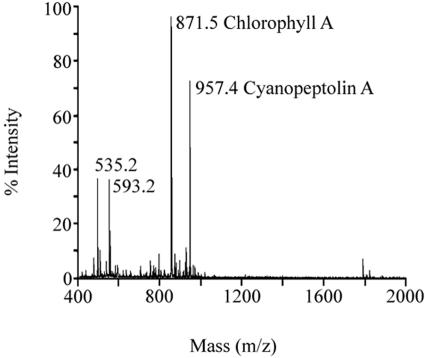
The putative role of McyH in microcystin biosynthesis and transport was investigated via deletion of part of the mcyH gene. To this end, two deletion constructs were engineered, p5K8BsmI-2 and p5K8BsmI-3 (Fig. 3). Following transformation with the deletion constructs, ΔmcyH mutant clones were screened via PCR. Of the 100 clones analyzed by PCR, four clones (two of each mutant strain) were selected for analysis. These were designated BsmI-2a, BsmI-2b, BsmI-3a, and BsmI-3b. These clones were analyzed by immunoblotting with an anti-McyH antibody. McyH could not be detected in ΔmcyH mutant extracts but was specifically detected in WT, ΔmcyA, and ΔmcyB strains (Fig. 4). MALDI-TOF mass spectrometry demonstrated that all four ΔmcyH mutant clones did not produce microcystins (m/z = 995). The other M. aeruginosa NRP, cyanopeptolin, however, was detected in the mutants (m/z = 957) (Fig. 5).
FIG. 4.
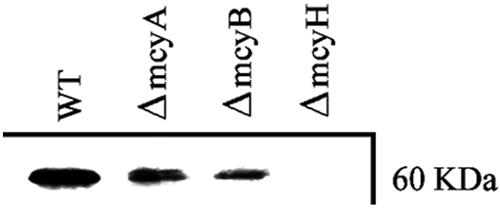
Analysis of McyH expression in 7806 WT and mutant strains grown under standard laboratory conditions. A section of a Western blot of WT 7806 and ΔmcyA, ΔmcyB, and ΔmcyH mutant total protein extracts with anti-McyH antibody is shown.
FIG. 5.
MALDI-TOF analysis of ΔmcyH mutant BsmI-2a. Major peaks include chlorophyll A (m/z 871.5) and cyanopeptolin A (m/z 957.4). Microcystin peaks (m/z 981 and 995) were absent in the mutant spectrograph (see reference 5 for a typical M. aeruginosa WT MALDI-TOF spectrograph).
To exclude possible polar effects of the mcyH deletion on other genes in the microcystin biosynthesis cluster, transcriptional analyses of several other mcy genes were performed by RPA. The transcript analyses confirmed the presence of mcyB, mcyF, mcyG, and mcyI transcripts in both ΔmcyH strains (Fig. 6B).
Immunodetection of other microcystin synthetase components.
The anti-McyF antibody was used to investigate the effect of the mcyH deletion on the expression of the aspartate racemase of the microcystin biosynthesis enzyme complex. A 28-kDa protein corresponding to McyF was specifically detected in the WT and in the ΔmcyA and ΔmcyB mutants. No signal at this size was detected in the ΔmcyH mutants (Fig. 7).
FIG. 7.
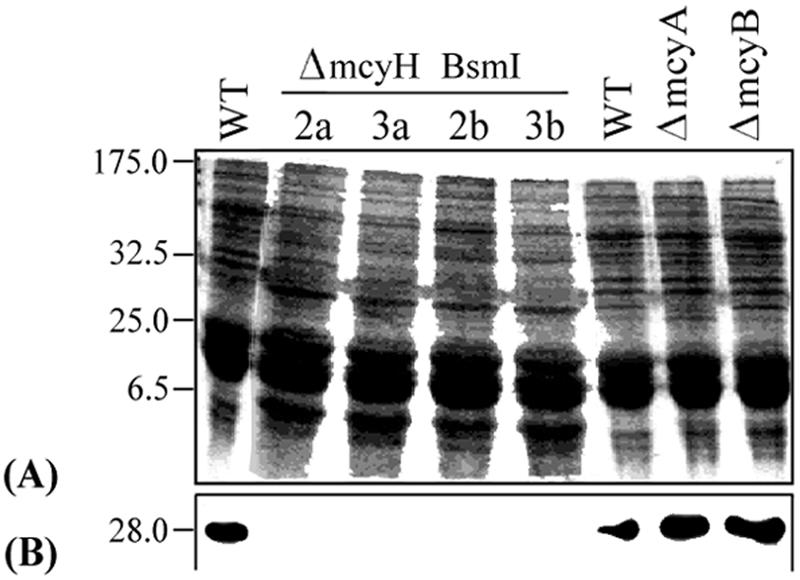
Translational analysis of mcyF in WT and mutant cells. (A) Coomassie blue-stained SDS-polyacrylamide gel of WT and ΔmcyH mutant total protein extracts. Molecular weight standards (in thousands) are shown on the left. (B) Section of corresponding immunoblot of panel A with anti-McyF antibody.
