Abstract
A real-time PCR was developed to quantify Leishmania infantum kinetoplast DNA and optimized to reach a sensitivity of 0.0125 parasites/ml of blood. In order to analyze the incidence of heterogeneity and number of minicircles, we performed comparative PCR by using the Leishmania DNA polymerase gene as a reporter. Assays performed in both promastigote and amastigote stages showed variations among different L. infantum and Leishmania donovani strains and the stability of the minicircle numbers for a particular strain. Analysis of blood samples from a patient who presented with Mediterranean visceral leishmaniasis confirmed the reliability of such an assay for Leishmania quantification in biological samples and allowed an estimation of positivity thresholds of classical tests used for direct diagnosis of the disease; positivity thresholds were in the range of 18 to 42, 0.7 to 42, and 0.12 to 22.5 parasites/ml for microscopic examination, culture, and conventional PCR, respectively. At the time of diagnosis, parasitemia could vary by a wide range (32 to 188,700 parasites/ml, with a median of 837 parasites/ml), while in bone marrow, parasite load was more than 100 parasites per 106 nucleated human cells. After successful therapy, parasitemia levels remain lower than 1 parasite/ml. In the immunocompromised host, relapses correlate with an increase in the level of parasitemia, sometimes scanty, justifying the need for assays with high sensitivity. Such sensitivity allows the detection of Leishmania DNA in the blood of 21% of patients with no history of leishmaniasis living in the Marseilles area, where leishmaniasis is endemic. This technique may be useful for epidemiologic and diagnostic purposes, especially for the quantification of parasitemia at low levels during posttherapy follow-up.
Leishmania infantum is the causative agent of visceral leishmaniasis in the Mediterranean area. Host-parasite relationships may vary from asymptomatic carriage of the parasite to the typical presentation of the disease.
The opportunistic character of this parasite is well demonstrated in immunosuppressed or immunocompromised patients (8); in cases of coinfection by human immunodeficiency virus and Leishmania, relapses are common even when a correct therapeutic scheme has been applied, and there is a need for biological indicators of evolution of the parasitosis in order to start earlier treatment of relapses or to prevent relapses by use of maintenance therapy (7). Asymptomatic carriage was recognized as the most common status in host-parasite relationships (25). In healthy humans, asymptomatic carriage of Leishmania infantum is diagnosed only by immunological tests because parasitic loads seem to be very low compared to those observed during the acute phase of the disease or in dog leishmaniasis (2).
In addition to the conventional microscopic, cultural, and serological methods, numerous DNA-based tests have been described, particularly the use of PCR technology (1, 4, 6, 9, 23). The sensitivities of such methods seem to be variable, depending on the choice of target sequence and the aim of the assay (11). These techniques improve the sensitivity of diagnosis, especially from blood specimens (1, 24), but are unable to quantify the parasitic load. Real-time PCR was designed for accurate quantification of the target sequence, but the trials performed in the leishmaniasis field were not sensitive enough for clinical applications, except in the acute phase of the disease or under experimental conditions (4, 21).
This paper describes a real-time PCR quantification of Leishmania based on the detection of kinetoplast DNA. Results showed a wide range of variation in the results, in good correlation with the clinical status of the subjects, thus allowing us to discriminate between symptomatic patients, cured patients, and asymptomatic carriers. The main point of interest seems to be the kinetic study of parasitemia in the immunocompromised host, allowing biological diagnosis of relapses and quantification of low levels of parasitemia in asymptomatic subjects. Comparative quantification experiments have been performed for parasite counting in tissue or bone marrow samples.
MATERIALS AND METHODS
Patients.
All samples tested came from patients whose Leishmania infection status was explored for different reasons such as exclusion or confirmation of the disease, survey after therapy, early diagnosis of relapse in immunocompromised patients, or biological survey of asymptomatic carriers. According to French legislation, these tests were qualified as assays with direct benefit for the patients.
Diagnosis of the disease relies on relevant clinical symptoms, a high level of specific antibodies (quantified by enzyme-linked immunosorbent assay with promastigote lysate as the antigen), and the presence of the parasite in bone marrow or blood, detected by conventional (microscopy and culture) or molecular techniques.
A survey of treated patients or at-risk patients relies on serological survey and a direct search for Leishmania performed on blood samples when one or more clinical symptoms occur.
In the Mediterranean area, where leishmaniasis is endemic, human asymptomatic carriage is still characterized in vitro on the basis of low levels of specific antibodies which are detectable only by immunoblot and directed against a reduced number of Leishmania antigens or in vivo by a leishmanin skin test (19, 20).
In this study, 11 blood and 10 bone marrow samples from visceral leishmaniasis patients at the time of diagnosis, 6 blood samples from immunocompetent cured patients, 40 samples from seven immunocompromised subjects taken during a survey after the initial cure of the disease, and 46 blood samples from subjects living in Marseilles who never presented with clinical symptoms of leishmaniasis were used.
Sample preparation and conventional techniques for direct diagnosis.
Generally, bone marrow was sampled at the first time of diagnosis, and blood was taken for the patient survey by means of serology and a direct search for the parasite. All blood and bone marrow samples were processed immediately as described below.
