Abstract
The prevalence of imipenem resistance among Pseudomonas aeruginosa isolates at a 195-bed tertiary care medical center in Cali, Colombia, rose from 2% in 1996 to 28% in 1997 and to over 40% in 2003. Many isolates showed high-level multiresistance, and phenotypic characterization suggested the spread of a predominant strain with minor variants. Sixty-six resistant isolates collected between February 1999 and July 2003 from hospitalized patients (n = 54) and environmental samples (n = 12) were subjected to a fuller analysis. Genetic fingerprints were compared by pulsed-field gel electrophoresis (PFGE) of SpeI-digested genomic DNA, and blaIMP and blaVIM genes were sought by PCR. PFGE and serotyping indicated that 52 of the 66 isolates belonged to a single strain, with 82% similarity; the PFGE pattern for this organism was designated pattern A. Two further pairs of isolates represented single strains; the remaining nine isolates were unique, and in the case of one isolate, no satisfactory PFGE profile could be obtained. The pattern A isolates were mostly of serotype O12 and were highly resistant to imipenem (MICs, 32 to >256 μg/ml), with this resistance decreased eightfold or more in the presence of EDTA. They yielded amplicons with blaVIM-specific primers, and sequencing of DNA from a representative isolate revealed blaVIM-8, a novel allele with three polymorphisms compared with the sequence of blaVIM-2. Two of these nucleotide changes were silent, but the third determined a Thr139Ala substitution. Only 4 of 13 resistant isolates (2 clinical isolates and 2 environmental isolates) assigned to other PFGE types carried blaVIM alleles, whereas the others were less multiresistant and mostly had lower levels of imipenem resistance (MICs, ≤32 μg/ml) which was not significantly reduced by EDTA. No blaIMP alleles were detected. During 2003, when the environmental study was undertaken, serotype O12 isolates with blaVIM were recovered from sinks and stethoscopes in the most-affected units, although not from the hands of staff; the problem declined once these reservoirs were disinfected and hygienic precautions were reinforced.
Pseudomonas aeruginosa is a frequent nosocomial pathogen that causes severe disease in many settings, particularly in compromised patients, including those with cancer, burns, and cystic fibrosis. Infections are frequently severe, and two recent studies indicated that the rate of mortality attributable to P. aeruginosa bacteremia is approximately 34% (10, 28). Many virulence factors may contribute to pathogenicity, including biofilm formation and the expression of adhesins, endotoxin, and hydrolytic exotoxins, which cause tissue destruction. Therapy is complicated by the organism's potent ability for adaptation, mutation, and gene acquisition. Resistance to antipseudomonal β-lactams may arise via hyperproduction of a chromosomal AmpC β-lactamase, acquisition of secondary β-lactamases, and/or upregulated efflux or impermeability (6, 12, 15, 17). Most carbapenem resistance is due to impermeability, which arises via the loss of the OprD (D2) porin, but carbapenem-hydrolyzing metallo-β-lactamases (MBLs) of the IMP, VIM, and SPM families are increasingly reported, with some of them coming from strains that have caused large outbreaks (8, 18, 19, 23, 31).
At present, 17 IMP β-lactamase variants, 10 VIM types, and 1 SPM have been catalogued, with most of the recorded MBL producers being P. aeruginosa or Acinetobacter spp. from East Asia or Europe (5, 30, 32, 33, 35). MBL producers are also emerging in the Americas, where SENTRY program data for Latin American in 2002 indicated 18% carbapenem resistance among Pseudomonas and Acinetobacter spp. and where it was estimated that 2% of all P. aeruginosa isolates from participating hospitals were MBL producers (M. Castanheria, R. E. Mendes, T. Murphy, M. Toleman, H. S. Sader, R. N. Jones, and T. R. Walsh, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2023, 2003). So far, five MBL types have been detected in the Americas: IMP-1, IMP-7, IMP-16, VIM-2, and SPM-1. The last four of these have exclusively been reported in P. aeruginosa. The only major outbreak of producers so far recorded in South America was in Brazil, where a P. aeruginosa strain with the SPM-1 enzyme spread among hospitals (7). We describe here an outbreak caused by a VIM-8-producing P. aeruginosa strain that lasted for 6 years in a 195-bed tertiary care medical center in Cali, Colombia.
MATERIALS AND METHODS
Epidemiological and clinical data.
The study was conducted at a 195-bed tertiary care center in Cali, southwest Colombia. This establishment is the medical reference center for the region, with 95 beds in adult, pediatric, and neonatal intensive care units (ICUs). Its transplantation unit undertakes kidney, liver, and heart transplants.
A total of 727 imipenem-resistant isolates of P. aeruginosa were identified from 422 patients between February 1997 and July 2003, based on routine identification and susceptibility tests performed with the MicroScan AutoScan system (Dade MicroScan Inc., Sacramento, Calif.) and the Kirby-Bauer disk diffusion method. Most of these isolates were multiresistant to extended-spectrum cephalosporins, including cefepime, aminoglycosides, and fluoroquinolones, but many were susceptible or only intermediately resistant to aztreonam and piperacillin. Sixty-six of these imipenem-resistant isolates were selected for detailed study. These comprised 36 clinical isolates collected between 1997 and 2003, which were analyzed retrospectively, and a further 18 clinical isolates and 12 environmental isolates collected prospectively in 2003.
