Abstract
Adherence to a stainless steel surface selected isolates of Listeria monocytogenes with enhanced surface colonization abilities and a change in phenotype from the common smooth colony morphology to a succession of rough colony morphotypes. Growth in broth culture of the best-adapted, surface-colonizing rough colony morphotype gave a smooth colony revertant. Comparative analysis revealed that the smooth and rough variants had similar phenotypic and biochemical characteristics (e.g., identical growth rates and tolerances to antibiotics and environmental stressors). Rough colony isolates, however, failed to coordinate motility or induce autolysis. The defect in autolysis of rough colony isolates, which involved impaired cellular localization of several peptidoglycan-degrading enzymes, including cell wall hydrolase A (CwhA), suggested a link to a secretory pathway defect. The genetic basis for the impairment was studied at the level of the accessory secretory pathway component SecA2. DNA sequencing of the secA2 gene in smooth and rough colony isolates found no mutations in the coding or promoter regions. Analysis of SecA2 expression with an integrated secA2-FLAG tag construct found the protein to be upregulated in the rough and revertant backgrounds compared to the parental smooth colony isolate. A compensatory mechanism involving the SecA2 secretion pathway components is postulated to control smooth to rough interconversion of L. monocytogenes. Such phenotypic variation may enhance the ability of this opportunistic pathogen to colonize environments as diverse as processing surfaces, food products, and animal hosts.
Changes in bacterial colony morphology often accompany microbial adaptation to new environments and ecological niches (12, 19, 32, 34). Understanding the molecular basis of morphotypic change during biofilm formation on abiotic surfaces and the subsequent seeding and adaptation to planktonic growth have important implications for the food industry and medicine. For example, the fouling of conveyor belts and prosthetic devices can lead to contamination of food products and the spread of infection, respectively (16, 36). Experimental models that are amenable to molecular methods are needed so that these problems can be circumvented. The readily cultured, intracellular, food-borne pathogen Listeria monocytogenes provides a practical model to explore morphotypic variation and its role in microbial adaptation.
Variant rough colony morphotypes were first described within a decade of the discovery of smooth-colony-forming L. monocytogenes (17). The rough colony morphotype was thought to occur spontaneously and irreversibly at low frequency during prolonged culture in the laboratory. Apart from obvious physical differences, such as the absence of a blue-green sheen upon Henry illumination and impaired cell separation that gave chaining cells without coordinated motility, the fermentative and biochemical profiles of rough and smooth colonies were considered identical (16, 17, 43).
Characterization of the L. monocytogenes secreted proteome (25) implicated a peptidoglycan hydrolase, CwhA (formerly termed invasion-associated protein or p60) (47), in the formation of the rough colony morphotype. These so-called type I rough colony isolates showed reduced CwhA secretion plus decreased attachment and invasion of certain nonphagocytic cell lines. In an intraperitoneal mouse infection model, a single type I rough isolate gave a 1,000-fold decrease in the 50% lethal dose (17). A CwhA null mutant was recently used to clarify the role of the CwhA protein during infection (39). The mutant had a chaining phenotype that was aberrantly septated during exponential growth. The accumulation and localization of the virulence factors ActA and InlA was abnormal and virulence in the mouse infection model was attenuated, but it retained a smooth colony morphotype. This result and the isolation of rough colony isolates (termed type II), which show wild-type levels of CwhA secretion and cellular invasion (28, 43), suggest that proteins other than CwhA determine the rough morphotype.
Truncations in the nonessential secretion-associated ATPase SecA2 were recently identified in a percentage of type I, but not type II, rough colony isolates (28). A secA2 deletion mutant was used to assess the significance of the secA2 truncations on protein secretion. The impaired secretion of a distinct subset of eight proteins, including CwhA, was similar to that found in secA2 deletion mutants of other bacterial species, including Streptococcus gordonii and Mycobacterium tuberculosis (1, 4). The attenuated virulence of the L. monocytogenes secA2 deletion mutant in an intravenous mouse model prompted speculation that proteins secreted via the SecA2 pathway have a role in the colonization of host tissues and that the SecA2 pathway may play a role in a cyclical transition between parasitic (smooth) and saprophytic (rough) growth (28). However, the authors were unable to determine the advantage afforded by the rough phenotype or obtain experimental evidence of the reversion from rough to smooth colony morphology.
Although L monocytogenes is generally regarded as a poor biofilm former, the development of multispecies biofilms in food-processing facilities (3, 22) is thought to be a major vehicle for the amplification and subsequent contamination of food products (44). This report describes the impact of phenotypic variation on the formation of pure-species biofilms on a stainless steel surface by a virulent smooth isolate of L. monocytogenes and the selection of a revertant smooth morphotype in broth culture. The findings of this research demonstrate a biological role for the rough colony morphotype in the colonization of abiotic surfaces, while perturbations of the SecA2 secretion pathway appear to be involved in transitions between the rough and smooth morphotypes.
MATERIALS AND METHODS
Bacterial strains.
The Listeria monocytogenes 1/2a isolate KM'92 (wild type) (NZRCC 92/870) (7) was obtained from the New Zealand Reference Culture Collection (Environmental Science and Research Ltd.). Working cultures of L. monocytogenes were routinely maintained on Trypticase soy agar containing 0.6% yeast extract (Merck KGaA, Darmstadt, Germany) (TSAYE) or in Trypticase soy broth containing 0.6% yeast extract (TSBYE) at 25°C for 48 h or 37°C for 24 h. Broth cultures were grown either at 25°C with agitation at 150 rpm or at 37°C with agitation at 200 rpm on a rotary shaker. The Escherichia coli K-12 derivatives XL1-Blue (Stratagene, La Jolla, Calif.) and DH5α (Invitrogen, Carlsbad, Calif.) were routinely cultured at 37°C on Luria-Bertani agar or in Luria-Bertani broth (LB) with agitation at 200 rpm. Ampicillin at 100 μg/ml or chloramphenicol (15 μg/ml for E. coli and 7.5 μg/ml L. monocytogenes) was added to broth or agar as required. Stock bacterial cultures were maintained at −80°C in growth medium supplemented with 15% (wt/vol) glycerol.
Development of pure-culture L. monocytogenes biofilms.
