Abstract
A preliminary investigation of the genetic biodiversity of Mycobacterium tuberculosis complex strains in Cameroon, a country with a high prevalence of tuberculosis, described a group of closely related M. tuberculosis strains (the Cameroon family) currently responsible for more than 40% of smear-positive pulmonary tuberculosis cases. Here, we used various molecular methods to study the genetic characteristics of this family of strains. Cameroon family M. tuberculosis strains (i) are part of the major genetic group 2 and lack the TbD1 region like other families of epidemic strains, (ii) lack spacers 23, 24, and 25 in their direct repeat (DR) region, (iii) have an identical number of repeats in 8 of 12 variable-number tandem repeats of mycobacterial interspersed repetitive unit (MIRU-VNTR) loci, (iv) have similar IS6110-restriction fragment length polymorphism (RFLP) multiband patterns (10 to 15 copies) with seven common IS6110 bands, (v) do not have an IS6110 element in their DR locus, and (vi) have four IS6110 elements in open reading frames (adenylate cyclase, phospholipase C, moeY, and ATP binding genes). Analysis by spoligotyping, MIRU-VNTR, and IS6110-RFLP typing methods revealed differences not observed in previous studies; polymorphism as assessed by MIRU-VNTR typing was lower than suggested by spoligotyping, and in rare cases, strains with identical IS6110-RFLP patterns had spoligotypes differing by as much as 15 spacers. Our findings confirm the recent expansion of this family in Cameroon and indicate that the interpretation of molecular typing results has to be adapted to the characteristics of the strain population within each setting. The knowledge of this particular genotype, with its large involvement in tuberculosis in Cameroon, allows greater refinement of tuberculosis transmission studies by interpreting data in the context of this geographic area.
Molecular epidemiology methods revolutionized the fields of research, prevention, and control of tuberculosis (TB), allowing the differentiation between strains, assessment of the overall diversity of Mycobacterium tuberculosis complex strains including differences by region and population, and measurement of the prevalence of endemic strains (28). However, few molecular epidemiological studies have been conducted in countries with a high incidence of TB. The available data suggest that families of closely related strains are common in these areas (12). The “Beijing family” is one of the most well-known families, highly prevalent in East Asia and widespread around the world (11).
Molecular analysis based on several variable genomic regions is required for a good definition of strains belonging to different families. Restriction fragment length polymorphism (RFLP) analysis based on the insertion sequence IS6110 results in a unique genotype since both the number of copies of this genetic element and its positions in the genome are variable (25, 26). Precise IS6110 insertion site mapping provides additional information on the fitness of the strain (1) given that IS6110 insertion can modify the expression of the gene involved. Another genetic element useful for characterizing tubercle bacilli is the direct repeat (DR) locus (13), a polymorphic insertion preferential locus (ipl) for IS6110. DR polymorphism can be analyzed by spoligotyping, a method involving PCR-reverse hybridization (14). The DR locus is likely to evolve more slowly than IS6110, making spoligotyping less adequate than IS6110-RFLP for discriminating strains but more convenient for investigating the biogeographic distribution of families of M. tuberculosis complex strains (32). Variable-number tandem repeats, named mycobacterial interspersed repetitive units (MIRU-VNTR), are another type of variable element in the M. tuberculosis complex genome showing extensive polymorphism (17) with a discrimination power close to that of IS6110-RFLP (6). Because of their stability, they can be used for a clear definition of families of tubercle bacilli as well.
Strains of the M. tuberculosis complex exhibit very little genome sequence diversity except in repeat sequences. Consequently, insignificant and rare genome alterations inherited and maintained through long-term evolution can be of phylogenetic value. On the basis of polymorphic nucleotides in katG codon 463 and gyrA codon 95, three genetic groups of M. tuberculosis have been identified (23). Group 1 isolates are evolutionarily the oldest, and after a loss of the region of difference TbD1, the modern lineage further evolved into two branches, group 2 and group 3 (2). Major epidemic strains including the Beijing family belong to the lineage lacking the TbD1 region.
Preliminary investigations of the genetic diversity of M. tuberculosis complex strains from the West region of Cameroon revealed that more than 40% of isolates belong to a closely related group of M. tuberculosis strains. This group was designated the “Cameroon family” (20). All strains of this family lacked spacers 23, 24, and 25 in their DR region and had closely related IS6110 ligation-mediated PCR (LM-PCR) patterns. Here, we report the use of typing methods involving various genetic markers for strains of the “Cameroon family” to more sharply define this group of M. tuberculosis strains.