Information on the patients and isolates was collected from clinical records and from the local Infectious Disease Committee's database. The data collected included the patient's age, sex, duration of ICU stay until isolation of imipenem-resistant P. aeruginosa, the source of the isolate, underlying disease, previous antibiotic treatment, type of infection, and clinical outcome. Patients were classified as infected or colonized according to the presence or absence of clinical symptoms.
Bacterial identification and antimicrobial susceptibility.
Bacterial identification was performed with a MicroScan AutoScan system (Dade MicroScan), according to the instructions of the manufacturer. The imipenem-EDTA disk method (36) was used to screen isolates for MBLs, taking an increase in zone diameter of more than 5 mm in the presence of EDTA as a predictor of likely MBL production. MBL Etests (AB Biodisk, Solna, Sweden) were used as a further test of MBL production, with MIC ratios (MIC of imipenem alone/MIC of imipenem plus EDTA) ≥8 taken as prima facie evidence of MBL production.
MICs initially were determined by broth microdilution assay on the MicroScan system and were interpreted according to the criteria of the NCCLS (22). The antimicrobials tested were amikacin, aztreonam, ceftazidime, cefepime, ciprofloxacin, gentamicin, imipenem, meropenem, and piperacillin. Since the MicroScan system recorded carbapenem MICs only up to 4 μg/ml, isolates with greater resistance were reexamined by agar dilution on Mueller-Hinton medium (Oxoid, Basingstoke, United Kingdom) with a final inoculum of 104 CFU/spot. Imipenem was obtained from Merck Sharp & Dohme (Hoddesdon, United Kingdom). Carbapenem resistance was further confirmed by the Kirby-Bauer agar disk diffusion method with imipenem (10 μg; Oxoid) and meropenem (10 μg; Becton Dickinson, Sparks, Md.) disks. P. aeruginosa ATCC 27853 was used as a control throughout.
Carbapenemase identification.
PCR amplification was performed with universal blaVIM and blaIMP primers (23). The isolates were also tested by PCR with primers specific for blaVIM-l-like or blaVIM-2-like subtypes (35). A selected blaVIM allele, amplified with subtype-specific primers, was sequenced on a CEQ8000 analyzer (Beckman Coulter, High Wycombe, United Kingdom) (18, 35).
Strain typing.
Isolates were compared by pulsed-field gel electrophoresis (PFGE) of SpeI-digested genomic DNA (3); banding patterns were analyzed with BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). The O serotypes of the isolates were determined by the slide agglutination method by comparison with International Antigenic Typing Scheme type strains (25).
Environmental screening.
From May to June 2003, environmental surveillance swab specimens (n = 70) were collected from nebulizers, ventilators, tubing devices, stethoscopes, and sink surfaces in each patient's room. Fingerprints from the hands of health care workers (n = 10) in the affected units, and particularly those in the ICUs, were sampled twice per day, at the beginning and the end of work shifts, before the final hand washing. Swab samples were taken from sinks in the morning to maximize bacterial recovery. All the swabs were inoculated into nutrient broth supplemented with 0.03% cetrimide (cetyltrimethyl ammonium bromide) and incubated at 37°C for 24 h. Subcultures were then performed on nutrient agar with 0.02% cetrimide, and the plates were incubated for 48 h at 37°C (based on the advice of Tyrone Pitt [Health Protection Agency], personal communication). Colonies producing a blue-green pigment were checked for oxidase reaction, and oxidase-positive isolates were subjected to identification and susceptibility testing with the MicroScan system, PFGE and serotyping were performed as described above, and blaVIM was sought by PCR.
Nucleotide sequence accession number. The blaVIM allele has been deposited in the GenBank database under accession number AY524987.
RESULTS
Description of outbreak.
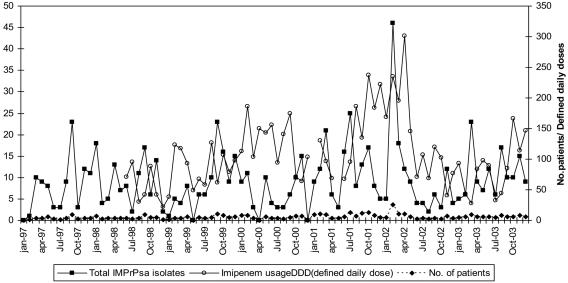
The overall prevalence of imipenem resistance among P. aeruginosa isolates in the hospital from 1997 to 2003 was 38%. Prevalence rates in individual years were as follows: 1997, 28%; 1998, 34%; 1999, 38%; 2000, 37%; 2001, 42%; 2002, 49%; and 2003, 37%. The prevalence rate was only 2% in 1996. The most striking rise was from 5% in February 1997 to 32% in March 1997; and during the next several years, resistant isolates were detected in multiple ICUs, although they were particularly detected in the adult unit. Their incidence peaked in February 2002, with 46 isolates from 26 patients (Fig. 1). This peak and a subsequent decline matched the peak and decline in imipenem usage (Fig. 1), but, more generally, the relationships between imipenem use and isolation of resistant P. aeruginosa were weak.
FIG. 1.
Isolation of imipenem-resistant P. aeruginosa isolates from 1997 to 2003: ▪, total number of imipenem-resistant P. aeruginosa isolates per month; ♦, total number of infected patients per month; ○, imipenem use defined as daily dose per 100 patient-days.