The method of Bremer et al. (6) was adapted for the development of biofilms in pure continuous-culture L. monocytogenes bioreactors. Bioreactors containing equally spaced stainless steel coupons were constructed as previously described. Bioreactor experiments were run at 25°C. Isolate inoculums were grown in 1/10 TSBYE at 25°C for 24 h with shaking. A 1-ml culture sample was inoculated into the bioreactor containing 250 ml of 1/10 TSBYE and stirred for 18 h (150 rpm) with a stir bar (5 by 30 mm). A continuous flow of fresh 1/10 TSBYE was then introduced into the bioreactor, and biofilms were allowed to develop on the stainless steel coupons for 72 h at flow rates equivalent to 1 reactor volume per 10, 1, and 0.5 times the generation time for each isolate for successive 24-h periods. A phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]) wash to remove remaining non-surface-adherent cells was then applied for 2 h at 1 reactor volume per 0.1 times the generation time. Coupons were removed and sampled as detailed by Bremer et al. (5). Variant colony morphologies obtained by coupon sampling are described in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Listeria monocytogenes KM'92 | ||
| Wild type (Wt) | Smooth colony containing single and paired regular-sized cells | 7 |
| SR (short-chaining rough) | Rough colony, undulate border containing irregular-sized cells in chains up to 10 cells in length | Wt bioreactor, this study |
| LR (long-chaining rough) | Rough colony, compact border containing irregular-sized cells in chains up to 100 cells in length | Wt bioreactor, this study |
| RS (revertant smooth) | Smooth colony containing singles, pairs, and short chains of up to 6 cells of regular sized cells | LR TSBYE this study |
| Wt-FLAG | Wt attB::pPL2secA2-FLAG | This study |
| SR-FLAG | SR attB::pPL2secA2-FLAG | This study |
| LR-FLAG | LR attB::pPL2secA2-FLAG | This study |
| RS-FLAG | RS attB::pPL2secA2-FLAG | This study |
| Wt-pPL2 | Wt attB::pPL2 | This study |
| SR-pPL2 | SR attB::pPL2 | This study |
| LR-pPL2 | LR attB::pPL2 | This study |
| RS-pPL2 | RS attB::pPL2 | This study |
| Escherichia coli | ||
| XL1-Blue | Wild-type E. coli | Stratagene |
| DH5α | Wild-type E. coli | Invitrogen |
| Plasmids | ||
| pPL2 | L. monocytogenes site-specific integrative vector | 26 |
| pQE30 | N-terminal hexahistidine tagging protein expression vector | Qiagen |
Induction of autolysis.
The rate of induced autolysis of KM'92 isolates was assessed with a technique adapted from Mani et al. (31) and Popowska et al. (40). Exponential phase cells at an optical density at 600 nm (OD600) of 0.7 were grown in 500 ml of TSBYE at 25°C with shaking. The cells were chilled on ice for 10 min, pelleted by centrifugation (7,000 × g for 12 min at 4°C), and resuspended in 40 ml of sterile, ice-cold, distilled H2O. The suspension was pelleted twice by centrifugation (3,000 × g for 15 min at 4°C) in a swinging-bucket rotor and resuspended in 50 mM Tris-HCl, pH 8, to an OD600 of 1.0. Duplicate cultures consisting of 25-ml samples in 100-ml conical flasks were incubated at 25°C with shaking, and the OD600 was recorded from duplicate samplings.
Antibiotic and environmental stress tolerance.
A broth microdilution method in 96-well microtiter plates (Nunc, Rochester, N.Y.) was used to determine the susceptibility of KM'92 isolates to ampicillin, chloramphenicol, rifampin, streptomycin, tetracycline, and vancomycin. Stocks of antibiotics were diluted in Trypticase soy broth (TSB), and the antibiotic susceptibility of each isolate was tested in triplicate. Overnight TSB cultures of wild-type KM'92 and its isogenic derivates SR, LR, and RS were grown with agitation at 37°C and diluted in TSB to give approximately 105 cells per well. After incubation at 37°C for 48 h, the concentration of antibiotic in the first well showing no growth was scored as the MIC. Samples of 5 μl from the last positive and all subsequent negative wells were drop plated onto TSAYE and PALCAM (Merck KGaA) plates that were incubated for 24 h at 37 and 30°C, respectively. The concentration of antibiotic in first well negative for growth on TSAYE plates was scored as the minimal bactericidal concentration. The PALCAM plates were used to confirm the presence of L. monocytogenes.
Environmental stress tolerance experiments were conducted in TSBYE medium supplemented with 0.5 to 12% (wt/vol) NaCl (Sigma, St. Louis, Mo.) or 0 to 12% ethanol (BDH, Darmstadt, Germany) or adjusted to pH values of 4 to 9.5 (adjusted with 1 M HCl or 1 M NaOH). Overnight TSBYE cultures of wild-type KM'92 and its isogenic derivates SR, LR, and RS were grown with agitation at 25°C. Samples of 105 CFU were inoculated in triplicate into 5 ml of TSBYE broth, the turbidity was scored visually over 144 h, and the morphology of single colonies recovered on TSBYE plates was recorded.
Subcellular fractionation of L. monocytogenes cells.
The method described by Jonquieres et al. (23) was adapted for the subcellular fractionation of L. monocytogenes cells. Cultures of KM'92 isolates were grown at 37°C to exponential phase (50-ml culture OD600 = 0.7) or early stationary phase (10-ml culture OD600 = 1.4) in filter-sterilized TSBYE (0.22 μM Millex-GP filter; Millipore, Billerica, Mass.). Cultures were centrifuged (7,000 × g for 10 min), and the supernatant filter was sterilized. Cell pellets were resuspended in 1 ml of wash buffer (10 mM Tris-HCl [pH 6.9], 10 mM MgCl2), centrifuged (7,000 × g for 10 min), and resuspended in 1 ml of wash buffer containing 500 mM sucrose (SWB). The cell pellet was recovered by centrifugation (7,000 × g for 10 min), and an additional 2-min centrifugation (7,000 × g) was used to remove the last traces of the supernatant.
Cell pellets were resuspended in 100 μl of SWB containing 1 mg of fresh lysozyme (Roche, Indianapolis, Ind.), 250 U of mutanolysin (Sigma) and 1 mM phenylmethylsulfonyl fluoride (Sigma) and incubated at 37°C for 2 h. Protoplasts were pelleted by centrifugation (15,000 × g for 10 min), and the supernatant (designated the cell wall fraction) was retained. The protoplasts were washed by centrifugation (15,000 × g for 10 min) in 1 ml of SWB, resuspended in 200 μl of protoplast lysis buffer (100 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM NaCl, 10 μg of DNase I per ml [Roche]) and frozen at −20°C. Three cycles of freeze-thawing were used to stimulate protoplast lysis, the mixture was centrifuged at 20,000 × g for 30 min at 4°C, and the supernatant (designated the cytoplasmic fraction) was retained. The pelleted cell membrane fraction was washed by centrifugation (20,000 × g for 10 min at 4°C) in 1 ml of protoplast lysis buffer and resuspended in 100 μl of TE buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA).
The filter-sterilized culture supernatant fraction was precipitated with ice-cold 16% trichloroacetic acid (BDH) for 1 h, pelleted by centrifugation (20,000 × g for 30 min at 4°C), and washed with 1 ml of ice-cold acetone. The pellet was air dried and resuspended in 50 μl of TE buffer. The supernatant concentrates were neutralized with 1 M Tris prior to protein separation. All fractions were stored at 4°C in their respective buffers containing 1 mM phenylmethylsulfonyl fluoride.
The protein concentrations of the cellular fractions were determined with the modified micro-Lowry protocol (DC protein assay; Bio-Rad, Hercules, Calif.) with bovine γ-globin (Sigma) as the protein standard. Plates were read at 750 nm (EL 340, BioTek Instruments Inc, Winooski, Vt.) and analyzed with DeltaSoft3 software (BioMetallics Inc., Princeton, N.J.).