MATERIALS AND METHODS
Mycobacterial strains and genomic extraction of DNA.
The analysis was based on 45 M. tuberculosis strains belonging to Cameroon family. These strains shared a spoligotype lacking spacers 23, 24, and 25 and had closely related IS6110 LM-PCR patterns (20). The mycobacteria were killed by heating at 90°C for 20 min, and the genomic DNA from the isolates was prepared by a standard cetyltrimethylammonium bromide-NaCl method (33).
IS6110-RFLP typing.
Southern blotting and hybridization with labeled IS6110 DNA were performed as previously described by using an internationally agreed-upon protocol (26). Bionumerics software version 2.5 (Applied Maths, Kortrijk, Belgium) was used to process autoradiographs of Southern blots. IS6110-RFLP patterns were analyzed for similarity with the Dice coefficient with an error of tolerance of 1%, and a dendrogram was constructed by the unweighted pair group method using arithmetic averages (UPGMA).
MIRU-VNTR typing.
MIRU loci 2, 4, 10, 16, 20, 23, 24, 26, 27, 31, 39, and 40 were individually amplified and analyzed as previously described (18). Results from each of the 12 loci were combined to form a 12-digit allele profile. MIRU pattern similarity was assessed by Pearson correlation, and a dendrogram was constructed by UPGMA.
DR-RFLP typing.
Southern blot hybridization with DR as a probe used the membranes blotted with PvuII-digested DNA that had previously been probed with the IS6110 fragment (13). The autoradiographs obtained were superimposed on those obtained with IS6110-RFLP to identify cohybridizing bands and to detect any IS6110 insertion into the DR region.
Left-right (LR) spoligotyping.
Primers Ris1 and Ris2 (10), corresponding to the 3′ and 5′ termini of IS6110, respectively, and the 5′-biotinylated Dra reverse primer for standard spoligotyping were used. Eighteen strains were analyzed, representative of all the different spoligotypes of strains previously included in the Cameroon family (20). The PCR products amplified with Ris1-Dra and Ris2-Dra primer pairs were subjected to the standard spoligotyping procedure (14). This procedure allowed amplification and detection of the particular DR spacers to the left and right of the IS6110 copy inserted in the DR region, depending on its orientation towards and within the DR locus (19).
Mapping of IS6110 insertion sites.
LM-PCR was performed as described previously (21) to obtain the left side of each IS6110 copy with a variable stretch of its flanking sequence. Eleven strains, representative of the 11 different IS6110 LM-PCR patterns of the Cameroon family strains previously described (20), were analyzed. The PCR product was separated on 2% (wt/vol) low-melting-temperature agarose gel (Gibco-BRL Life Technologies). A 100-bp ladder served as the external molecular size marker. Sixteen different bands were excised from the gel and then purified on columns (QIAquick gel extraction kit; QIAGEN S.A., Courtaboeuf, France). The primer IS2 (21), corresponding to the left side of IS6110, was used for direct sequencing of the purified PCR products. The flanking sequences were analyzed by using Tuberculist (http://genolist.pasteur.fr/TubercuList/), a genome browser for M. tuberculosis H37Rv databases, with gapped BLAST analysis to determine the exact insertion site of each IS6110 element.
katG463, gyrA95, and TbD1 analysis.
The polymorphisms at katG codon 463 and gyrA codon 95 were studied by sequencing (23). The presence or absence of the TbD1 region was analyzed by PCR as previously described (2).
RESULTS
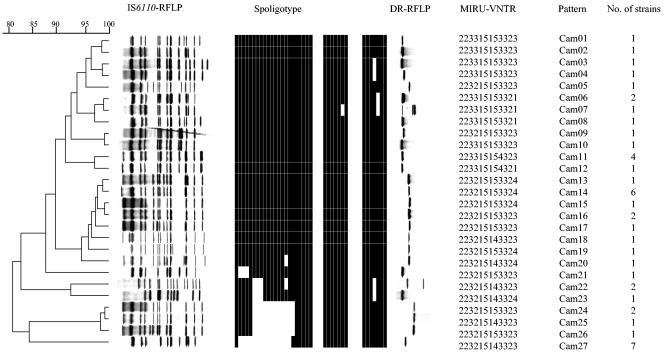
Twenty-seven different combined patterns were found among the 45 M. tuberculosis strains with the various typing methods (IS6110-RFLP, DR-RFLP, MIRU-VNTR, and spoligotyping) (Fig. 1).