In total, 727 imipenem-resistant P. aeruginosa isolates were cultured from various specimens and sites (Table 1), including respiratory tract specimens (n = 165; 22.7%), abdominal swabs (n = 122; 16.8%), catheters (n = 110; 15.1%), abscesses (n = 100; 15%), blood (n = 105; 14.4%), urine (n = 46; 6.3%), surgical wounds (n = 32; 4.4%), and other specimens (n = 47; 5.3%). The adult ICU accounted for 40.3% of the isolates, the neonatal ICU accounted for 10.4%, and the pediatric ICU accounted for 4.7%; other adult medical wards accounted for 24.5%, the pediatric medical ward accounted for 6.6%, the emergency unit accounted for 4.4%, and other units accounted for 9.1%.
TABLE 1.
Characteristics of 54 P. aeruginosa clinical isolates from Cali, Colombia, 1999 to 2003a
| Isolate | Isolation date (mo-yr) | Source | Ward | Phenotype | PFGE type/serotype | Zone diam (mm)
|
Etest MIC for MBL producers (mg/liter)
|
blaVIM-2-like subtype by PCR | ||
|---|---|---|---|---|---|---|---|---|---|---|
| IMP | IMP + EDTA | IMP | IMP + EDTA | |||||||
| 001 | Feb-99 | Surgical wound | AICU | 1 | A/O12 | 6 | 11 | >256 | 32 | + |
| 2/16 | Mar-99 | Surgical wound | Medical ward | 1 | A/O12 | 6 | 13 | ND | ND | + |
| 2/19 | May-99 | Secretion | Medical ward | Other | A/O12 | 6 | 13 | ND | ND | + |
| 2/20 | May-99 | Catheter | Medical ward | Other | A/O12 | 6 | 14 | ND | ND | + |
| 2/18 | Jun-99 | Urine | Medical ward | Other | A/O12 | 13 | 18 | ND | ND | + |
| 002 | Sep-99 | Urine | Medical ward | Other | A/O12 | 6 | 13 | >256 | 8 | + |
| 004 | Sep-99 | Blood | Medical ward | 6 | A/O12 | 6 | 12 | >256 | 8 | + |
| 2/15 | Sep-99 | Peritoneal swab | AICU | Other | A/O12 | 10 | 15 | ND | ND | + |
| 005 | Oct-99 | Catheter | AICU | 6 | A/O12 | 6 | 11 | >256 | 8 | + |
| 008 | Dec-99 | Catheter | AICU | Other | A/O12 | 6 | 15 | >256 | 4 | + |
| 023 | Dec-99 | Catheter | AICU | Other | A/O12 | 12 | 19 | 64 | 4 | + |
| 003 | Jan-00 | Catheter | AICU | 6 | A/O12 | 6 | 12 | >256 | 4 | + |
| 2/21 | Feb-00 | Catheter | AICU | Other | A/O12 | 13 | 17 | ND | ND | + |
| 018 | Feb-02 | ET swab | NICU | 1 | A/O12 | 6 | 11 | >256 | 8 | + |
| 021 | Feb-02 | Blood | NICU | 1 | A/O12 | 6 | 11 | >256 | 2 | + |
| 022 | Feb-02 | Eye secretion | NICU | 1 | A/O12 | 6 | 11 | >256 | 8 | + |
| 025 | Feb-02 | Blood | NICU | 4 | A/O12 | 6 | 11 | >256 | 8 | + |
| 026 | Feb-02 | Blood | NICU | 1 | A/O12 | 6 | 11 | >256 | 4 | + |
| 2/17 | Feb-02 | Catheter | Pediatric ward | 1 | A/O12 | 6 | 12 | ND | ND | + |
| 013 | Apr-02 | Abdominal fluid | AICU | 4 | A/O12 | 6 | 11 | >256 | 16 | + |
| 017 | Apr-02 | Wound | Medical ward | Other | A/O12 | 6 | 11 | >256 | 8 | + |
| 019 | Apr-02 | Blood | NICU | 1 | A/O12 | 6 | 12 | >256 | 8 | + |
| 014 | Jul-02 | Nasal Swab | AICU | 1 | A/O12 | 6 | 13 | >256 | 16 | + |
| 020 | Jul-02 | Blood | NICU | 1 | A/O12 | 6 | 12 | >256 | 8 | + |
| 024 | Aug-02 | Urine | AICU | 1 | A/O12 | 11 | 21 | 128 | 2 | + |
| 015 | Sep-02 | Blood | Medical ward | 1 | A/O12 | 6 | 14 | >256 | 8 | + |
| 016 | Sep-02 | Blood | Medical ward | 2 | A/O12 | 6 | 10 | >256 | 8 | + |
| 006b | Nov-02 | Oral swab | AICU | 6 | A/O12 | 6 | 14 | >256 | 8 | + |
| 009 | Nov-02 | Urine | AICU | 1 | O12c | 6 | 12 | >256 | 8 | + |
| 010 | Nov-02 | BAL | AICU | 2 | A/O12 | 6 | 11 | >256 | 8 | + |
| 031 | Nov-02 | Catheter | AICU | Other | A/O12 | 10 | 21 | 32 | 2 | + |
| 011 | Dec-02 | BAL | AICU | 4 | A/O12 | 6 | 16 | >256 | 16 | + |
| 012 | Jan-03 | Urine | Medical ward | 4 | A/O12 | 6 | 11 | >256 | 16 | + |
| 2/25 | Mar-03 | Abdominal swab | PICU | 1 | A/O12 | 6 | 12 | ND | ND | + |
| 2/26 | Mar-03 | Secretion | AICU | Other | A/O12 | 10 | 16 | ND | ND | + |
| 2/27 | Mar-03 | ET swab | Medical ward | Other | A/O12 | 16 | 19 | ND | ND | + |
| 2/22 | Apr-03 | Blood | NICU | 1 | A/O12 | 6 | 12 | ND | ND | + |