Cell fractions at the indicated equivalent protein loading were separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 10% acrylamide gels (Mini Protean; Bio-Rad) with Benchmark or SeeBlue Plus2 (Invitrogen) prestained protein standards. Western blots were electrotransferred to nitrocellulose (Hybond-C extra, Amersham BioSciences, Uppsala, Sweden), probed with the PepD anti-CwhA (8) or anti-Flag tag M2 monoclonal antibody (Sigma), detected with the anti-mouse immunoglobulin G-alkaline phosphatase conjugate secondary antibody (Sigma), and developed with the Sigma Fast nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) tablet system (Sigma).
Peptidoglycan hydrolase activity gels were prepared as described by Wuenscher et al. (47), except 1.5% (wt/vol) Micrococcus luteus cells (Sigma) were incorporated into the separating gel.
Complementation of rough isolates with secA2.
The L. monocytogenes integrative plasmid pPL2secA2 (28) contains the full-length secA2 gene from 1/2a strain 10403S with a FLAG tag sequence incorporated at the 3′ end of the open reading frame. A control plasmid (pPL2) was obtained by excising the secA2 gene from the multiple cloning site of pPL2secA2 with PstI (New England Biolabs). L. monocytogenes was electroporated as described by Park and Stewart (38), with an additional lysozyme treatment (I. Glomski, personal communication). In brief, ampicillin (10 μg/ml) was added to cultures (at OD600 = 0.2) grown in 100 ml of BHI containing 500 mM sucrose and then incubated at 37°C with shaking for a further 2 h. The wild-type, SR, and RS cells were pelleted in an angle rotor (7,000 × g for 10 min at 4°C), while LR required centrifugation in a swinging bucket rotor (2,000 × g for 10 min at 4°C). Cells were resuspended in 100 ml of ice-cold, filter-sterilized HSB (1 mM HEPES [pH 7], 500 mM sucrose) and then washed with 35 ml of HSB and 10 ml of HSB containing 1 mg of lysozyme. The suspensions were incubated at 37°C for 20 min, and the cells were pelleted (5,000 × g for 10 min at 4°C) and resuspended in 10 ml of HSB and then 500 μl of HSB. Samples of 100 μl were either used immediately or snap frozen at −80°C. The samples were mixed on ice with 1 to 2 μg of plasmid DNA and electroporated (10 kV/cm, 400 Ω, 25 μF) (Gene Pulser II; Bio-Rad). After a 1-h static regeneration in 1 ml of BHI containing 500 mM sucrose at 37°C, the cells were plated on TSAYE containing 7.5 μg of chloramphenicol per ml and incubated at 37°C for 48 h. The integration of pPL2 or pPL2secA2 was confirmed by PCR assay (26) with primers PL95 and PL102 (Table 2).
TABLE 2.
Oligonucleotides used in this studya
| Primer | Oligonucleotide sequence 5′-3′ |
|---|---|
| secA2 SphI Fwd | TAT AGC ATG CAG ACA GAA TTA TGA TGA TCG |
| secA2 KpnI Rev | TAA GGT ACC CAT AAA TTT GGA AAA AAT CAG |
| Seq Fwd#1 | GTA AAC CTT TAT AGT GTA AGT GGG |
| Seq Fwd#2 | GAG TAT TTA GCT AGA CGT GAT CG |
| Seq Fwd#3 | GAA GTA AAA GAA GAA TCA CGT ACG |
| Seq Fwd#4 | AAA CTT TCC GCG AAA CTT AA |
| Seq Rev#4 | GCT GTA TAA ATG GCC TTT TTT TGG |
| Seq Rev#3 | GTT TTA GCT GTT CCT GTC ATT CC |
| Seq Rev#2 | TAC GAA TAT CGT AGT TTG CTC C |
| PL102 | TAT CAG ACC TAA CCC AAA CCT TCC |
| PL95 | ACA TAA TCA GTC CAA AGT AGA TGC |
Underlining denotes a novel restriction enzyme site.
Sequencing and N-terminal hexahistidine tagging of secA2.
Genomic DNA was isolated from L. monocytogenes as described by Pospiech et al. (41), with both lysozyme (10 mg/ml) and mutanolysin (2,500 U/ml) included in the lysis buffer. The secA2 gene from KM'92 isolates were PCR amplified from genomic DNA with the high-fidelity polymerase PfuTurbo (Stratagene). The optimized conditions used 100 μM each deoxynucleoside triphosphate (Roche), 500 nM each secA2-specific primer (Invitrogen) (Table 2), 1 mM MgSO4 and 15 ng of genomic DNA. The PCR product for DNA sequencing was amplified with the primer combination Seq Fwd#1 and secA2 KpnI Rev. For the construction of a hexahistidine tag at the 5′ end of secA2, a second PCR product was obtained with the primer combination secA2 SphI Fwd and secA2 KpnI Rev. Tubes were initially incubated for 10 min at 95°C before the addition of PfuTurbo, followed by 2 min at 95°C. PCR used 24 cycles of denaturation (95°C for 30 s), annealing (60°C for 15 s) and extension (72°C for 150 s), with a final extension at 72°C for 300 s. Amplimers for DNA sequencing reactions (2.55 kb) and cloning (2.35 kb) were purified with QIAquick column purification (Qiagen, Valencia, Calif.). Samples were sequenced with the primers described in Table 2, with the MegaBACE DNA analysis system (Amersham BioSciences) at the University of Waikato DNA Sequencing Facility.
The PCR products for hexahistidine tagging of SecA2 from each KM'92 isolate were ligated into a complementary double-digested pQE30 (Qiagen) and used to transform E. coli XL1-Blue. Transformants, confirmed by restriction analysis of their plasmids, were inoculated into LB broth preculture containing 100 μg of ampicillin per ml. The hexahistidine-tagged SecA2 proteins were isolated from inclusion bodies under denaturing conditions as described in the QIAexpressionist handbook (Qiagen) from a 20-ml isopropylthiogalactopyranoside (IPTG)-induced culture.
Nucleotide sequence accession numbers.
The DNA sequence for the secA2 gene described in this paper has been deposited in the GenBank/EMBL/DDBJ database under accession number AY072791.
RESULTS
Surface-adherent growth selects for rough isolates of L. monocytogenes.
Inoculation of the bioreactor with the smooth KM'92 wild-type colony morphology, previously implicated in an outbreak of listeriosis in 1992 (7), gave three distinct colony morphologies when surface-adherent growth from the stainless steel coupons was plated on TSAYE. These included the smooth wild-type (2 × 106 CFU/cm2) and two new rough colony morphologies, SR (short-chaining rough) (7 × 103 CFU/cm2) and LR (long-chaining rough) (5 × 104 CFU/cm2). The morphotypes of SR and LR were clearly different. The LR colonies were more compact than their SR counterparts (Fig. 1A), with chaining cells forming structures that were 10 times longer (about 100 cells in length in TSBYE) than SR colonies (Fig. 1B). While the LR cells clumped in broth, the SR cells tended to sediment (Fig. 1C). All KM'92 isolates appeared stable, because they maintained their colony and cellular morphology through several serial passages on nutrient-rich agar. In contrast to SR, plating of stationary-phase LR cells grown in nutrient-rich broth (either TSBYE or BHI) revealed three colony morphologies, the rough morphotypes LR and SR plus a revertant smooth colony termed RS. Inoculation of the bioreactor system with EGD-e, a second smooth strain of L. monocytogenes (14), also selected rough isolates during surface colonization (data not shown), indicating that the morphotypic variation observed in the bioreactor was not unique to KM'92.