FIG. 1.
Patterns of the 45 M. tuberculosis strains obtained by various typing methods. The genetic distances between strains depicted in the dendrogram are based upon combined IS6110-RFLP, MIRU-VNTR, and spoligotyping analysis using the same weight for each method. The dendrogram was constructed by UPGMA. Spoligotypes were published previously (20).
IS6110-RFLP analysis.
The IS6110-RFLP analysis revealed 20 different patterns with about 80% similarity (Fig. 1). Twelve isolates had unique patterns, and 33 isolates belonged to eight groups of identical patterns, each containing two to seven strains. The number of IS6110 DNA-containing PvuII fragments was high in all strains, indicating that these strains contain 10 to 15 copies of IS6110. Forty-two strains, that is, all but three (patterns Cam22 and Cam23), shared seven insertion element-containing PvuII fragments, of lengths 7.2, 5, 2.4, 2.2, 2.1, 1.5, and 1.3 kb.
MIRU-VNTR analysis.
The MIRU-VNTR loci were studied (Fig. 1 shows the 12-digit designations). Eight MIRU loci did not display variation in their copy numbers (MIRU loci 2, 4, 10, 20, 23, 24, 31, and 39). MIRU loci 16, 26, and 27 contained two alleles each, which differed by only one repeat unit. MIRU locus 40 was also polymorphic and contained three alleles with 1, 3, and 4 repeat units, respectively. At the polymorphic loci, some alleles were more frequent than others. Thus, most of the strains (40 out of 45) contained alleles with three repeat units in MIRU locus 27, and only five strains (patterns Cam11 and Cam12) had one additional copy. In total, eight different MIRU patterns were observed in this set of isolates. One isolate had a unique pattern, and the 44 other isolates fell into seven groups of 2 to 11 strains. MIRU pattern analysis indicates 95% similarity for all the strains, except for one group of five strains (patterns Cam6, Cam7, Cam8, and Cam12) with 85% similarity. These five strains contained only one repeat unit in their MIRU locus 40, whereas the other 40 strains contained three or four units.
DR locus analysis by RFLP and LR spoligotyping.
Most of strains (42 out of 45) had a single DR-RFLP band, of which there were four variants of different sizes (Fig. 1). The presence of a single DR-RFLP band indicated that IS6110 was not inserted into the DR region of these 42 strains. Two other strains (pattern Cam22) had a two-band DR-RFLP pattern. The superimposition of DR- and IS6110-RFLP films showed that one of these bands at about 5.5 kb hybridized with both the IS6110 probe and the DR probe; thus, there was an IS6110 copy in the DR. One other strain (pattern Cam7) had a three-band DR-RFLP pattern, and two of these bands, at about 7.2 and 4.4 kb, hybridized with both the IS6110 probe and the DR probe, indicating that there were two IS6110 copies in the DR.
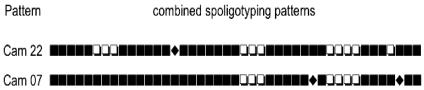
We tested whether the insertion of IS6110 copies into the DR region of strains with patterns Cam22 and Cam7 had resulted in hybridization signals in spoligotypes being lost. We compared the three spoligotypes obtained for each pattern with the primer pairs Dra-Drb, Ris1-Dra, and Ris2-Drb (Fig. 2). In the strain with pattern Cam22, Ris2-Dra spoligotyping evidenced spacer 15, a spacer that was absent from the Dra-Drb spoligotype. Thus, spacer 15 was the DR insertion site of one copy of IS6110 as detected by superimposition of IS6110- and DR-RFLP films. In the strain with pattern Cam7, Ris1-Dra and Ris2-Dra spoligotyping detected spacers 31 and 41, neither of which were detected by Dra-Drb spoligotyping. These two spacers were the DR insertion sites of the two IS6110 copies detected by superimposition of IS6110- and DR-RFLP banding patterns. No missed signals were detected for the strains with the other patterns or for spacers 23, 24, and 25, which are deletions characterizing this family of strains.
FIG. 2.