| 2/30 | Apr-03 | Feces | NICU | 4 | A/O12 | 6 | 12 | ND | ND | + |
| 2/23 | May-03 | Urine | Medical ward | 1 | A/O12 | 6 | 12 | ND | ND | + |
| 2/29 | May-03 | Catheter | AICU | 4 | K/O12 | 6 | 13 | ND | ND | + |
| 2/28 | May-03 | Bronchial secretion | AICU | 4 | A/O12 | 6 | 13 | ND | ND | + |
| 2/24 | Jun-03 | Peritoneal swab | AICU | Other | K/O12 | 6 | 12 | ND | ND | + |
| 2/31 | Jul-03 | Blood | Medical ward | Other | A/O12 | 14 | 18 | ND | ND | + |
| 2/32 | Jul-03 | Urine | AICU | 1 | A/O12 | 6 | 12 | ND | ND | + |
| 007 | NK | NK | 6 | A/O12 | 6 | 12 | >256 | 8 | + | |
| 027 | Nov-02 | Peritoneal fluid | AICU | 3 | B/O6 | 6 | 10 | 16 | 8 | − |
| 028 | Nov-02 | Catheter | AICU | Other | C/O1 | 11 | 12 | 32 | 16 | − |
| 029 | Dec-02 | ET swab | AICU | Other | D/O6 | 12 | 16 | 32 | 8 | − |
| 030 | Dec-02 | Secretion | AICU | 3 | D/O6 | 9 | 15 | 32 | 8 | − |
| 032 | Mar-02 | Blood | NICU | Other | E/O6 | 17 | 18 | 16 | 8 | − |
| 033 | Feb-99 | Biopsy | AICU | 4 | F/OPAd | 26 | 26 | 16 | 4 | − |
| 034 | Sep-99 | Blood | AICU | 3 | G/O6 | 23 | 24 | <4 | 1 | − |
| 035 | Nov-02 | Blood | Medical ward | Other | H/O6 | 25 | 26 | <4 | 2 | − |
| 036 | Nov-02 | Urine | AICU | Other | I/OII | 27 | 27 | <4 | >1 | − |
Data for the outbreak type A strain are in boldface. Abbreviations: IMP, imipenem; ND, not determined; ET, endotracheal; AICU, adult ICU; NICU, neonatal ICU; PICU, pediatric ICU; NK, not known; Other, phenotype other than 1 to 6 (for definitions of phenotypes, see text).
Isolate used for sequencing of blaVIM allele.
A satisfactory PFGE profile was not obtained.
Polyagglutinating strain.
A total of 128 different antibiotic resistance phenotypes were seen among the imipenem-resistant isolates, although many of these were minor variants. Six phenotypes were predominant, as follows: phenotype 1, resistant to imipenem, meropenem, ceftazidime, aminoglycosides, and quinolones but susceptible to aztreonam and piperacillin (32.5%); phenotype 2, similar to phenotype 1 but resistant also to aztreonam and piperacillin (7.9%); phenotype 3, resistant only to imipenem (3.9%); phenotype 4, similar to phenotype 1 but susceptible only to piperacillin (10.5%); phenotype 5, resistant to imipenem and meropenem but susceptible to the other antimicrobials tested (2.7%); and phenotype 6, similar to phenotype 1 but susceptible also to cefepime (2.7%). The remaining 122 phenotypes accounted for 39.8% of the imipenem-resistant isolates.
Typing of clinical isolates.
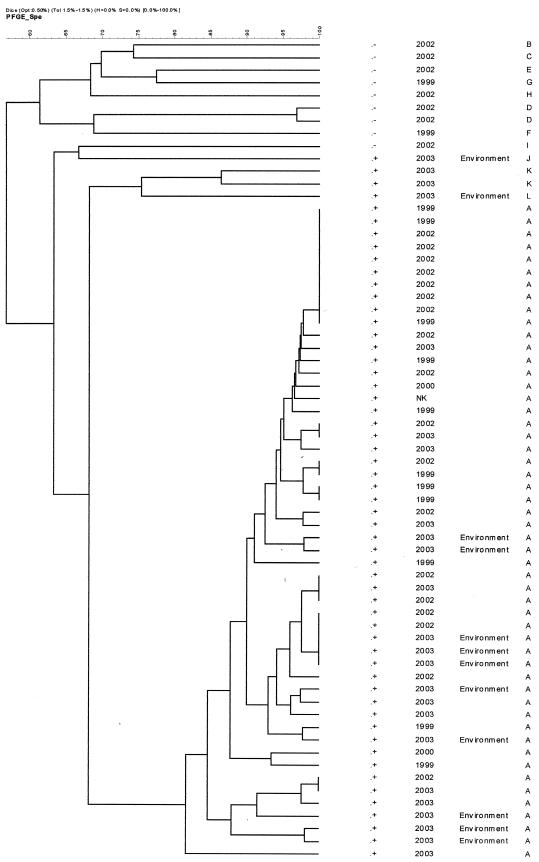
Fifty-four imipenem-resistant clinical isolates were retained for analysis. These isolates were collected in the following years: 1999, 13 isolates; 2000, 2 isolates; 2002, 25 isolates; 2003, 13 isolates; and unknown, 1 isolate. They included representatives of phenotypes 1 (predominantly), 2, 4, and 6, as well as minor types. Of these 54 isolates, 42 clustered as the same type, designated type A, with >82% similarity by PFGE (Fig. 2). All 42 of these isolates were of serotype O12, and the combination of imipenem and EDTA exhibited synergy against these isolates, with imipenem MICs reduced eightfold or more by the presence of EDTA. All these isolates were positive for blaVIM by PCR but were negative for blaIMP. More than 90% of the isolates were multiresistant to β-lactam antimicrobials except aztreonam and piperacillin, to which 63.6 and 98% of the isolates, respectively, appeared to be susceptible on the basis of the criteria of the NCCLS (22). For many of the isolates (ca. 40%) piperacillin MICs were ≤16 μg/ml, and thus, they would be considered susceptible on the basis of, for example, the criteria of the British Society for Antimicrobial Chemotherapy (1, 2), which are stricter than those of the NCCLS (susceptible MIC ≤ 64 μg/ml).