FIG. 1.
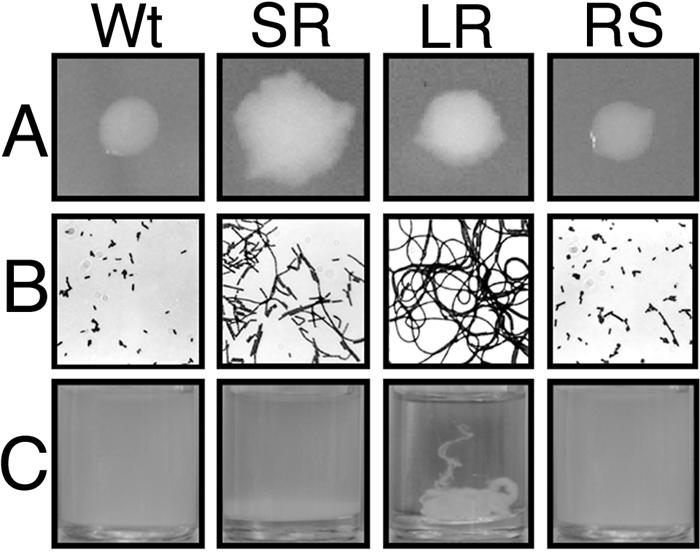
Macroscopic and microscopic morphology of KM'92 bioreactor-derived isolates. (A) Colonies of KM'92 bioreactor-derived isolates were grown on TSAYE for 48 h at 25°C. Wt, wild type. (B) Gram-stained KM'92 bioreactor-derived isolates are shown at 1,000 × magnification. (C) Cultures of bioreactor-derived isolates were grown in TSBYE shaken at 150 rpm for 24 h at 25°C.
The growth characteristics and susceptibilities of the four isolates of KM'92 in common food-processing conditions and to a variety of antibiotics were determined. No differences were found in growth rates or growth yields in acidified (to pH 4.2) or ethanol- (up to 8%) or NaCl- (up to 12%) containing TSBYE. The morphotypes appeared equally susceptible to growth inhibition and the cidal effects of individual antibiotics, including ampicillin, chloramphenicol, rifampin, streptomycin, tetracycline, and vancomycin. Routine biochemical tests, which included sugar fermentation profiles, esculin hydrolysis, hemolytic activity, and catalase production, confirmed all four isolates as L. monocytogenes. Negatively stained transmission electron microscopy images revealed that all four strains were flagellated (data not shown). Tests with tryptone swim agar (9) at 25°C showed that the two rough isolates lacked coordinated motility. The revertant smooth morphotype RS gave growth zones indicating coordinated motility, but these were smaller than for the wild-type morphotype (7.5 mm versus 11 mm after 48 h).
Reversion of LR is dependent on nutrient concentration and growth phase.
The contrasting intrinsic stability of KM'92 morphotypes on agar and the novel morphotypic conversion of LR to both SR and RS in broth led us to consider the effects of culture conditions on the reversion process. Cultures of LR grown in full-strength TSBYE or BHI under static conditions or with shaking at either 25 or 37°C gave the morphotypic conversion. The conversion was observed only once cultures had reached the late exponential or early stationary phase. The LR colonies gave separate colonies of SR or RS rather than a sectoring outgrowth. The frequency of morphotypic conversion ranged between 6.8 × 10−1 and 2.6 × 10−4 (detection limit, 5 × 10−4), and the conversion was found in only 50% of all full-strength TSBYE or BHI cultures sampled in stationary phase. The formation of RS and SR cells was severely repressed in Listeria enrichment broth (Merck KGaA) and completely inhibited in 1/10th strength TSBYE or BHI. Reversion was not observed in acidified medium or medium supplemented with ethanol or NaCl. These results suggest that some stresses inhibit the morphotypic conversion.
Rough isolates of KM'92 give enhanced biofilm formation.
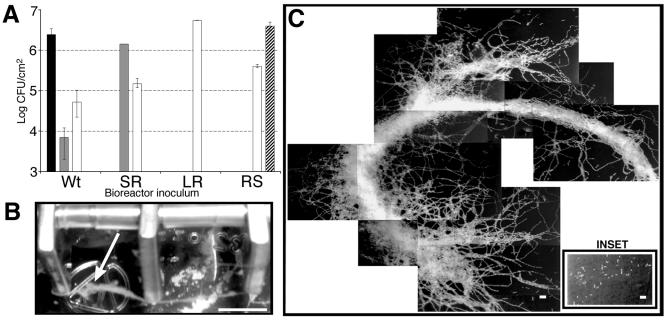
The ability of the four KM'92 isolates to colonize the stainless steel coupons within the bioreactor system was tested individually. The number of cells colonizing the stainless steel coupons was estimated by a swabbing and plating technique. The cell numbers on coupons were independently verified by visual inspection of acridine orange-stained surface-adherent cells. The analysis revealed increased surface colonization by the rough isolates SR and LR compared with the two smooth isolates (Fig. 2A). The viable cell count data underestimated surface growth because chains of cells can give rise to single colonies. Serial dilution of stationary-phase broth cultures indicated that a further correction factor for cell chain length of 10 and 100 should be applied to SR and LR, respectively. Surface-adherent growth in the bioreactor further increased the cell chain length of rough cells. This was most prominent in the LR-inoculated bioreactor, with alga-like streamers projecting from the stainless steel surface (Fig. 2B and 2C). In addition, the majority of the visible surface-adherent streamer biomass was lost during the phosphate-buffered saline wash step. This physical loss, together with the increased cell chain length, suggests a gross underestimate of surface-adherent cell numbers when plating techniques are used.
FIG. 2.
Biofilm-forming characteristics of KM'92 bioreactor-derived isolates. (A) Stainless steel colonization and isolation of morphotypic variants. Mean cell densities attached to the stainless steel coupons in bioreactors inoculated with the single morphotypic variant of KM'92 are indicated on the x axis. The morphologies of the resultant colonies are indicated: wild type (Wt), black; SR, grey; LR, white; RS, striped. Viable cells were estimated by the swabbing technique and plating serial dilutions onto TSAYE. Standard deviations were calculated from mean viable cell numbers from triplicate coupons. (B) Visible cell growth (indicated by the arrow) emanating from the surface of stainless steel coupons within a bioreactor inoculated with KM'92 morphotype LR. Bar, 0.75 cm. (C) Epifluorescence microscopy of L. monocytogenes cells attached to the surface of a stainless steel coupon. The coupon was removed from a bioreactor inoculated with the LR morphotype. Inset: Colonization of a stainless steel surface from a bioreactor inoculated with the wild-type morphotype. Bar. 10 μm.