Schematic representation of combined analysis of the three kinds of spoligotyping patterns (Dra-Drb, Ris1-Dra, and Ris2-Drb). Open squares indicate the absence of a particular spacer, solid squares indicate the presence of a particular spacer, and diamond shapes indicate particular spacers not detected by spoligotyping but shown to be present by LR spoligotyping.
Combined analysis with the various molecular markers.
The classifications obtained by the three different approaches (IS6110, DR region, and MIRU-VNTR) were similar. Overall, the similarity among the strains was more than 70%. The greatest pattern diversity was with IS6110-RFLP analysis, and the lowest diversity was with MIRU typing (Table 1). Groups of strains shared common patterns for two markers but differed according to the third marker, the third marker being IS6110, MIRU, or DR. Thus, five strains with patterns Cam11 and Cam12 sharing the same spoligotype and IS6110-RFLP patterns had MIRU 40 patterns differing by two repeats. Interestingly, these five strains were the only strains with an additional repeat in MIRU locus 27. Strains with patterns Cam16 and Cam24 shared both IS6110-RFLP and MIRU patterns but had spoligotypes differing at 12 contiguous spacers. Seven strains (pattern Cam27) lacking a block of 15 spacers (spacers 2 to 16) presented identical IS6110-RFLP and MIRU patterns. These observations suggested that these deletions of numerous adjacent direct variable repeat (DVR) sequences in the DR region of this family of M. tuberculosis strains were recent single events rather than a series of sequential deletion events.
TABLE 1.
Pattern diversity and similarity with the three typing methods for characterization of 45 Cameroon family M. tuberculosis strains
| Typing method | No. of patterns | No. of unique strains | No. of clusters | Pattern diversity (%) | Pattern similarity (%) |
|---|---|---|---|---|---|
| IS6110-RFLP | 20 | 12 | 8 | 44.5 | 80 |
| Spoligotyping | 10 | 4 | 6 | 22.2 | 90 |
| MIRU-VNTR | 8 | 2 | 6 | 17.8 | 95 |
| Combined analysis | 27 | 20 | 7 | 60 | 75 |
IS6110 insertion sites.
We determined the insertion sites of IS6110 corresponding to the six LM-PCR bands shared by all the strains of the Cameroon family (CSIP-1 [Cameroon strain insertion position 1] to CSIP-6) (Table 2). CSIP-7 to CSIP-15 corresponded to IS6110 copies present in only some of the strains. Six of the 15 sites were in intergenic regions (CSIP-1, CSIP-6, CSIP-7, CSIP-8, CSIP-12, and CSIP-15), and nine were in various open reading frames (ORFs), two of them being in unknown genes (CSIP-9 and CSIP-11). The insertion site CSIP-2 was located in the upstream region of the plcB (phospholipase C) gene, a probable virulence factor implicated in intracellular survival of the bacilli; it may act by altering cell signaling events or by direct cytotoxicity (5, 30). CSIP-3 was an IS6110 insertion site in the moeY gene (Rv1355c) involved in the biosynthesis of molybdoenzymes. The site of insertion CSIP-4 was located 57 bp upstream from Rv3377c, which is a possible cyclase similar to those involved in steroid biosynthesis, intermediary metabolism, and respiration (9). CSIP-5 was located in Rv3179, which contains an ATP-GTP binding site motif. CSIP-10, found only in strains with pattern Cam27, was located in cutI, a gene encoding a probable cutinase-like precursor that is a membrane component (5). Cutinases are typically extracellular enzymes produced by fungi and involved in the breakdown of cutin, the insoluble biopolyester that covers plant surfaces. In pathogenic fungi, cutinase allows penetration through the host plant cutin barrier during the initial stage of fungal infection (3, 27). CSIP-13, found in strains with pattern Cam20, was located in a gene of the PPE family, PPE55, implicated in virulence and in the host immune response (4). Several IS6110 insertion sites were in intergenic regions. CSIP-7 was in the ipl locus, which is part of the IS1547 insertion sequence (Rv3327) (7). It was found in strains with pattern Cam01 to Cam12, Cam15, Cam16, and Cam23. CSIP-12 was located in the 537-bp region between the genes dnaA and dnaN (16). The DnaA protein plays an important role in the initiation and regulation of chromosomal replication. The DnaN protein is a DNA polymerase III, a complex multichain enzyme responsible for most of the replicative synthesis in bacteria. CSIP-12 was found in the strain with pattern Cam18.
TABLE 2.