FIG. 2.
Dendrogram of PFGE data (SpeI digests) showing the relatedness of P. aeruginosa isolates. Fifty-two isolates belonged to the type A strain and carried blaVIM. Four other isolates also produced a VIM carbapenemase; +, blaVIM present; −, blaVIM absent.
The remaining 12 clinical isolates yielded nine different profiles by PFGE, belonged to various serotypes, and included 1 isolate for which no satisfactory profile could be obtained. Two were positive for blaVIM, but the remainder were negative. The latter isolates were much less multiresistant than type A isolates to non-β-lactam antibiotics, including gentamicin and ciprofloxacin (7% resistant versus 70% resistant in strain A to both these antimicrobials).
Identification of VIM-type enzymes.
Sequencing of the blaVIM allele from a representative PFGE type A isolate revealed a blaVIM- 2 gene variant, designated blaVIM-8 (the number was allocated by G. Jacoby, personal communication). The novel gene differed from blaVIM-2 by an A-to-G polymorphism at nucleotide 4.5, giving a threonine-to-alanine substitution at amino acid 139. The blaVIM-8 allele also had two silent polymorphisms, A492 to G492 and G693 to A693.
Morbidity and mortality of patients harboring type A strain isolates.
Case notes were reviewed for 40 patients from whom type A strain isolates were obtained. These comprised adults in the adult ICU (n = 20), infants in the neonatal ICU (n = 6), children in the pediatric ICU (2), children in the pediatric medical ward (n = 2), and patients in other adult wards (n = 10). The largest groups were those aged >60 years (n = 11) or <1 year (n = 8). Thirty-one patients were considered to be infected, whereas nine were considered colonized. The most common isolation sites were blood (n = 12), respiratory secretions (n = 8), central venous catheters (n = 7), urinary tract infections (n = 6), peritoneal fluid samples (n = 5), and surgical wounds (n = 4). Multiple isolates were obtained from three patients with septicemia: simultaneously from the blood and feces from one patient and the blood and respiratory tract from the other two patients. The average hospital stay to the time of isolation was 57 days, and 35 of the isolates were recovered from patients who had been hospitalized for 40 days or longer. Of these 40 patients, 33 (82.5%) were undergoing mechanical ventilation, 35 (87.5%) had previously received antibiotics, 9 (22.5%) had received imipenem therapy for 2 to 15 days in the previous 8 to 21 days, and 13 (32.5%) had previously received meropenem therapy; 1 patient had received both of these carbapenems. The most frequent underlying conditions or histories were trauma, cirrhosis, transplantation, chronic renal insufficiency, lupus, and peritonitis. Thirteen (37.5%) of the 31 infected patients died, with P. aeruginosa infection judged to have contributed to death in 10 cases. Three colonized patients also died, and one of them was coinfected with a non-type A P. aeruginosa strain.
Patients infected with the type A strain were variously given high doses of meropenem together with an aminoglycoside (generally gentamicin) or were given piperacillin-tazobactam (4.5 g every 8 h [q8h]) plus an aminoglycoside. Later, piperacillin-tazobactam (4.5 g q8h) plus aztreonam (2 g q8h) was the therapy used. The infections were insidious, and isolates of the type A strain showed minor variations in susceptibilities in vitro; moreover, the general health and nutrition of the patients improved with antimicrobial therapy, precluding a rigorous outcome analysis in relation to therapy.
Environmental screening.
In May and June 2003, screening of the hospital environment for imipenem-resistant P. aeruginosa isolates was undertaken in the most-affected areas, comprising the adult, neonatal, and pediatric ICUs. Samples were also taken from operating rooms. In total, 70 culture swab specimens were taken: 33 from sinks, 5 from stethoscopes, 22 from tubing devices, and 10 from the hands of health care workers. The stethoscope samples were taken from inside the metallic inner ring by using swabs prerinsed in cetrimide broth medium.
Twelve samples were positive for imipenem-resistant P. aeruginosa: nine isolates were recovered from the adult ICU, eight of which were from sinks and one of which was from a stethoscope, and three isolates were recovered from sinks in the neonatal ICU. Imipenem and EDTA were synergistic against these 12 isolates by the disk method, and the isolates were positive for blaVIM by PCR; 10 clustered with the type A strain (Fig. 2), although one of these did not agglutinate with type O12 antiserum; one was serotype O12 but did not have the PFGE pattern typical of type A; and the last one was distinct both by its PFGE pattern and by its serotype (Table 2). No carriers were found among health care workers, and no imipenem-resistant P. aeruginosa strains were recovered from environmental samples from the operating room or the pediatric ICU.