DISCUSSION
The combined sequence analyses for McyH strongly suggested that it belonged to the ABC transporter superfamily. A Blastp analysis of the inferred primary peptide sequence of McyH identified several similar protein sequences from a wide range of organisms. Interestingly, the 15 sequences of highest similarity belonged to both toxic and nontoxic species of cyanobacteria. Other homologs from plants, bacteria, nematodes, and mammals were also identified, including the eukaryotic peroxisomal membrane proteins. These ABC transporters are known to transport long-chain fatty acids (27) which resemble the dienoic acid side chain (Adda) of microcystin. These bioinformatic analyses support the hypothesis that McyH may be responsible for the active transport of microcystin, as ABC transporters of similar primary structure (amino acid sequence) have previously been shown to interact with similar allocrites (24).
In addition to sharing extensive sequence similarity with other ABC transporters, McyH possessed the diagnostic Walker A and B motifs and the upstream linker consensus sequence (Fig. 1). The molecular mass of the putative ATP-binding domain of McyH (26.99 kDa) is also typical for an ABC ATPase, which average 27 kDa (14). Several other lines of evidence indicated that McyH functions as an ABC exporter in M. aeruginosa. First, it lacked the EAA motif characteristic of bacterial import systems (4). Second, hydropathy profiling and secondary structure predictions indicated that McyH is a fusion protein with membrane and ABC domains encoded within a single polypeptide. This suggested an export function for the protein, since the membrane and ABC domains of exporters are usually fused, while the domains of importers are always separately encoded within individual polypeptides (24). As the general structure of ABC transporters consists of two membrane-spanning domains and two ABC domains (12), the putative McyH transporter may function as a homodimer. Phylogenetically, McyH clusters with ABC-A1 proteins on the same branch as the experimentally characterized bleomycin transporter and putative exochelin and amphotericin transporters. These results also support the argument that McyH plays a role in exporting microcystin, as these compounds are structurally similar to microcystin and are also synthesized via nonribosomal peptide pathways. Furthermore, the amphotericin and exochelin transporters, like McyH, are encoded within the biosynthetic gene clusters of their putative allocrites (2, 37).
The clustering of McyH within the ABC-A1 group supported initial structural predictions for the transporter (i.e., homodimeric), as subgroup ABC-A1 is comprised of proteins with N-terminal membrane-spanning modules fused to C-terminal ABC modules (24). The cyanobacterium-specific ABC proteins used in the phylogenetic analysis did not form a distinct cluster but were dispersed evenly across the ABC-B1 and -B2 branches (Fig. 2). These results supported the theory that ABC proteins partition primarily according to polarity of transport (i.e., import verses export) rather than according to the species of origin (24).
While the majority of the proteins implicated in the transport of nonribosomally synthesized compounds (NRPs and PKs) cluster together on the ABC-A1 branch, those hypothesized to transport rapamycin, frenolicin, and bacitracin are scattered across the lower branches (import section) of the tree (Fig. 2). This finding suggests that these proteins may not transport the aforementioned compounds. An alternative function for these proteins could be the import of structural precursors for NRP and PK biosyntheses.
The mutation and inactivation of putative ABC transporter genes constitute the traditional method for determining the function of their encoded proteins. To this end, mcyH deletion mutant strains were engineered for M. aeruginosa PCC 7806. Surprisingly, disruption of the mcyH gene resulted in the loss of expression of at least one of the components of the Mcy complex, McyF (Fig. 7), and in the complete abolition of microcystin production under standard laboratory growth conditions (Fig. 5). These nontoxic mutant strains were therefore not useful for further functional characterization of the McyH protein. The previous insertional inactivation of other individual genes in the mcy gene cluster, mcyA, -B, -D, -E, and -F, has similarly led to the production of mutant strains with nontoxic phenotypes (5, 19, 33). Furthermore, it has been shown that by knocking out one protein component of the mcy complex (McyB), all mcy peptide and polyketide synthetases in the resulting mutants were lost, including those transcribed in the opposite direction (33). This phenomenon is unlikely to be the result of polar effects on the transcription of microcystin biosynthesis genes, as mcy transcripts were detected in all mutant strains (ΔmcyA, ΔmcyB, and ΔmcyD [data not shown] and ΔmcyH [Fig. 6]) investigated that bear the Cmr insertional inactivation cassette.