We conservatively estimate that SR and LR showed at least 5- and 100-fold greater colonization of the stainless steel surface, respectively, than the wild type or RS. Conditions within the bioreactor selected for the LR morphotype, as LR was detected in bioreactors inoculated with the wild type, SR, and RS. Furthermore, when LR was used as the inoculum, it was stably maintained throughout the bioreactor run.
Localization of CwhA and other peptidoglycan hydrolases is impaired in rough colony isolates.
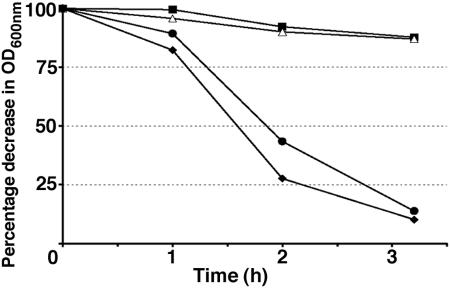
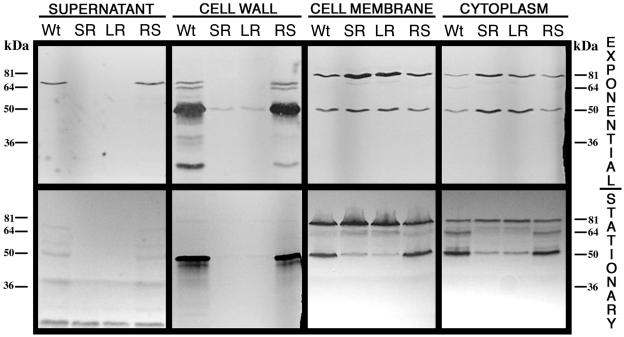
Autolysis experiments with exponential-phase cultures showed that the rough isolates were impaired in peptidoglycan hydrolase induction (Fig. 3). In contrast to the wild-type and RS smooth morphotypes, which gave a 90% decrease in optical density in 3 h, the SR and LR rough isolates gave only a 10% decrease. These results suggested that peptidoglycan hydrolases were affected in the rough isolates. The production and the subcellular localization of CwhA and other peptidoglycan hydrolases in the KM'92 isolates were assessed with renaturing SDS-PAGE and Western blot analysis of subcellular protein fractions. Western blots of SDS-PAGE-separated cellular proteins from supernatant, cell wall, cell membrane, and cytoplasmic fractions were probed with the CwhA-specific monoclonal antibody. This analysis showed reduced secretion of CwhA from the rough isolates in both exponential and stationary phase (Fig. 4A) and identified SR and LR as type I rough isolates. The impaired secretion in both rough isolates correlated with an association of CwhA with the cell membrane fraction (Fig. 4C).
FIG. 3.
Artificially induced autolysis in KM'92 isolates. Exponential-phase cells were harvested at an OD600 of 0.7 and induced to autolyze with a cold shock treatment as described in Materials and Methods. ♦, wild type; ▪, SR; ▵, LR; •, RS.
FIG. 4.
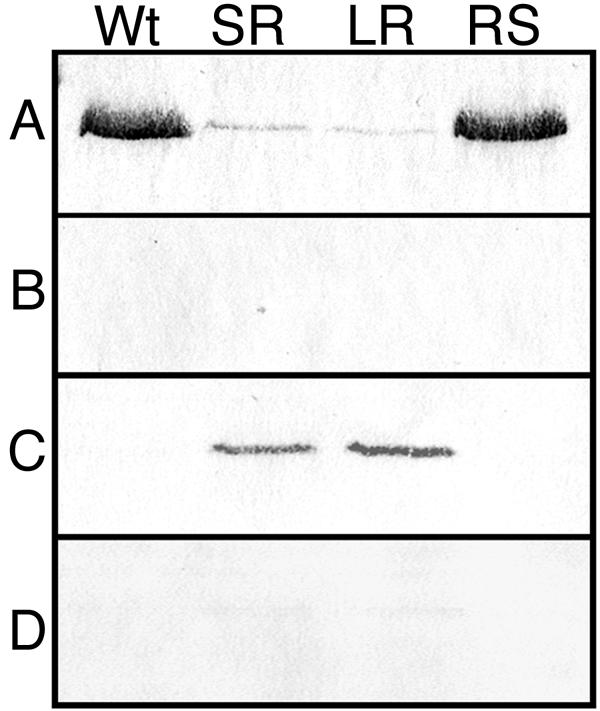
Anti-CwhA immunoblot of cellular fractions from KM'92 isolates. Proteins from exponential-phase (OD600 = 0.7) wild-type (Wt), SR, LR, and RS cells were fractionated as described in Materials and Methods into (A) extracellular supernatant, (B) cell wall, (C) cell membrane, and (D) cytoplasm. Equivalent loadings (50 μg of protein) from the extracellular supernatant, cell membrane, and cytoplasm, and 20 μl, corresponding to one-fifth of the total cell wall fraction, were separated by SDS-PAGE on 10% acrylamide gels. The separated proteins were electrotransferred to nitrocellulose and probed with the PepD-CwhA monoclonal antibody as described in Materials and Methods. The antigenic band detected in the preparations had an apparent molecular size of ≈55 kDa. Stationary-phase cells gave identical results.
Renaturing SDS-PAGE with M. luteus cells as a cell wall substrate gave insight into the role of peptidoglycan hydrolases in the formation of the KM'92 type I rough morphotype. The gels identified multiple bands with peptidoglycan hydrolase activity. The smooth isolates gave identical gel profiles, with the peptidoglycan hydrolases predominantly secreted or located in the cell wall fraction. In comparison, the gel profiles of the rough isolates indicated various impairments in the secretion, localization, activity, and/or production of peptidoglycan hydrolases (Fig. 5). The impaired cellular localization was most pronounced in the exponential phase of growth, with obvious defects in the localization of peptidoglycan hydrolases to the cell wall fraction. A single faint 50-kDa band of activity was visualized in the cell walls of the rough isolates, whereas seven bands, ranging from 20 and 66 kDa, were observed in the cell walls of the wild-type and revertant smooth isolates. In addition to the expected CwhA secretion defect, a prominent secreted band at 66 kDa in the wild-type and RS isolates was missing in the supernatant fraction of exponential-phase SR and LR cells. The secretion defect correlated with increased activity of a 66-kDa band in the membrane fraction of the rough isolates. These data show that transport across the cell membrane was rate limiting on the correct localization of at least two peptidoglycan hydrolases (CwhA and the 66-kDa band). The similarity of these data to the secA2 deletion mutant analysis of Lenz and Portnoy (28) led us to test for mutations in the secA2 genes of the KM'92 morphotypes.
FIG. 5.