TABLE 2.
Characteristics of imipenem-resistant P aeruginosa isolates from environmental sourcesa
| Isolate | Isolation date (day/mo/yr) | Source | Phenotype | PFGE profile/serotype | Zone diam (mm)
|
blaVIM-2-like subtype by PCR | |
|---|---|---|---|---|---|---|---|
| IMP | IMP + EDTA | ||||||
| 2/01 | 25/05/03 | Stethoscope AICU 7 | Other | A/O12 | 6 | 11 | + |
| 2/02 | 27/03/03 | Sink NICU | Other | A/O12 | 6 | 18 | + |
| 2/03 | 17/06/03 | Sink AICU 17 | 2 | A/O12 | 6 | 16 | + |
| 2/04 | 17/06/03 | Sink AICU 18 | 2 | A/O12 | 6 | 15 | + |
| 2/05 | 17/06/03 | Sink AICU 19 | 4 | A/Non O12 | 6 | 13 | + |
| 2/06 | 17/06/03 | Sink AICU 20 | 4 | A/O12 | 6 | 12 | + |
| 2/07 | 17/06/03 | Sink AICU 12 | 4 | A/O12 | 6 | 12 | + |
| 2/08 | 17/06/03 | Sink AICU 13 | 4 | A/O12 | 6 | 11 | + |
| 2/09 | 20/06/03 | Sink AICU 15 | 4 | A/O12 | 6 | 11 | + |
| 2/10 | 20/06/03 | Sink NICU 192 | 1 | A/O12 | 6 | 21 | + |
| 2/11 | 20/06/03 | Sink NICU 195 | 4 | L/O12 | 6 | 12 | + |
| 2/12 | 20/06/03 | Sink AICU 1 | Other | J/indeterminate | 6 | 12 | + |
Abbreviations: IMP, imipenem; NICU, neonatal ICU; AICU, adult ICU.
Infection control measures.
Inspection revealed that the design of the sinks was not appropriate; in particular, their joints to the walls were not sealed, facilitating colonization with and the persistence of P. aeruginosa. Control measures were taken to eradicate the strain. First, the ICUs were intensively cleaned with hypochlorite, with overnight treatment of the sinks and their drains; moreover, use of these sinks was restricted pending replacement. Stethoscopes were disassembled and cleaned with 70% ethanol, this being the standard procedure.
All ICU staff were strongly reminded to follow isolation procedures and to wash their hands with alcohol glycerol. Follow-up studies in September 2003 reisolated imipenem-resistant P. aeruginosa isolates from only two of nine of the sinks previously found to be contaminated. The water supply into these sinks was then suspended, preventing their further use. Subsequent testing in November 2003 did not reveal the environmental presence of the epidemic strain; nevertheless, the strain continued to be isolated from patients, although more often as a colonist than as a pathogen. Its isolation remains related to very long stays, surgical procedures, peritonitis, and extensive prior antimicrobial therapy.
DISCUSSION
Carbapenems are potent agents against multiresistant gram-negative bacilli, including P. aeruginosa, but survey data show emerging resistance, principally in nonfermenters (9, 10, 34; M. D. Obritsch, D. N. Fish, R. MacLaren, and R. Jung, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1970, 2003; M. K. Weaver, D. Styers, M. E. Jones, C. Thornsberry, and D. F. Sahm, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1962, 2003). In the case of P. aeruginosa, the most frequent mechanism of resistance is loss of the porin OprD (24), but carbapenem-hydrolyzing β-lactamases are increasingly reported. They spread first in Japan and Korea and more recently in Europe (4, 26, 27) and the Americas (7, 11). Large outbreaks by MBL-producing P. aeruginosa strains have been described in hospitals in Italy, Greece, and Korea (4, 14, 16, 18, 20, 29), as well as in Latin America, notably, including one caused by a P. aeruginosa strain with an SPM-1 β-lactamase in Brazil (7). The SENTRY surveillance program for 1999 to 2002 reported MBLs in P. aeruginosa and/or Acinetobacter spp. from Argentina, Chile, Brazil, and Venezuela, but not Colombia (8); nevertheless, the prevalence of imipenem resistance among P. aeruginosa isolates at the hospital in Cali described in the present study rose from 2% in 1996 to 28% in 1997 and 49% in 2002. At least from 1998 onwards, this rise in resistance was substantially due to the type A outbreak-related P. aeruginosa strain, which produced blaVIM-8. Retrospective analysis suggested that the possible source of this strain was a patient transferred from another hospital in 1997. The isolates obtained from this patient in the adult ICU were highly resistant to ceftazidime, imipenem, and meropenem, as were most of the subsequent outbreak isolates. Unfortunately, these isolates were not recoverable for analysis in the present study.
The type A strain was mostly isolated from groups conventionally seen as being at high risk for infection with multiresistant isolates, such as ICU patients, patients with long hospital stays, patients of early or advanced age, and patients with severe underlying conditions. An environmental reservoir was found, especially in the adult ICU, where several sinks were contaminated. This contamination was exacerbated by design flaws in the sinks, and the outbreak was reduced following cleaning and decommissioning of these sinks. Contamination of stethoscopes was also noted.
Isolates belonging to the type A strain showed minor phenotypic variability but mostly had high-level multiresistance to β-lactams, aminoglycosides, and fluoroquinolones, suggesting the presence of aminoglycoside-modifying enzymes and DNA gyrase mutations, as well as the MBL. Many integrons determining the VIM or the IMP β-lactamase also encode aminoglycoside-modifying enzymes.