Blood and bone marrow were treated similarly in order to perform direct microscopic examination, isolation of parasite by culture, and detection of Leishmania DNA by PCR.
First, nucleated cells were purified by density gradient (histopaque [d = 1.119]; Sigma-Aldrich, St. Quentin Fallavier, France). Cells were then washed three times in RPMI medium by centrifugation before subsequent analysis.
Approximately 5% of the cells were processed by cytocentrifugation and May-Grunwald Giemsa staining for microscopic direct examination.
From bone marrow samples, half of the cells were inoculated onto Novi, McNeal, and Nicolle (NNN) medium, and the other part was used for DNA extraction. From blood samples (initial volume of 5 ml), the cells corresponding to 2 ml of the initial volume of blood were used for DNA isolation and the other part was inoculated onto NNN medium. The tubes were incubated at 24°C, and an aliquot of the liquid phase of the medium was transferred to fresh medium every week. Parasite detection was performed by microscopy every week for 1 month.
DNA extraction.
DNA extraction was performed on the pellet of nucleated cells by using a QIAamp DNA mini kit (reference number 51306; QIAGEN). In order to achieve maximum yield, digestion was performed overnight with proteinase K in Qiagen lysis buffer before the QIAamp DNA mini kit was used. DNA was eluted in 50 μl of distilled water and stored at −80°C. This protocol was chosen after comparison between the classical sodium dodecyl sulfate-proteinase K-phenol-chloroform extraction according to Sambrook et al. (26) and the QIAGEN method with and without modifications.
Qualitative PCR detecting Leishmania DNA.
We routinely performed a nested PCR, according to the method of Fisa et al., for diagnosis and survey of Mediterranean visceral leishmaniasis (9). Amplicons were analyzed on 2% agarose gels in the presence of ethidium bromide.
Detection of kinetoplast DNA by qPCR.
A real-time quantitative PCR (qPCR) for detection and quantification of Leishmania infantum DNA in biological samples was developed. For accurate sensitivity, kinetoplast DNA was chosen as the molecular target. External primers were derived from RV1 and RV2, described previously by Lachaud et al. (11). Detection was performed by means of a hybridization probe based on TaqMan chemistry (Applera, Courtaboeuf, France) designed with Primer 3 software.
A Stratagene (La Jolla, Calif.) MX 4000 system was used for amplification and detection. Optimization experiments led us to use the Stratagene qPCR master mix (catalog number 600549-51), 15 pmol of direct primer (CTTTTCTGGTCCTCCGGGTAGG), 15 pmol of reverse primer (CCACCCGGCCCTATTTTACACCAA), and 50 pmol of TaqMan probe (FAM-TTTTCGCAGAACGCCCCTACCCGC-TAMRA). Assays were performed with a 25-μl final volume with 1 μl of sample DNA. The standard curve was established from Leishmania DNA extracted from 5 × 106 parasites; 1 μl of serial dilutions, ranging from 50,000 to 0.0001 parasites, was introduced into reaction tubes. TaqMan chemistry allowed two-step temperature (94 and 55°C) cycling over 45 cycles.
Quantification of the Leishmania DNA polymerase gene.
In order to analyze the variability of the number of minicircles detected by means of the qPCR described above, comparative quantification was performed by using a single copy gene, the DNA polymerase gene, as a normalizer (4). Primers and a probe described previously by Bretagne et al. (TGTCGCTTGCAGACCAGATG [200 pmol], GCATCGCAGGTGTGAGCAC [200 pmol], and VIC-CCAGGCTCGAAGTTGTTGCTGCCC-TAMRA [200 pmol]) and the same working conditions as previously described for kinetoplast DNA amplification were used.
Quantification of the human albumin gene in tissue samples.
Housekeeping genes from host cells were quantified in order to perform comparative quantification in tissue samples (bone marrow and biopsy). We used primers (GCTGTCATCTCTTGTGGGCTGT [100 pmol] and ACTCATGGGAGCTGCTGGTTC [30 pmol]) and a probe specific for the human albumin gene (VIC-GGAGAGATTTGTGTGGGCATGACA-TAMRA [100 pmol]) according to the method of Laurendeau et al. (16) The thermal profile was identical to those of kinetoplast DNA amplifications. The standard curve was established from DNA extracted from the THP1 human monocytic cell line.
Expression of the results.
For blood, results were expressed as the number of Leishmania parasites present in 1 ml of blood, taking in account the concentration and dilution rates introduced during the extraction and amplification process. In order to allow a comparison of parasitic loads between the different bone marrow samples, we quantified the number of nucleated human cells among the aliquot analyzed and the number of Leishmania parasites in the same aliquot; results were expressed as the number of parasites for 106 host cells.
RESULTS
Sensitivity of qPCR.
Sensitivity was dependent on the DNA extraction method, and we optimized a commercial kit based on the adsorption-elution method for routine diagnosis purposes. We performed experiments in order to compare the yield of the QIAamp DNA extraction kit to that of standard phenol-chloroform extraction after lysis by sodium dodecyl sulfate and digestion with proteinase K (Table 1). The QIAamp method requires digestion of the sample overnight in Qiagen lysis buffer to reach an 80% yield. Isolation of nucleated cells before digestion of the sample is also critical for better results in quantitative experiments compared to direct extraction from whole blood (Table 1).