An alternative explanation for the lack of toxin production in the ΔmcyH mutants relates to the stability of the microcystin synthetase complex. Both ΔmcyH mutants show the presence of WT mcy transcript levels but the absence of microcystin, its partial products, or the McyF translation product. Previous in situ studies using immunogold hybridization with an anti-McyB antibody have revealed a subcellular localization of the microcystin synthetase complex close to the inner side of the cytoplasmic membrane (M. Hisbergues, data not shown), similar to the distribution shown for microcystin (28). Being a putative membrane-bound protein, McyH may play a role in stabilizing the microcystin biosynthesis multienzyme complex by anchoring it to the membrane. Therefore, disruption of the transporter via deletional mutagenesis is likely to lead to dissociation of the complex and degradation of its individual protein components, and hence the observed lack of McyF in both ΔmcyH mutant strains. Similar results have been recorded for other bacterial secondary metabolite pathways. For example, in a study investigating the biosynthesis of the lantibiotic nisin, Siegers et al. (29) used coimmunoprecipitation and the yeast two-hybrid system to demonstrate that the lanthionine synthetase complex (NisB-NisC) is anchored to the membrane by the nisin exporter NisT. Subsequent inactivation of nisT via insertional mutagenesis resulted in the complete abolition of nisin biosynthesis. Further investigation of the intracellular localization of both the Mcy complex and the ABC transporter by using similar techniques may confirm the involvement of McyH in the overall structure of the microcystin biosynthetic multienzyme.
Analysis of the McyH protein in WT and mutant strains of M. aeruginosa PCC 7806 revealed that expression of the hypothetical ABC transporter is reduced in the nontoxic ΔmcyA and ΔmcyB mutants (Fig. 4). Instability and degradation of McyH following the disruption of other Mcy components (i.e., McyA and McyB) similarly may be the cause of this reduced expression. Alternatively, in the absence of allocrite (microcystin), expression of the transporter could be down regulated. Similar observations have been recorded for ABC proteins spanning several different classes. For example, expression of the BcrABC transporter, responsible for the extrusion of the nonribosomal peptide antibiotic bacitracin, is increased severalfold when cells are exposed to bacitracin (18). Similarly, ABC transporters belonging to the multidrug resistance classes have been demonstrated to be inducible by their own allocrites. For example, expression of the human Mrp2 protein is significantly up regulated following treatment of patients with the antibacterial drug rifampin (8).
Taken together, the data here suggest a role for McyH in both toxin biosynthesis and export. If McyH is in fact responsible for the active transport of microcystin in M. aeruginosa, as is suggested by the bioinformatic analyses performed in this study, then this ABC transporter will be the first toxin exporter to be identified in a cyanobacterium. Such a finding raises numerous questions regarding the ecophysiological role of microcystin and other nonribosomally synthesized secondary metabolites.
Acknowledgments
This work was funded by the Australian Research Council, the Australian Cooperative Research Centre for Water Quality and Treatment, and the Deutscher Akademischer Austausch Dienst.
REFERENCES
- 1.Bartsevich, V. V., and H. B. Pakrasi. 1995. Molecular identification of an ABC transporter complex for manganese—analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 14:1845-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caffrey, P., S. Lynch, E. Flood, S. Finnan, and M. Oliynyk. 2001. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem. Biol. 8:713-723. [DOI] [PubMed] [Google Scholar]
- 3.Calcutt, M. J., and F. J. Schmidt. 1994. Gene organization in the bleomycin-resistance region of the producer organism Streptomyces verticillius. Gene 151:17-21. [DOI] [PubMed] [Google Scholar]
- 4.Dassa, E., and M. Hofnung. 1985. Homologies between integral proteins of the inner membrane of binding protein transport systems in enterobacteria. Ann. Inst. Pasteur Microbiol. 136A:281-288. [DOI] [PubMed] [Google Scholar]
- 5.Dittmann, E., B. A. Neilan, M. Erhard, H. von Döhren, and T. Börner. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26:779-787. [DOI] [PubMed] [Google Scholar]
- 6.Fiedler, G., M. Arnold, S. Hannus, and I. Maldener. 1998. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 27:1193-1202. [DOI] [PubMed] [Google Scholar]
- 7.Forchhammer, K., and N. Tandeau de Marsac. 1994. The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J. Bacteriol. 176:84-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fromm, M., H. Kauffmann, P. Fritz, O. Burk, H. Kroemer, R. Warzok, M. Eichelbaum, W. Siegmund, and D. Schrenk. 2000. The effect of rifampin treatment on intestinal expression of human MRP transporters. Am. J. Pathol. 157:1575-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehring, A. M., I. Mori, R. D. Perry, and C. T. Walsh. 1998. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry 1:17104. [DOI] [PubMed] [Google Scholar]