Many type A isolates appeared to be susceptible to piperacillin (with or without tazobactam) and aztreonam. Such susceptibility has been described in other VIM- and IMP-producing P. aeruginosa strains (5, 11, 35) and is not surprising in the case of aztreonam, which is stable to MBLs. The behavior of piperacillin partly reflects the NCCLS's very high breakpoints (susceptible, MIC ≤ 64 μg/ml), which are 16-fold above the modal MIC for susceptible P. aeruginosa isolates and well within the range expected for strains with acquired resistance. Nevertheless, it is surprising that nearly half of the isolates were susceptible to piperacillin at the much lower breakpoints used in the United Kingdom and Europe (1). Combinations of aztreonam with piperacillin-tazobactam were used clinically for the infected patients, many of whom recovered. It was not, however, possible to identify the relative combinations of therapy and general healthcare to the outcome.
This is the first evidence of a VIM-type enzyme in Colombia and only the second account of a major outbreak caused by an MBL producer in South America. The enzyme, VIM-8, was a novel variant, differing from VIM-2 only by a Thr139Ala substitution. Thr139 is also modified (to Ile) in VIM-9 (GenBank accession number AY524988), a variant from P. aeruginosa in the United Kingdom (J. M. Coelho, N. Woodford, J. Turton, and D. Livermore, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1965, 2003). The producer strain belonged to serotype O12, an uncommon (≤1%) serotype among P. aeruginosa isolates as a whole but one that has a long-recognized association with multiresistance (23). Outbreaks of VIM β-lactamase-producing, serotype O12 P. aeruginosa strains have occurred in Italy (5), Greece (20, 32), and Korea (18). Fifteen years ago, O12 was the major serotype of P. aeruginosa strains hosting the PSE-1 β-lactamase, at least in Europe (26). It may be that serotype O12 strains, which form a tight cluster in terms of DNA relatedness (21, 25), are particularly adept at acquiring or hosting the integrons that encode the IMP, VIM, and PSE enzymes, but this hypothesis is speculative.
This study illustrates the importance of phenotypic and genotypic surveillance to guide infection control. Initially, in 1997, the outbreak was attributed to the overuse of imipenem, and unsuccessful control measures whose aim was to replace imipenem use with cefepime and piperacillin-tazobactam use were taken. Nevertheless, the rate of imipenem use rose (Fig. 1), and imipenem-resistant P. aeruginosa strains continued to be isolated, with little obvious relationship to the rate of imipenem use (Fig. 1). Only much later, when phenotypic surveillance and molecular typing were performed, were appropriate infection control measures taken, and these achieved a reduction in prevalence from the peak in 2002 (Fig. 1). These environmental studies revealed persistent contamination of the adult and neonatal ICUs by type A strains. Although the affected patients were exposed to many antibiotics, previous imipenem consumption was uncommon (only nine patients). Similar findings were also reported in Japan and Italy, where IMP and VIM β-lactamase-producing P. aeruginosa strains spread as hospital pathogens without specific selection by carbapenems (5, 18).
REFERENCES
- 1.Andrews, J. M. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl. 1):5-16. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, J. M. 2004. BSAC standarized susceptibility routing. Version 3. J. Antimicrob. Chemother. 53:713-728. [DOI] [PubMed] [Google Scholar]
- 3.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 4.Cornaglia, G., et al. 1999. Appearance of IMP-1 metallo-β-lactamase in Europe. Lancet 353:899-900. [DOI] [PubMed] [Google Scholar]
- 5.Cornaglia, G., A. Mazzariot, L. Lauretti, G. M. Rossolini, and R. Fontana. 2000. Hospital outbreak of carbapenem resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-β-lactamase. Clin. Infect. Dis. 31:1119-1125. [DOI] [PubMed] [Google Scholar]
- 6.De Freitas, A. L., and A. L. Barth. 2002. Antibiotic resistance and molecular typing of Pseudomonas aeruginosa: focus on imipenem. Braz. J. Infect. Dis. 6:1-7. [DOI] [PubMed] [Google Scholar]
- 7.Gales, A. C., L. C. Menezes, S. Silbert, and H. Sader. 2003. Dissemination in distinct Brazilian regions of an epidemic carbapenem-resistant Pseudomonas aeruginosa producing SPM metallo-β-lactamase. J. Antimicrob. Chemother. 52:699-702. [DOI] [PubMed] [Google Scholar]
- 8.Gales, A. C., R. N. Jones, J. Turnidge, R. Rennie, and R. Ramphal. 2003. Emergence of an IMP-like metallo-enzyme in an Acinetobacter baumannii clinical strain from a Brazilian teaching hospital. Diagn. Microbiol. Infect. Dis. 45:77-79. [DOI] [PubMed] [Google Scholar]
- 9.Gales, A. C., R. N. Jones, J. Turnidge, R. Rennie, and R. Ramphal. 2001. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance program, 1997-1999. Clin. Infect. Dis. 32:S146-S155. [DOI] [PubMed] [Google Scholar]
- 10.Giamarellou, H. 2002. Prescribing guidelines for severe Pseudomonas infections. J. Antimicrob. Chemother. 49:229-233. [DOI] [PubMed] [Google Scholar]