TABLE 1.
Quantification of Leishmania in whole blood (1 ml) or Ficoll-purified samples after the addition of known amounts of parasitesa
| Sample and parasite addition (no. of parasites) |
Leishmania DNA (parasites/ml) detected byc:
|
||
|---|---|---|---|
| QIAamp (PK 15 min) | QIAamp (PK overnight) | Alcohol precipitation | |
| Whole blood + 100 | 0 | 6 | 62 |
| WBC + 10,000 | 2,350 | 9,890 | 8,500 |
| WBC + 1,000 | 103 | 929 | 724 |
| WBC + 100 | 7 | 97 | 71 |
| WBC + 10 | 0.03 | 8.2 | 6 |
| WBC + 1 | 0 | 0.80 | 0.03 |
Three DNA extraction methods were tested: the QIAamp DNA mini kit procedure, the same protocol with an extended proteinase K digestion time, and the classical phenol-chloroform method. Each test was performed in duplicate.
WBC, white blood cells.
PK, proteinase K digestion.
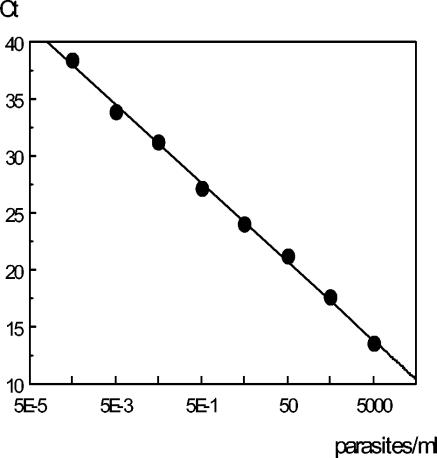
The sensitivity of the qPCR reaction was tested by using serial dilutions of parasite DNA extracted from a known number of parasites. Detection of the kinetoplast DNA of L. infantum reached the level of 0.0005 parasites per reaction tube with a dynamic range of 107. This sensitivity allowed a detection limit of 0.0125 parasite DNA/ml of blood, taking into account the amount of biological sample (1 μl of sample DNA) and the elution volume of the extracted DNA (50 μl). In order to verify the accuracy of the assay, PCR experiments were performed with negative blood spiked with culture promastigotes. Figure 1 presents the standard curve, slope, and efficacy of a typical experiment, and Table 1 summarize the results obtained with spiked samples tested by three extraction protocols. Once the QIAamp protocol with extended proteinase K digestion was selected, we tested the reproducibility of qPCR at 100 parasites/ml and 1 parasite/ml by testing each sample DNA 15 times, and the coefficients of variation were 3.8 and 5.2%, respectively.
FIG. 1.
Standard curve obtained from serial dilutions of Leishmania DNA expressed as the number of parasites per reaction tube. Each point was tested in duplicate. Slope, −3.40; PCR efficacy, 96.7%; R square, 0.996.
Linearity of the whole process.
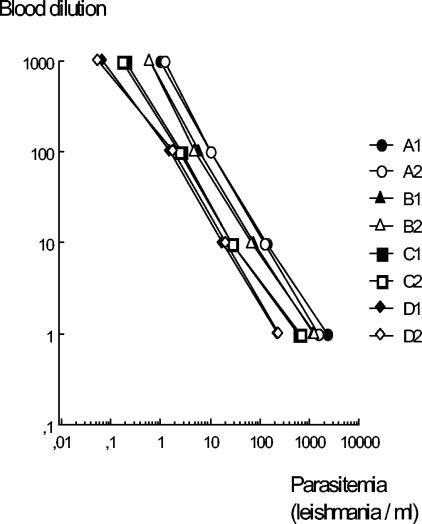
In order to verify that this method is a reliable means of quantification, we took blood from four patients with clinical symptoms and performed serial dilutions with a negative sample (rate, 1 to 10) in duplicate experiments. After DNA extraction and quantification of the parasites, we plotted the results against the different dilutions. Results (presented in Fig. 2) showed that quantification was linear over parasitemia levels ranging from 0.05 to 3,000 Leishmania parasites/ml. Curves obtained from the different samples had identical slopes, and correlation coefficients were greater than 0.998.
FIG. 2.
Linearity of extraction and quantification process. Positive blood samples (from donors A to D) were diluted with negative blood, DNA was extracted in duplicate, and Leishmania DNA was quantified. The coefficient of correlation between dilutions and parasitemia was greater than 0.998 in the eight assays.
Relative stability of the number of kinetoplast DNA targets among promastigote and amastigote stages of L. infantum and Leishmania donovani.