Renaturing SDS-PAGE analysis of the peptidoglycan hydrolase profiles from cellular fractions of KM'92 isolates. Proteins from exponential-phase (OD600 = 0.7) and early-stationary-phase cells of wild-type (Wt), SR, LR, and RS isolates were separated into four fractions: extracellular supernatant, cell wall, cell membrane, and cytoplasm, as described in Materials and Methods. Equivalent loadings (50 μg protein) from the supernatant, cell membrane, and cytoplasm and 20 μl, corresponding to one-fifth of the total cell wall fraction, were separated by SDS-10% PAGE in gels that contained 1.5% (wt/vol) Micrococcus luteus cells. Gels were renatured in buffer. The zones of clearing (black bands) which developed indicate the action of peptidoglycan hydrolases.
Sequencing, complementation, and hexahistidine tagging of secA2 from KM'92 isolates.
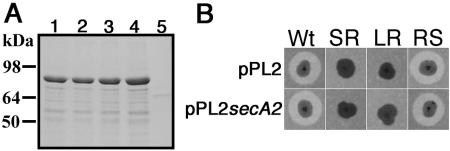
The promoter and entire coding region of the secA2 genes from the four isolates of KM'92 were sequenced. All four sequences were identical, with each predicted to encode full-length SecA2. This was confirmed by hexahistidine tagging the N terminus of each secA2 gene and expressing the recombinant protein in E. coli (Fig. 6A). In each case, a full-length His-tagged protein with an apparent molecular mass of about 85 kDa was produced. Furthermore, rough isolates that had integrated the single-copy, site-specific vector pPL2secA2 (26) failed to restore the secretion defect, and the transformants retained their rough morphotype (Fig. 6B). As no antibody is presently available for SecA2, its expression in the morphotypes was compared with a C-terminal FLAG tag epitope on the integrated second copies of secA2 in the pPL2secA2-transformed isolates. A band corresponding to SecA2-FLAG was present in each of the strains containing pPL2secA2 but not in the untransformed or plasmid-only controls (Fig. 7). Expression of SecA2-FLAG was significantly less in the wild-type FLAG background than in the two rough and the revertant smooth secA2-FLAG backgrounds.
FIG. 6.
N-terminal hexahistidine tagging of SecA2 and complementation of KM'92 isolates. (A) The secA2 gene amplified from genomic DNA of each KM'92 morphotype was cloned into the N-terminal hexahistidine tagging vector pQE30 and induced with IPTG as described in Materials and Methods. The majority of the insoluble fraction was analyzed by SDS-PAGE and Coomassie blue R250 staining. The migration of the major overexpressed band indicated a molecular mass of ≈85 kDa, consistent with SecA2. Lanes: 1, wild type; 2, SR; 3, LR; 4, RS; 5, pQE30 only. (B) KM'92 isolates were electroporated with the L. monocytogenes integrative vector pPL2 or pPL2secA2. Transformants containing the integrated plasmid were confirmed by PCR, patched onto TSAYE containing 0.2% M. luteus cells, and incubated at 37°C for 48 h. Zones of clearing surrounding the colonial growth indicate the action of secreted peptidoglycan hydrolases.
FIG. 7.
Anti-FLAG immunoblot of protoplasts from wild-type and pPL2secA2-FLAG- and pPL2-complemented isolates of KM'92. Equivalent loadings of 50 μg of protein from the combined cytoplasmic and cell membrane fractions were separated by SDS-PAGE. The separated proteins were electrotransferred to nitrocellulose and probed with the anti-Flag tag M2 monoclonal antibody as described in Materials and Methods. No SecA2-FLAG was detected in the cell wall fraction of pPL2secA2-FLAG-complemented strains.
DISCUSSION
Phenotypic variation has a pivotal role in the adaptation of numerous bacterial species to environmental niches (12, 19, 32, 34). Until now, the inability to detect conversion between smooth and rough morphotypes has precluded systematic analysis of the effects of phenotypic variation on the rough morphotype of L. monocytogenes. In this study, surface-adherent growth has been identified as a factor that stimulates the proliferation of multiple rough morphotypes. The analysis of biofilms formed by pure cultures of rough morphotypic variants showed that these isolates were better able to foul abiotic surfaces due to superior surface colonization abilities. Furthermore, while surface-adherent growth stimulated a smooth-to-rough transition, growth into the stationary phase of the best-adapted biofilm-forming rough colony isolate LR produced the revertant smooth morphotype RS.
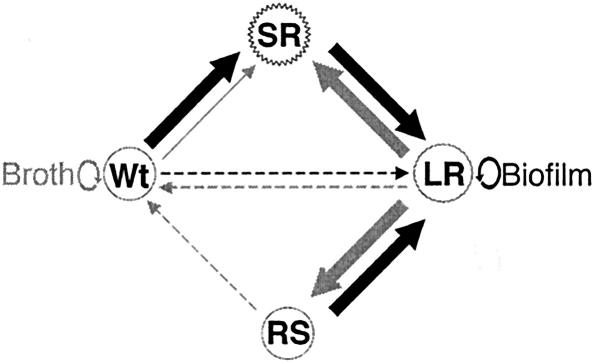
Proteomic analysis of the smooth and rough morphotypes showed impaired localization of multiple peptidoglycan hydrolases (including CwhA) in the rough isolates, while the rough-to-smooth (LR to RS) conversion restored wild-type levels of protein secretion. Unlike the type I rough isolates described by Lenz and Portnoy (28), no mutations were identified in the secA2 gene, which is involved in extracellular protein localization. Instead, measurement of the expression of FLAG-tagged SecA2 indirectly demonstrated increased expression of SecA2 in the rough isolates SR and LR compared with the parental wild type. While the wild-type and RS morphotypes appear indistinguishable, they showed two measurable differences. RS recovered a significantly lower level of coordinated motility and retained much higher levels of SecA2 expression than the wild type. Thus, RS and the wild type are unlikely to be genetically identical, and the pathway required to revert to the wild-type phenotype is not yet known. The cycling between morphotypes is schematically illustrated in Fig. 8, showing the relative stability of the wild-type and LR morphotypes in broth and biofilm, respectively.
FIG. 8.
Schematic of morphotypic cycling of KM'92 during biofilm and broth growth. Arrows indicate the direction and the condition that mediates morphotypic change (biofilm formation, black arrows; broth culture, grey arrows). The thickness of the arrow denotes the relative frequency of morphotypic conversion (thin, rarely observed; thick, commonly observed). The conditions for the direct conversion between wild type and LR or RS to wild type (dotted arrows) have yet to be identified. Circular arrows show the relative stability of the wild type in broth and of LR in biofilms.