Insertion sites of IS6110 in Cameroon family M. tuberculosis strains
| IS6110 insertion sitea | Nucleotide no.b | Disrupted regionc | Gene name(s) or description | First descriptiond (source or reference) |
|---|---|---|---|---|
| CSIP-1 | 2265111 | Rv2017:Rv2018 | Unknown/unknown | This study |
| CSIP-2 | 2629008 | Rv2350 | plcB | 30 |
| CSIP-3 | 1522171 | Rv1355 | moeY | This study |
| CSIP-4 | 3790908 | Rv3377c | Cyclase | 22 |
| CSIP-5 | 3548751 | Rv3179 | ATP-GTP binding motif | This study |
| CSIP-6 | 2581467 | Rv2308:Rv2309c | Unknown/metV | This study |
| CSIP-7 | 3710391 | Rv3324c:Rv3327 | ipl site (IS1547) | 7 |
| CSIP-8 | 2364849 | Rv2104c:Rv2107 | Unknown/PE | 1 |
| CSIP-9 | 3493662 | Rv3128c | Unknown | 1 |
| CSIP-10 | 1989048 | Rv1758 | cutL cutinase | 22 |
| CSIP-11 | 1906551 | Rv1682 | Unknown | This study |
| CSIP-12 | 1880 | Rv0001:Rv0002 | dnaA/dnaN | 16 |
| CSIP-13 | 3746050 | Rv3347c | PPE55 | This study |
| CSIP-14 | 2784805 | Rv2476c | Transposase | This study |
| CSIP-15 | 3547517 | Rv3178:Rv3179 | Unknown/ATP-GTP binding motif | 1 |
Each IS6110 copy was named according to the band position in the LM-PCR pattern.
Insertion position of each IS6110 is according to the numbering of the M. tuberculosis H37Rv genome map (5).
The Rv number and the gene names are from the M. tuberculosis H37Rv genome map (5).
Reference of the first study to describe an IS6110 insertion in the same Rv.
katG463, gyrA95, and TbD1 analysis.
The katG and gyrA genes were sequenced, and all the strains were katG463 CGG (Arg) and gyrA95 ACC (Ser). Thus, this family of M. tuberculosis strains is part of genetic group 2, as defined by Sreevatsan et al. (23). The TbD1 region was absent from all the 45 strains.
DISCUSSION
Cameroon is a country in which the prevalence of TB is high. During the last three decades, the population structure of the tubercle bacilli causing TB has changed substantially in Cameroon. Mycobacteria africanum was previously widespread but is now less common, and a particular clade of M. tuberculosis has expanded and is currently responsible for more than 40% of smear-positive pulmonary TB cases (20). Other dominant M. tuberculosis families have been previously reported in other high-prevalence TB settings, i.e., the Beijing family in East Asia (29) and the F11 genotype M. tuberculosis in Western Cape, South Africa (31). An extensive molecular analysis is necessary to define the specific characters of these predominant families of strains. Such analysis would be an important step in the fight against TB since it can help us to understand why any particular clade is so successful.
In this study, we genetically characterized M. tuberculosis strains of the Cameroon family. Cameroon family M. tuberculosis strains, like Beijing and Haarlem family strains responsible for major epidemics, lack the TbD1 region (2). On the basis of their katG and gyrA gene sequences, they are part of the major genetic group 2, considered to be the principal cause of clustered TB cases (23).
We also analyzed polymorphism in Cameroon family strains by studying three repetitive DNA sequences, DR, MIRU-VNTR, and IS6110. These elements are the most polymorphic of the known markers in the M. tuberculosis complex. The strains lacked spacers 23, 24, and 25 in the DR region as assessed by standard spoligotyping. Using LR spoligotyping, we confirmed that the absence of hybridization was indeed due to the absence of the three spacers and not to the signals being abolished by IS6110 insertion. Other genetic features common to the Cameroon family are an IS6110-RFLP pattern sharing seven IS6110 bands and an identical number of repeats in 8 of 12 MIRU-VNTR loci. Because it is a straightforward technique, spoligotyping has been adopted for initial identification of strains of this genetically close family in Cameroon.