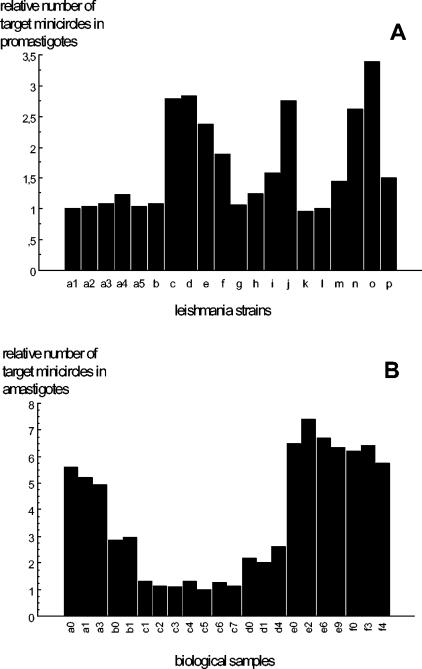
Comparative quantification of minicircles, after standardization by means of DNA polymerase gene quantification, showed that the relative number of targets varies among the promastigote strains in the range of 1 to 3.4 (Fig. 3A). For one particular line, we extracted DNA from promastigotes harvested from the primo culture and the 16th, 32nd, and 64th in vitro passages. Results showed the stability of the number of minicircles.
FIG. 3.
Variations of the relative number of kinetoplast DNA minicircles among culture promastigotes and amastigotes in human samples. DNA from 13 L. infantum (a to m) and 3 L. donovani (n, o, p) culture promastigotes was tested (A). Samples a1 to a5 correspond to different in vitro passages of the same strain. For the amastigote stage (B), relative quantification was performed on positive biological samples from six patients (a to f); the number next to the patient designation indicates the delay (in months) between each sampling. The b0,e0, and f0 samples were from bone marrow, and the others were blood samples.
Concerning the amastigote stage, the same experiments were performed on 22 samples (19 blood samples and 3 bone marrow samples) from six patients during the survey. Results confirmed the variations observed between isolates and the stability of the kinetoplast copy number in amastigotes infecting a patient at different times (Fig. 3B). These observations validate the possibility of quantifying Leishmania through kinetoplast DNA amplification; in order to prevent underestimation of the number of parasites, we chose the strain that presented the lowest number of minicircles as the reference strain (MCAN/82/GR/MON497) for quantification.
Evaluation of the sensitivity of the other diagnosis assays.
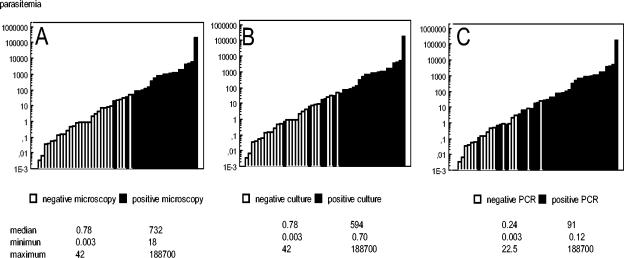
A comparison between qPCR parasitemia and the results obtained from direct examination, culture, and standard PCR allowed us to estimate the sensitivity threshold of these techniques (Fig. 4). Direct examination or culture was positive in all samples whose level of parasitemia was more than 42 parasites/ml. The minimal amount of parasites detected by microscopic examination was 18 parasites/ml, while in one case, culture detected a parasitemia of 1 Leishmania parasite/ml. Nested PCR, routinely used for the diagnosis of the disease, could detect parasite load as weak 0.12 parasites/ml and was positive in all cases where the parasitemia level was greater than 22 parasites/ml.
FIG. 4.
Statistics of 51 blood samples from proven visceral leishmaniasis patients found to be positive by qPCR. Blood was sampled at the time of diagnosis and during the survey of the patients. Histograms show distribution of results with respect to those of microscopic examination (A), culture (B), and qualitative PCR (C).
Symptomatic disease is correlated with high parasitic load.
At the time of diagnosis, Leishmania DNA was present in all blood samples. Parasitemia levels ranged from between 32 and 188,700 parasites/ml, with a median of 837 parasites/ml. In all cases except one, conventional means (direct examination and cultures) were also positive. In the last case, in which the blood sample was negative for parasitemia, bone marrow was sampled at the same time and was found positive by both techniques.
In 10 cases, bone marrow was tested at the initial step of diagnosis; the parasite count was expressed relative to 106 host cells by means of a comparative PCR quantifying both the parasite number and the host cell number (by means of qPCR for human albumin). In these samples, the number of human cells varied over 2 orders of magnitude (from 28,800 to 2,370,000). Results are presented in Fig. 5; at time of diagnosis, all bone marrow samples from visceral leishmaniasis patients had a parasitic load superior or equal to 100 parasites per 106 nucleated host cells.
FIG. 5.
Quantification of Leishmania DNA per million nucleated host cells in bone marrow. Samples a to e: survey after treatment. Samples f to p were taken at diagnosis time or to confirm clinical relapses. Samples f and l were taken at 2-day intervals from a child with visceral leishmaniasis; the first was hemodiluted and found negative by conventional techniques.
Parasitemia decreased rapidly during Ambisome treatment.
After treatment, parasitemia decreased rapidly and remained lower than 1 parasite/ml, whatever the initial level. In two immunocompetent subjects, we observed that parasitemia fell to an undetectable level 10 days after Ambisome therapy. Figure 6A shows the survey of parasitemia following the treatment of subjects who do not relapse. In bone marrow samples, levels observed in five cured patients were under 1 parasite per 1 million cells (Fig. 5).