This report provides the first experimental evidence of the enhanced biofilm-forming abilities of naturally occurring clonal L. monocytogenes isolates in a continuous-flow bioreactor. Comparisons of biofilm formation by L. monocytogenes strains have produced conflicting views on the relationships between division or serotype (46) and biofilm formation (2, 11, 24, 29, 36). Dickson and Siragusa (10) demonstrated the increased hydrophobicity of a rough isolate in comparison with nonisogenetic smooth isolates. Thus, changes in cell wall protein composition may affect attachment by modifying the net negative charge present on the cell surface. Vatanyoopaisarn et al. (45) showed the importance of flagella in the initial stages of surface attachment by L. monocytogenes. Peptidoglycan hydrolase production in the closely related lactococci was found to have a critical role in enhancement of surface colonization by exposing proteins, polysaccharides, and or lipoteichoic acids (35). These data appear at odds with the enhanced biofilm formation ability of the KM'92 rough isolates with impaired motility and mislocalized peptidoglycan hydrolase activity. L. monocytogenes biofilm formation has usually been analyzed with microplate assay methods, which allow high-throughput analysis of batch cultures. Biofilm formation is, however, a complex process involving multifactorial interactions; properties that are important in batch culture biofilm formation may be less important in the bioreactor, and vice versa. For example, batch culture biofilm systems are unlikely to identify multigenerational traits because nutrient limitation occurs before positive selection pressures amplify the most successful surface-adherent cells.
Shear forces have also been shown to affect the extent and strength of bacterial attachment (18). These forces occur in the bioreactor under continuous culture conditions but will be less prominent in static microtiter plate cultures. Therefore, attached cells capable of forming chains can be expected to have greater advantage under such bioreactor culture conditions. Thus, biofilm formation in static culture and the bioreactor continuous-culture system are unlikely to be comparable processes. Furthermore, biofilm formation in the bioreactor is more likely to mimic the conditions found in food processing facilities and in some important biological contexts such as the gall bladder.
In gram-positive bacteria, more than one SecA secretion system has evolved. Core components of the essential SecA pathway are found in single copy (except for SecA2) in the genomes of L. monocytogenes strains EGD-e and 4b (tblastn searches of the Pasteur and TIGR web sites with SecY, -E, -G, -D, and -F of EGD-e), whereas several gram-positive bacteria containing a second secA gene also have a second secY (tblastn searches found two copies of secY in Bacillus anthracis, Staphylococcus epidermidis, and Staphylococcus aureus) (33). In Streptococcus gordonii, a second secY is required for secretion of the glycosylated platelet binding protein GspB (1). The individual components and the mechanistic interactions required for secretion via the SecA2 pathway have yet to be defined.
Protein secretion deficiencies have been documented for secA2 deletion mutants from a variety of pathogens. The shared feature of these secreted protein subsets is their role in the virulence of these organisms (1, 4, 28). Analysis of the secretome from an L. monocytogenes secA2 deletion mutant revealed impaired localization of 19 of 49 surface-extractable and 7 of 25 secreted proteins (27), including a second peptidoglycan hydrolase (NamA) with a molecular mass of 66 kDa. The similarity of our data for the rough isolates with the renaturing SDS-PAGE profiles of Lenz et al. (27) obtained with a ΔsecA2 strain suggested that the SecA2 secretion pathway is involved in the morphotypic conversion of biofilm-conditioned rough isolates.
The incorrect localization of multiple peptidoglycan hydrolases and the elevated levels of SecA2 protein expression suggest that secA2 mRNA and/or SecA2 protein is misregulated in rough isolates. Furthermore, mutations in other genes can suppress the rough morphotype without altering SecA2 expression. Although the exact role of the SecA2 secretion pathway in the interconversion between the rough and smooth morphotypes has yet to be determined, compensatory changes in functional interactions between SecA2 and other components of the Sec secretory pathway are prime candidates for investigation. In-depth analysis with transposon mutagenesis, transcriptome profiling, and mapping of SecA2's protein partners should help unravel the role of the SecA2 secretion pathway in the environment-dependent interconversion between the parasitic smooth and biofilm-adapted saprophytic rough morphologies displayed by this opportunistic pathogen.
The rough morphotype may have a biological role in the colonization of gastrointestinal tissues such as the gall bladder. L. monocytogenes has been shown to colonize the murine gall bladder through a delayed mechanism, with the majority of cells contained within this organ observed microscopically as forming chains (20). Gall bladder colonization by the opportunistic intracellular pathogen Salmonella enterica has been associated with biofilm formation (42). We also suggest that rough isolates might specifically colonize the gall bladder by entering a low-immunogenicity, quiescent state. CwhA is a highly immunogenic protein, with processed CwhA peptides able to stimulate a protective immune response (13, 21). Decreased secretion of CwhA and other peptidoglycan hydrolases by rough isolates may help these morphotypic variants evade detection during gastrointestinal passage and facilitate colonization of the gall bladder via enhanced biofilm formation. With the availability of an isogenic set of morphotypes, these ideas about the consequences of phenotypic variation can now be tested in animal models.
This research has important implications for industry. Contamination of minimally processed foods by L. monocytogenes can be a significant health problem and is of major economic importance (30, 37, 44). Because rough isolates have enhanced biofilm-forming abilities, the subsequent risk of product contamination is increased. This risk could be exacerbated by the possibility of gross, sporadic product contamination with streamers of rough L. monocytogenes isolates. The potential for reversion from the rough “avirulent” morphotype to the smooth “virulent” form under certain conditions compounds the contamination problem and warrants identification of the factors that determine the reversion. A similar reversion was reported to occur with rough cells passaged through the gastrointestinal tract of mice (48). If a parallel can be drawn with the ingestion of grossly contaminated ready-to-eat food products, the adaptive advantage conferred by selective interconversion between rough and smooth morphotypes within the multiple environments during L. monocytogenes infection could be an important risk factor for listeriosis.
Acknowledgments
This research was supported by the New Zealand Foundation for Research, Science and Technology under contract CO-2406.
We thank the following people: W. Goebel for the kind gift of the CwhA monoclonal antibody, L. Lenz for very generously providing the pPL2secA2 vector, and W. Mitchell for supplying L. monocytogenes strain EGD-e. We also thank Annalee O'Rourke, C. Gahan, and C. Hill for critical review of the manuscript.