- 10.Hagemann, M., S. Richter, and S. Mikkat. 1997. The ggtA gene encodes a subunit of the transport system for the osmoprotective compound glucosylglycerol in Synechocystis sp. strain PCC 6803. J. Bacteriol. 179:714-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, 488-489. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Higgins, C. F. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152:205-210. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann, D., J. M. Hevel, R. E. Moore, and B. S. Moore. 2003. Sequence analysis and biochemical characterization of the nostopeptolide A biosynthetic gene cluster from Nostoc sp. GSV224. Gene 311:171-180. [DOI] [PubMed] [Google Scholar]
- 14.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 22:381-399. [DOI] [PubMed] [Google Scholar]
- 15.Kaebernick, M., B. A. Neilan, T. Börner, and E. Dittmann. 2000. Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl. Environ. Microbiol. 66:3387-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konz, D., A. Klens, K. Schorgendorfer, and M. A. Marahiel. 1997. The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10716: molecular characterization of three multi-modular peptide synthetases. Chem. Biol. 4:927-937. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumuller, A., D. Konz, and M. Marahiel. 2001. The two-component regulatory system BacRS is associated with bacitracin ′self-resistance' of Bacillus licheniformis ATCC 10716. Eur. J. Biochem. 268:3180-3189. [DOI] [PubMed] [Google Scholar]
- 19.Nishizawa, T., M. Asayama, and M. Shirai. 2001. Cyclic heptapeptide microcystin biosynthesis requires the glutamate racemase gene. Microbiology 147:1235-1241. [DOI] [PubMed] [Google Scholar]
- 20.Novy, R., D. Drott, K. Yaeger, and R. Mierendorf. 2001. Overcoming the codon bias of. E. coli for enhanced protein expression. Innovations 12:1-3. [Google Scholar]
- 21.Omata, T., G. D. Price, M. R. Badger, M. Okamura, S. Gohta, and T. Ogawa. 1999. Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc. Natl. Acad. Sci. USA 9:13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulsen, I. T., M. K. Sliwinski, and S. M. Saier, Jr. 1998. Microbial genome analysis: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 3:573-592. [DOI] [PubMed] [Google Scholar]
- 23.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 24.Saurin, W., M. Hofnung, and E. Dassa. 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 48:22-41. [DOI] [PubMed] [Google Scholar]
- 25.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1-20. [DOI] [PubMed] [Google Scholar]
- 26.Schwecke, T., J. F. Aparicio. I. Molnar, A. Konig, L. E. Khaw, S. F. Haydock, M. Oliynyk, P. Caffrey, J. Cortes, J. B. Lester, et al. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. USA 15:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shani, N., and D. Valle. 1998. Peroxisomal ABC transporters. Methods Enzymol. 292:753-776. [DOI] [PubMed] [Google Scholar]
- 28.Shi, L., W. W. Carmichael, and I. Miller. 1995. Immuno-gold localization of hepatotoxins in cyanobacterial cells. Arch. Microbiol. 163:7-15. [DOI] [PubMed] [Google Scholar]
- 29.Siegers, K., S. Heinzmann, and K. D. Entian. 1996. Biosynthesis of lantibiotic nisin. Posttranslational modification of its prepeptide occurs at a multimeric membrane-associated lanthionine synthetase complex. J. Biol. Chem. 24:12294-12301. [DOI] [PubMed] [Google Scholar]
- 30.Sielaff, H., E. Dittmann, N. Tandeau de Marsac, C. Bouchier, H. von Döhren, T. Börner, and T. Schwecke. 2003. The mcyF gene of the microcystin biosynthetic gene cluster from Microcystis aeruginosa encodes an aspartate racemase. Biochem. J. 373:909-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivonen, K., and. G. Jones. 1999. Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management. E & FN Spoon, London, United Kingdom.
- 32.Theiss, W. C., W. W. Carmichael, J. Wyman, and R. Bruner. 1988. Blood pressure and hepatocellular effects of the cyclic heptapeptide toxin produced by the freshwater cyanobacterium (blue-green alga) Microcystis aeruginosa strain PCC 7820. Toxicon 26:603-613. [DOI] [PubMed] [Google Scholar]
- 33.Tillett, D., E. Dittmann, M. Erhard, H. von Döhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC 7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 34.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the a- and b-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, S. 1989. Drinking water and primary liver cancer., p. 30-37. In Z. Tang, and S. Xia (ed.), Primary liver cancer. China Academic Publishers, New York, N.Y.
- 36.Yu, S., E. Fiss, and W. R. Jacobs, Jr. 1998. Analysis of the exochelin locus in Mycobacterium smegmatis: biosynthesis genes have homology with genes of the peptide synthetase family. J. Bacteriol. 180:4676-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, W., J. Arceneaux, M. Beggs, B. Byers, K. Eisenach, and M. Lundrigan. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two nonribosomal peptide synthetase genes. Mol. Microbiol. 29:629-639. [DOI] [PubMed] [Google Scholar]