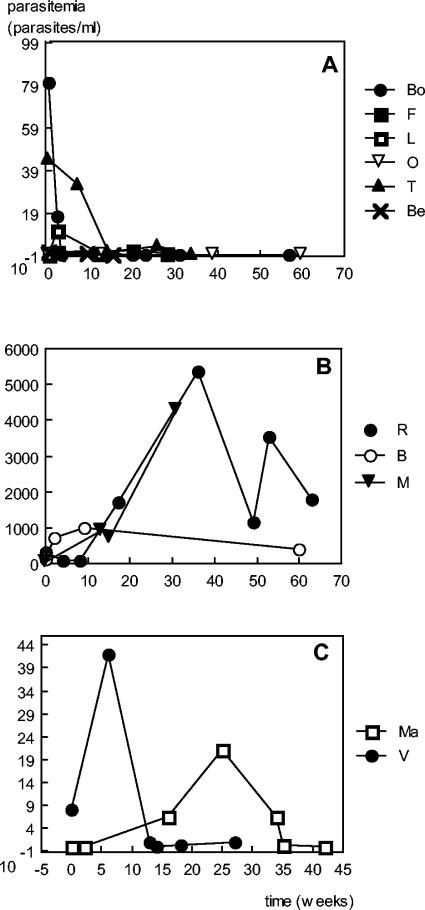
FIG. 6.
Kinetics of parasitemia. (A) Subjects without relapse after Ambisome therapy. (B) Patients who do not control parasite development. R was a coinfected patient who refused any antileishmanial treatment, B was a coinfected patient resistant to treatment, and M was an older subject resistant to meglumine and Ambisome treatment. (C) Typical relapses in coinfected patients; peaks of parasitemia coincide with clinical symptoms.
Relapses correlate with medium parasitemia levels.
After the initial cure of the disease, elevation of parasitemia is linked to clinical relapse. Figure 6 shows kinetic studies of parasitemia in different clinical presentations.
For three human immunodeficiency virus patients, a survey of relapse of leishmaniasis was performed; in two cases (Fig. 6C), maximum parasitic loads were 20 and 42 parasites/ml, lower than those observed at the initial diagnosis of leishmaniasis. During the periods of regression of clinical symptoms, the level of parasitemia was lower than 0.8 parasites/ml. In the third case, after initial successful treatment, higher parasitemia levels were observed for months despite antileishmanial therapy (Fig. 6B [patient B]). In the last case (Fig. 6B [patient R]), parasitemia remained high because the patient refused any antileishmanial treatment; this evolution was identical to that observed for an old patient who resisted any treatment (Fig. 6B patient M]).
Asymptomatic subjects present with low parasitemia.
Parasitemias of 46 asymptomatic subjects living in the Marseilles area, where leishmaniasis is endemic, were undetectable in 36 cases and lower than 0.2 parasites/ml in 10 cases (mean, 0.051; median, 0.015; range, 0.0001 to 0.21). Six out of these 10 subjects had no Leishmania-specific antibodies when tested by immunoblot, the most sensitive technique. In three cases, three blood samples taken at 2-week intervals were tested, and the results were in the same order of magnitude as the initial ones. Out of 36 qPCR-negative blood samples, 14 were positive by immunoblot.
DISCUSSION
We developed a real-time qPCR in order to quantify Leishmania loads in blood or tissues with the best sensitivity. To achieve this goal, we chose kinetoplast DNA as the molecular target. Many authors have published papers showing that a method based on kinetoplast DNA amplification was the most sensitive because this molecular target is present at about 104 copies per cell (15, 21, 22). Lachaud et al. compared six PCR methods for diagnosis of canine leishmaniasis; they found a sensitivity of 10−3 parasites per tube when kinetoplast DNA-based methods were used, identical to our results (14). As a consequence, the positive predictive value for diagnosis of the disease was poor because this conventional PCR detected asymptomatic carriage of the parasite. As a result, it follows that many authors described assays that were less sensitive and detected only the high parasitic loads observed during the acute phase of the disease (23). The ability of this qPCR assay to discriminate between 0.001 and 10,000 Leishmania genomes in a single test tube reverses this disadvantage.
The heterogeneity of kinetoplast minicircles could be a problem for accurate quantification; in order to analyze the impact of this heterogeneity among the isolates, we first improved the sensitivity of detection of L. infantum by analyzing the available kinetoplast sequences from databases and designing primers and probes able to hybridize to the most consensual sequence. Consequently, some Leishmania species are not detected by means of these reagents, particularly the cutaneous leishmaniasis agents (data not shown). We then analyzed the ratio between the count of minicircles and the number of DNA polymerase genes in 13 L. infantum and 3 L. donovani promastigotes. As DNA polymerase is a single-copy gene, this ratio reflects the variations in minicircle numbers among the isolates. Results showed ratios ranging from 1 to 3.4. We performed the same experiments directly on biological samples in order to analyze these variations in the amastigote stage; ratios varied from 1 to 7.4 among the L. infantum-infected cells but remained stable among the parasites sampled from a single patient during the survey. These variations have an impact on quantification results but a negligible consequence on their biological interpretation when the wide range of variations observed in the different clinical situations is taken into account. The stability of the number of minicircles observed for a particular isolate allows the comparison of parasitemia levels during the survey of the patient, particularly for relapse detection. Other repetitive molecular targets do not present this inconvenience and could lead to better accuracy, but their sensitivities are too low for survey and epidemiologic purposes. Hassan et al. (10) proposed the use of mini-exon-derived RNA genes, present at 200 to 250 copies per cell, to improve sensitivity to 0.1 Leishmania parasites after amplification, electrophoresis, and hybridization; this technique was found to be 10-fold less sensitive than a kinetoplast DNA-based method. Piarroux et al. (24) detected Leishmania DNA by targeting a repeated genomic sequence, and this method reached a sensitivity of 1 parasite per reaction; this assay was sufficient for the diagnosis of the disease but not for survey after therapy. Nicolas and colleagues (21) developed a real-time PCR for monitoring experimental Leishmania infections in mice, but sensitivity was too low for application to human disease. Schulz and colleagues (27) developed a quantitative assay that was able to discriminate different Leishmania species, but accuracy was weak when parasitemia was under 100 parasites/ml (coefficient of variation of 60.89% for parasitemia of 100 Leishmania parasites/ml). As a consequence, such a test cannot be used for follow-up testing of treated patients, while the present test allows the possibility of quantification at 1 parasite/ml (coefficient of variation of 5.2%).