REFERENCES
- 1.Bensing, B. A., and P. M. Sullam. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081-1094. [DOI] [PubMed] [Google Scholar]
- 2.Borucki, M. K., J. D. Peppin, D. White, F. Loge, and D. R. Call. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 69:7336-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourion, F., O. Cerf, and T. Meylheuc. 1996. Disinfection efficacy against pure-culture and mixed-population biofilms of Listeria innocua and Pseudomonas aeruginosa on stainless steel, Teflon and rubber. Sci. Aliment. 16:151-166. [Google Scholar]
- 4.Braunstein, M., B. J. Espinosa, J. Chan, J. T. Belisle, and W. R. Jacobs, Jr. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48:453-464. [DOI] [PubMed] [Google Scholar]
- 5.Bremer, P. J., I. Monk, and C. M. Osborne. 2001. Survival of Listeria monocytogenes attached to stainless steel surfaces in the presence or absence of Flavobacterium spp. J. Food Prot. 64:1369-1376. [DOI] [PubMed] [Google Scholar]
- 6.Bremer, P. J., I. Monk, and R. Butler. 2002. Inactivation of Listeria monocytogenes/Flavobacterium spp. biofilms using chlorine: impact of substrate, pH, time and concentration. Lett. Appl. Microbiol. 35:321-325. [DOI] [PubMed] [Google Scholar]
- 7.Brett, M. S., P. Short, and J. McLauchlin. 1998. A small outbreak of listeriosis associated with smoked mussels. Int. J. Food Microbiol. 43:223-229. [DOI] [PubMed] [Google Scholar]
- 8.Bubert, A., P. Schubert, S. Kohler, R. Frank, and W. Goebel. 1994. Synthetic peptides derived from the Listeria monocytogenes p60 protein as antigens for the generation of polyclonal antibodies specific for secreted cell-free L. monocytogenes p60 proteins. Appl. Environ. Microbiol. 60:3120-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson, J. S., and G. R. Siragusa. 1994. Cell surface charge and initial attachment characteristics of rough strains of Listeria monocytogenes. Lett. Appl. Microbiol. 19:192-196. [Google Scholar]
- 11.Djordjevic, D., M. Wiedmann, and L. A. McLandsborough. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68:2950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 13.Geginat, G., M. Lalic, M. Kretschmar, W. Goebel, H. Hof, D. Palm, and A. Bubert. 1998. Th1 cells specific for a secreted protein of Listeria monocytogenes are protective in vivo. J. Immunol. 160:6046-6056. [PubMed] [Google Scholar]
- 14.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 15.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 16.Gray, M. L., H. J. Stafseth, and F. Thorp, Jr. 1957. Colonial dissociation of Listeria monocytogenes. Zentralbl. Bakteriol. Parasitenkd. Abt. I Orig. 169:378-391. [PubMed] [Google Scholar]
- 17.Gutekunst, K. A., L. Pine, E. White, S. Kathariou, and G. M. Carlone. 1992. A filamentous-like mutant of Listeria monocytogenes with reduced expression of a 60-kilodalton extracellular protein invades and grows in 3T6 and Caco-2 cells. Can. J. Microbiol. 38:843-851. [DOI] [PubMed] [Google Scholar]
- 18.Hall-Stoodley, L., and P. Stoodley. 2002. Developmental regulation of microbial biofilms. Curr. Opin. Biotechnol. 13:228-233. [DOI] [PubMed] [Google Scholar]
- 19.Han, B., A. Pain, and K. Johnstone. 1997. Spontaneous duplication of a 661 bp element within a two-component sensor regulator gene causes phenotypic switching in colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol. Microbiol. 25:211-218. [DOI] [PubMed] [Google Scholar]
- 20.Hardy. J., K. P. Francis, M. DeBoer, P. Chu, K. Gibbs, and C. H. Contag. 2004. Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303:851-853. [DOI] [PubMed] [Google Scholar]
- 21.Harty, J., and E. G. Pamer. 1995. CD8 T lymphocytes specific for secreted p60 antigen protect against Listeria monocytogenes. J. Immunol. 154:4642-4650. [PubMed] [Google Scholar]
- 22.Jeong, D., and J. F. Frank. 1994. Growth of Listeria monocytogenes at 21°C in biofilms with micro-organisms isolated from meat and dairy processing environments. Lebensmittel-Wiss. Technol. 27:415-424. [DOI] [PubMed] [Google Scholar]
- 23.Jonquieres, R., H. Bierne, F. Fiedler, P. Gounon, and P. Cossart. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34:902-914. [DOI] [PubMed] [Google Scholar]
- 24.Kalmokoff, M. L., J. W. Austin, X. D. Wan, G. Sanders, S. Banerjee, and J. M. Farber. 2001. Absorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J. Appl. Microbiol. 91:725-734. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn, M., and W. Goebel. 1989. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect. Immun. 57:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenz, L. L., S. Mohammadi, A. Geissler, and D. A. Portnoy. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. USA 100:12432-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenz, L. L., and D. A. Portnoy. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45:1043-1056. [DOI] [PubMed] [Google Scholar]
- 29.Lunden, J. M., M. K. Miettinen, T. J. Autio, and H. J. Korkeala. 2000. Persistent Listeria monocytogenes strains show enhanced adherence to food contact surfaces after short contact times. J. Food Prot. 63:1204-1207. [DOI] [PubMed] [Google Scholar]
- 30.Mafu, A. A., D. Roy, J. Goulet, and L. Savoie. 1991. Characterization of physicochemical forces involved in adhesion of Listeria monocytogenes to surfaces. Appl. Environ. Microbiol. 57:1969-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 175:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massey, R. C., A. Buckling, and S. J. Peacock. 2001. Phenotypic switching of antibiotic resistance circumvents permanent costs in Staphylococcus aureus. Curr. Biol. 11:1810-1814. [DOI] [PubMed] [Google Scholar]
- 33.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 34.Medina, E., M. Rohde, and G. S. Chhatwal. 2003. Intracellular survival of Streptococcus pyogenes in polymorphonuclear cells results in increased bacterial virulence. Infect. Immun. 71:5376-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercier, C., C. Durrieu, R. Briandet, E. Domakova, J. Tremblay, G. Buist, and S. Kulakauskas. 2002. Positive role of peptidoglycan breaks in lactococcal biofilm formation. Mol. Microbiol. 46:235-243. [DOI] [PubMed] [Google Scholar]
- 36.Norwood, D. E., and A. Gilmore. 1999. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 86:576-582. [DOI] [PubMed] [Google Scholar]
- 37.Olier, M., F. Pierre, S. Rousseaux, J. P. Lemaitre, A. Rousset, P. Piveteau, and J. Guzzo. 2003. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene. 94:129-132. [DOI] [PubMed] [Google Scholar]
- 39.Pilgrim, S., A. Klob-Mäurer, I. Gentschev, W. Goebel, and M. Kuhn. 2003. Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect. Immun. 71:3473-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popowska, M., M. Kloszewska, S. Gorecka, and Z. Markiewicz. 1999. Autolysis of Listeria monocytogenes. Acta Microbiol. Pol. 48:141-152. [PubMed] [Google Scholar]
- 41.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 42.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowan, N. J., A. A. Candlish, A. Bubert, J. G. Anderson, K. Kramer, and J. McLauchlin. 2000. Virulent rough filaments of Listeria monocytogenes from clinical and food samples secreting wild-type levels of cell-free p60 protein. J. Clin. Microbiol. 38:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlech, W. F., 3rd. 2000. Epidemiology and clinical manifestations of Listeria monocytogenes infection, p. 473-479. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. A. Rood. (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 45.Vatanyoopaisarn, S., A. Nazli, C. E. Dodd, C. E. Rees, and W. M. Waites. 2000. Effect of flagella on initial attachment of Listeria monocytogenes to stainless steel. Appl. Environ. Microbiol. 66:860-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widemann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wuenscher, M. D., S. Kohler, A. Bubert, U. Gerike, and W. Goebel. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J. Bacteriol. 175:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zachar, Z., and D. C. Savage. 1979. Microbial interference and colonization of the murine gastrointestinal tract by Listeria monocytogenes. Infect. Immun. 23:168-174. [DOI] [PMC free article] [PubMed] [Google Scholar]