- 11.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. I. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP-7. Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins, P. G., A. C. Fluit, D. Milatovic, J. Verhoef, and F. J. Schmitz. 2002. Antimicrobial susceptibility of imipenem-resistant Pseudomonas aeruginosa. J. Antimicrob. Chemother. 50:299-301. [DOI] [PubMed] [Google Scholar]
- 13.Hirakata, Y., K. Izumikawa, T. Yamaguchi, H. Takamura, H. Tanaka, R. Yoshida, J. Jatsuda, M. Nakano, K. Tomono, S. Maesaki, M. Kaku, Y. Yamada, S. Kamihira, and S. Kohno. 1998. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug resistant gram-negative rods carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 42:2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirakata, Y., T. Yamaguchi, M. Nakano, K. Izumikawa, M. Mine, S. Aoki, A. Kondoh, J. Matsuda, M. Hirayama, K. Yanagihara, Y. Miyazaki, K. Tomono, Y. Yamada, S. Kamihira, and S. Kohno. 2003. Clinical and bacteriological characteristics of IMP type metallo-β-lactamase producing Pseudomonas. Clin. Infect. Dis. 37:26-32. [DOI] [PubMed] [Google Scholar]
- 15.Howard, D. H., R. D. Scott II, R. Packard, and D. Jones. 2003. The global impact of drug resistance. Clin. Infect. Dis. 36:S4-S10. [DOI] [PubMed] [Google Scholar]
- 16.Hsueh, P. R., L. J. Teng, P. C. Yang, Y. C. Chen, S. W. Ho, and K. T. Luh. 1998. Persistence of a multidrug-resistant Pseudomonas aeruginosa clone in an intensive care burn unit. J. Clin. Microbiol. 36:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jean, S. S., L. J. Teng, P. R. Hsueh, S. W. Ho, and K. T. Luh. 2002. Antimicrobial susceptibilities among clinical isolates of extended-spectrum cephalosporin-resistant gram-negative bacteria in a Taiwanese university hospital. J. Antimicrob. Chemother. 49:69-76. [DOI] [PubMed] [Google Scholar]
- 18.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 20.Mavroidi, A., A. Tsakris, E. Txelepi, S. Pournaras, V. Loukova, and L. S. Tzouvelekis. 2000. Carbapenem-hydrolysing VIM-2 metallo-β-lactamase in Pseudomonas aeruginosa from Greece. J. Antimicrob. Chemother. 46:1041-1042. [DOI] [PubMed] [Google Scholar]
- 21.Mifsud, A. J., J. Watine, B. Picard, J. C. Charet, C. Solignac- Bourrel, and T. L. Pitt. 1997. Epidemiologically related and unrelated strains of Pseudomonas aeruginosa serotype O12 cannot be distinguished by phenotypic and genotypic typing. J. Hosp. Infect. 36:105-116. [DOI] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 24.Pai, H., J. W. Kim, J. Kim, J. H. Lee, K. W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitt, T. L. 1988. Epidemiological typing of Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 7:238-247. [DOI] [PubMed] [Google Scholar]
- 26.Pitt, T. L., D. M. Livermore, G. Miller, A. Vatopoulos, and N. J. Legakis. 1990. Resistance mechanism of multiresistant serotype O12 Pseudomonas aeruginosa isolated in Europe. J. Antimicrob. Chemother. 26:319-328. [DOI] [PubMed] [Google Scholar]
- 27.Pournaras, S., M. Maniati, E. Petinaki, L. S. Tzouvelekis, A. Tsakris, N. J. Legakis, and A. N. Maniatis. 2003. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-β-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 51:1409-1414. [DOI] [PubMed] [Google Scholar]
- 28.Rello, J. P., P. Jubert, J. Valles, A. Artigas, M. Rue, and M. S. Niederman. 1996. Evaluation of outcome for intubated patients with pneumonia due to Pseudomonas aeruginosa. Clin. Infect. Dis. 23:973-978. [DOI] [PubMed] [Google Scholar]
- 29.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toleman, M., D. Biedenbach, D. Bennett, R. N. Jones, and T. Walsh. 2003. Genetic characterization of a novel metallo-β-lactamase gene, blaIMP-13, harboured by a novel Tn5051-type transposon disseminating carbapenemase genes in Europe: report from the SENTRY worldwide antimicrobial surveillance programme. J. Antimicrob. Chemother. 52:583-590. [DOI] [PubMed] [Google Scholar]
- 31.Toleman, M. A., A. M. Simm, T.A. Murphy, A. C. Gales, K. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY Antimicrobial Surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 32.Tsakris, A., S. Porunaras, N. Woodford, M. F. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tysall, L., M. W. Stockdale, P. R Chadwick, M. F. I. Palepou, K. J. Towner, D. M. Livermone, and N. Woodford. 2002. IMP-1 carbapenemase detected in an Acinetobacter clinical isolates from the UK. J. Antimicrob. Chemother. 49:217-218. [DOI] [PubMed] [Google Scholar]
- 34.Woodford, N., M. F. Palepou, G. S. Babini, J. Bates, and D. M. Livermore. 1998. Carbapenemase-producing P. aeruginosa in UK. Lancet 352:546-547. [DOI] [PubMed] [Google Scholar]
- 35.Yan, J. J., P. R. Hsueh, W. C. Ko, K. T. Luh, S. H. Tsai, H. M. Wu, and J. J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yong, D., K. Lee, J. H. Yum, H. B. Shin, G. M. Rossolini, and Y. Chong. 2002. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. Clin. Infect. Dis. 10:3798-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]