The pre-PCR steps, sample processing, and DNA extraction also need to be optimized for better quantification; these experiments were performed with the same strain that we used for standard DNA preparation in order to eliminate the incidence of strain-to-strain variations. As described in previous work (12, 13), we observed that purification of nucleated cells prior to DNA extraction increased the yield about 10-fold. Assays performed on dilutions of positive blood confirmed the capability of the assay to produce a linear response, notably for the lowest levels of parasitemia.
The results obtained from patients who presented with the disease showed continuous variations over a wide range of parasitemia levels and allowed us to compare the sensitivity of the different assays used under routine conditions. It is noteworthy that the examination of stained leukocytes after Ficoll separation had a positivity threshold equivalent to that of NNN culture, corresponding to a parasitemia level of more than 42 parasites/ml, since our qualitative nested PCR had a lower threshold of between 0.12 and 22.5 parasites/ml.
The results also illustrate the diversity of host-Leishmania relationships and the need to quantify parasites for correct interpretation of PCR results.
When the high parasitic loads at the time of diagnosis are taken into consideration, qPCR doesn't offer a decisive advantage for initial diagnosis of visceral leishmaniasis. In bone marrow samples, quantification of the human albumin gene showed that the number of host nucleated cells could vary over 2 orders of magnitude, justifying the adjustment of the enumeration of parasites to the number of host cells analyzed. The design of comparative multiplex experiments allows the performance of the two determinations in the same tube. This possibility would also be of great interest when performing drug sensitivity tests on Leishmania-infected cells, replacing the fastidious microscopic counts by a duplex PCR.
In some cases, samples are taken during the course of Ambisome treatment. In 3 to 5 days, an important decrease of parasitemia can be observed, such that in some cases, blood taken at the end of the course contained no detectable parasites. If primary resistance to treatment is suspected, the kinetics of Leishmania clearance, assessed by daily blood sampling, should be of interest.
After cure, nonimmunocompromised patients present with no detectable parasitemia or levels equivalent to those of asymptomatic carriers of Leishmania; it is well known that relapses are exceptional in cured subjects except in case of severe immunodepression. Immunological studies pointed out the fact that, after cure, visceral leishmaniasis patients develop mechanisms of control of parasite growth which do not need a polarized Th1 response (18). The nature of such mechanisms remains unknown, but they are able to maintain the parasite load near the detection limit.
In an immunocompromised host, the level of parasitemia falls under 1 parasite/ml after the initial treatment but in most cases remains detectable throughout the asymptomatic phase preceding the relapse. Clinical relapse is accompanied by an increase of parasitemia, discrete in some cases (10 to 40 parasites/ml). This fact indicates that a survey of at-risk patients needs a technique sensitive enough to give accurate measurements at low ranges. In a recent study, Bossolasco and colleagues described such a method, with a sensitivity of 0.625 parasites/ml, in which the small subunit rRNA gene was amplified (3). During posttreatment follow-up of coinfected patients, those researchers estimated that an increase of parasitemia above 10 parasites/ml preceded the clinical relapse. Our results are in agreement with those observations. Due to the positive and negative results provided, standard PCR cannot constitute a good criterion of evolution of the disease, especially since a sensitive assay could be positive in the case of asymptomatic carriage. For two patients, we observed an increase of parasitemia during relapse that remained under the positivity threshold of direct examination and culture, so qPCR seems to be the more suitable biological test predictive or confirmative of clinical relapse in at-risk patients and would be adequate for follow-up after therapy.
The detection of asymptomatic carriage of Leishmania in the Mediterranean region, where the disease is endemic, is difficult because parasitic loads and immune responses are weaker than those described in other areas where visceral leishmaniasis is endemic (5, 28). Previous studies showed that 56% of healthy dogs presented with antibodies to L. infantum when detected by immunoblot, while PCR was positive in 80% of the cases (2). In humans, host-parasite relationships are different because only 5 to 38% of the healthy population present with a positive immunoblot (20), and standard PCR performed on blood samples failed to reveal the presence of the parasite in most cases (17). We tested samples from subjects without any clinical symptoms of leishmaniasis and found weak levels of Leishmania DNA in blood in 10 subjects, 6 of them being immunoblot negative. In such cases, parasitemia is in the range of 10−1 to 10−3 parasites/ml. These results were reproducible, but levels are inferior to the theoretical threshold corresponding to one parasite present in the sample submitted to extraction. Two hypotheses could explain this observation: either the extraction yield could be impaired when parasitemia is very low or the phagocytic cells do not contain live parasites because the phagocytic cells have killed them and PCR may detect a small part of the genetic material of the parasites which persists for a time inside the phagocytic cell after parasite destruction. The possibility of detection of Leishmania DNA in healthy humans living in the Mediterranean area could lead to better epidemiological findings; some persons could be infected by L. infantum and control parasite development without detectable specific antibodies. This fact was pointed out by Costa et al., who studied Leishmania chagasi cryptic infections (5). The advantage of PCR in this field relies upon the possibility of obtaining direct proof of the parasitism and indicates that the frequency of human asymptomatic carriage in the Mediterranean area is underestimated when only detection of specific antibodies is taken into account. Moreover, detection of asymptomatic carriage of Leishmania in subjects who risk developing an opportunistic infection could lead to a better and faster diagnosis when the first symptoms occur.
The most important result of this study is that parasite load could be correlated to clinical status. A single test, performed under well-standardized conditions, could be helpful for diagnosis of the disease, use of monitoring survey during therapy, diagnosis of relapses, comparisons of drug efficacy or prophylactic schemes, and epidemiological studies. As PCR has been previously held to be the “gold standard, ” real-time qPCR will certainly become the reference technique in the future.
REFERENCES
- 1.Adhya, S., M. Q. Hassan, S. Mukherjee, P. P. Manna, A. Basu, S. Sen, and S. Bandyopadhyay. 2002. Visceral leishmaniasis in India: promises and pitfalls of a PCR-based blood test. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S179-S183. [DOI] [PubMed] [Google Scholar]
- 2.Berrahal, F., C. Mary, M. Roze, A. Berenger, K. Escoffier, D. Lamouroux, and S. Dunan. 1996. Canine leishmaniasis: identification of asymptomatic carriers by polymerase chain reaction and immunoblotting. Am. J. Trop. Med. Hyg. 55:273-277. [DOI] [PubMed] [Google Scholar]
- 3.Bossolasco, S., G. Gaiera, D. Olchini, M. Gulletta, L. Martello, A. Bestetti, L. Bossi, L. Germagnoli, A. Lazzarin, C. Uberti-Foppa, and P. Cinque. 2003. Real-time PCR assay for clinical management of human immunodeficiency virus-infected patients with visceral leishmaniasis. J. Clin. Microbiol. 41:5080-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretagne, S., R. Durand, M. Olivi, J.-F. Garin, A. Sulahian, D. Rivollet, M. Vidaud, and M. Deniau. 2001. Real-time PCR as a new tool for quantifying Leishmania infantum in liver in infected mice. Clin. Diagn. Lab. Immunol. 8:828-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa, C. H., J. M. Stewart, R. B. Gomes, L. M. Garcez, P. K. S. Ramos, M. Bozza, A. Satoskar, S. Dissanayake, R. S. Santos, M. R. B. Silva, J. J. Shaw, J. R. David, and J. H. Maguire. 2002. Asymptomatic human carriers of Leishmania chagasi. Am. J. Trop. Med. Hyg. 66:334-337. [DOI] [PubMed] [Google Scholar]
- 6.Cruz, I., C. Canavate, J. M. Rubio, M. A. Morales, C. Chicharro, F. Laguna, M. Jimenez-Mejias, G. Sirera, S. Videla, and J. Alvar. 2002. A nested polymerase chain reaction (Ln-PCR) for diagnosing and monitoring Leishmania infantum infection in patients co-infected with human immunodeficiency virus. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S185-S189. [DOI] [PubMed] [Google Scholar]
- 7.Deniau, M., C. Canavate, F. Faraut-Gambarelli, and P. Marty. 2003. The biological diagnosis of leishmaniasis in HIV-infected patients. Ann. Trop. Med. Parasitol. 97(Suppl. 1):115-133. [DOI] [PubMed] [Google Scholar]
- 8.Desjeux, P., and J. Alvar. 2003. Leishmania/HIV co-infections: epidemiology in Europe. Ann. Trop. Med. Parasitol. 97(Suppl. 1):3-15. [DOI] [PubMed] [Google Scholar]
- 9.Fisa, R., C. Riera, E. Ribera, M. Gallego, and M. Portus. 2002. A nested polymerase chain reaction for diagnosis and follow-up of human visceral leishmaniasis patients using blood samples. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S191-S194. [DOI] [PubMed] [Google Scholar]
- 10.Hassan, M. Q., A. Ghosh, S. S. Ghosh, M. Gupta, D. Basu, K. K. Mallik, and S. Adhya. 1993. Enzymatic amplification of mini-exon-derived RNA gene spacers of Leishmania donovani: primers and probes for DNA diagnosis. Parasitology 107(Pt. 5):509-517. [DOI] [PubMed] [Google Scholar]
- 11.Lachaud, L., E. Chabbert, P. Dubessay, J. Dereure, J. Lamothe, J. P. Dedet, and P. Bastien. 2002. Value of two PCR methods for the diagnosis of canine visceral leishmaniasis and the detection of asymptomatic carriers. Parasitology 125:197-207. [DOI] [PubMed] [Google Scholar]
- 12.Lachaud, L., E. Chabbert, P. Dubessay, J. Reynes, J. Lamothe, and P. Bastien. 2001. Comparison of various sample preparation methods for PCR diagnosis of visceral leishmaniasis using peripheral blood. J. Clin. Microbiol. 39:613-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lachaud, L., J. Dereure, E. Chabbert, J. Reynes, J.-M. Mauboussin, E. Oziol, J.-P. Dedet, and P. Bastien. 2000. Optimized PCR using patient blood samples for diagnosis and follow-up of visceral leishmaniasis, with special reference to AIDS patients. J. Clin. Microbiol. 38:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachaud, L., S. Marchergui-Hammami, E. Chabbert, J. Dereure, J. P. Dedet, and P. Bastien. 2002. Comparison of six PCR methods using peripheral blood for detection of canine visceral leishmaniasis. J. Clin. Microbiol. 40:210-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambson, B., A. Smyth, and D. C. Barker. 2000. Leishmania donovani: development and characterisation of a kinetoplast DNA probe and its use in the detection of parasites. Exp. Parasitol. 94:15-22. [DOI] [PubMed] [Google Scholar]
- 16.Laurendeau, I., M. Bahuau, N. Vodovar, C. Larramendy, M. Olivi, I. Bieche, M. Vidaud, and D. Vidaud. 1999. TaqMan PCR-based gene dosage assay for predictive testing in individuals from a cancer family with INK4 locus haploinsufficiency. Clin. Chem. 45:982-986. [PubMed] [Google Scholar]
- 17.le Fichoux, Y., J.-F. Quaranta, J.-P. Aufeuvre, A. Lelievre, P. Marty, I. Suffia, D. Rousseau, and J. Kubar. 1999. Occurrence of Leishmania infantum parasitemia in asymptomatic blood donors living in an area of endemicity in southern France. J. Clin. Microbiol. 37:1953-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mary, C., V. Auriault, B. Faugere, and A. J. Dessein. 1999. Control of Leishmania infantum infection is associated with CD8+ and gamma interferon- and interleukin-5-producing CD4+ antigen-specific T cells. Infect. Immun. 67:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mary, C., D. Lamouroux, S. Dunan, and M. Quilici. 1992. Western blot analysis of antibodies to Leishmania infantum antigens: potential of the 14-kD and 16-kD antigens for diagnosis and epidemiologic purposes. Am. J. Trop. Med. Hyg. 47:764-771. [DOI] [PubMed] [Google Scholar]
- 20.Marty, P., A. Lelievre, J. F. Quaranta, A. Rahal, M. Gari-Toussaint, and Y. Le Fichoux. 1994. Use of the leishmanin skin test and Western blot analysis for epidemiological studies in visceral leishmaniasis areas: experience in a highly endemic focus in Alpes-Maritimes (France). Trans. R. Soc. Trop. Med. Hyg. 88:658-659. [DOI] [PubMed] [Google Scholar]
- 21.Nicolas, L., E. Prina, T. Lang, and G. Milon. 2002. Real-time PCR for detection and quantitation of Leishmania in mouse tissues. J. Clin. Microbiol. 40:1666-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noyes, H. A., H. Reyburn, J. W. Bailey, and D. Smith. 1998. A nested-PCR-based schizodeme method for identifying Leishmania kinetoplast minicircle classes directly from clinical samples and its application to the study of the epidemiology of Leishmania tropica in Pakistan. J. Clin. Microbiol. 36:2877-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nuzum, E., F. White III, C. Thakur, R. Dietze, J. Wages, M. Grogl, and J. Berman. 1995. Diagnosis of symptomatic visceral leishmaniasis by use of the polymerase chain reaction on patient blood. J. Infect. Dis. 171:751-754. [DOI] [PubMed] [Google Scholar]
- 24.Piarroux, R., F. Gambarelli, H. Dumon, M. Fontes, S. Dunan, C. Mary, B. Toga, and M. Quilici. 1994. Comparison of PCR with direct examination of bone marrow aspiration, myeloculture, and serology for diagnosis of visceral Leishmaniasis in immunocompromised patients. J. Clin. Microbiol. 32:746-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riera, C., R. Fisa, M. Udina, M. Gallego, and M. Portus. 2004. Detection of Leishmania infantum cryptic infection in asymptomatic blood donors living in an endemic area (Eivissa, Balearic Islands, Spain) by different diagnostic methods. Trans. R. Soc. Trop. Med. Hyg. 98:102-110. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schulz, A., K. Mellenthin, G. Schonian, B. Fleischer, and C. Drosten. 2003. Detection, differentiation, and quantitation of pathogenic Leishmania organisms by a fluorescence resonance energy transfer-based real-time PCR assay. J. Clin. Microbiol. 41:1529-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma, M. C., A. K. Gupta, V. N. Das, N. Verma, N. Kumar, R. Saran, and S. K. Kar. 2000. Leishmania donovani in blood smears of asymptomatic persons. Acta Trop. 76:195-196. [DOI] [PubMed] [Google Scholar]