The chromosomal sites of integration of IS6110 can affect pathogenic behavior and drive genome evolution (1). Indeed, IS6110 frequently disrupts coding regions in clinical isolates (22). The mapping of IS6110 in two epidemic strains of M. tuberculosis (1) has suggested that IS6110 is involved in the regulation of gene expression; it is thus interesting to establish the location of each site of insertion in the chromosome. We mapped six IS6110 elements common to all the Cameroon family strains (CSIP-1 to CSIP-6). The following four of these IS6110 elements were in ORFs: (i) adenylate cyclase, involved in energy metabolism (although its role in M. tuberculosis is unclear, it is secreted from other bacterial pathogens such as Bacillus anthracis and invades a variety of eukaryotic cells where it affects both metabolism and immune response) (9); (ii) phospholipase C, a known virulence factor involved in macromolecule metabolism (30); (iii) moeY; and (iv) an ATP binding fragment. Thus, several of the ORFs disrupted in Cameroon family M. tuberculosis strains could be involved in virulence, but their roles, if any, are uncertain. Note that the pathogenesis of M. tuberculosis is not attributable to any single gene product, unlike other bacterial pathogens, where a single gene product can be critical.
We also mapped the other IS6110 elements present in some strains of the Cameroon family (CSIP-7 to CSIP-15). Two of these elements disrupted ORFs thought to be involved in virulence: (i) a PPE family gene and (ii) the gene for a cutinase-like cell wall component; cutinases can hydrolyze fatty acid esters, including mycolic acids, and they may be involved in in vivo survival and virulence (5). However, their role in M. tuberculosis remains unknown. One strain had IS6110 inserted into the 537-bp intergenic region in the origin of replication. Integration of IS6110 in the dnaA-dnaN region has previously been reported only in strains of the major genetic group 1 (16), considered to be the ancestor of groups 2 and 3. We demonstrate that strains other than those of the major genetic group 1 harbor IS6110 in the origin of replication. Overall, we report seven new insertion sites of IS6110 in the M. tuberculosis genome.
The DR locus is an IS6110 ipl in the M. tuberculosis complex genome, and most strains harbor at least one IS6110 copy in this region. The DR may have been the original point of entry for IS6110, and other copies arose from subsequent transposition events. The putative ancestral IS6110 insertion is in DVR 24, at position 15600 in the H37Rv genome sequence (5, 32). Homologous recombination between repeat sequences leading to DVR deletion seems to be the most likely mechanism for the loss of IS6110 from the DR locus (8, 32). The Cameroon family of strains lack spacers 23, 24, and 25 and have no IS6110 elements in the DR region; it is therefore possible that their common ancestor lost the ancestral IS6110 and the two DVRs adjacent to DVR 24 (DVRs 23 and 25); all the other spoligotypes in the family could have subsequently diverged from this common ancestor. The loss of spacers additional to spacers 23, 24, and 25 may have been a recent event in this family because we found strains with identical IS6110-RFLP patterns and spoligotypes differing only in these additional spacers. These observations are consistent with the Cameroon family of M. tuberculosis strains having expanded only recently in Cameroon (20).
Previous studies of the polymorphism of M. tuberculosis complex strains from various areas or from just one geographical setting led to the adoption of certain rules for the interpretation of fingerprints to assess the relationships among the isolates (Table 3), and these rules are as follows: (i) IS6110-RFLP is the fingerprint method with the highest discriminatory power for M. tuberculosis with five or more IS6110 copies, and secondary typing with another molecular marker cannot differentiate among isolates with identical IS6110-RFLP patterns (15), and (ii) MIRU-VNTR typing has a discriminatory power greater than that of spoligotyping (24). Our study of strains from one country using three molecular markers suggests that different rules for the interpretation of molecular typing results may be appropriate. Although IS6110-RFLP was indeed the most discriminatory method, in some cases spoligotyping or MIRU-VNTR typing differentiated strains with identical IS6110-RFLP patterns. This result may be due to the close genetic relationship of Cameroon family M. tuberculosis strains. Differences in spoligotypes involved blocks of as much as 15 spacers. As they were contiguous, these patterns are presumably the consequences of single genetic deletion events, and thus they have the same significance as concerns time as the loss of a single spacer. Differences in the MIRU-VNTR genotype between strains with identical IS6110-RFLP patterns were limited to a single locus, usually MIRU 40. MIRU 40 was also the most polymorphic of these loci in this study, consistent with previous studies (18). Because the Cameroon family strains probably expanded recently, our observations support the previously suggested faster molecular clock for MIRU 40 (18). MIRUs 16 and 26, two other polymorphic MIRUs in Cameroon family strains, were also previously found among the MIRUs with the highest allelic diversity (18). In contrast, MIRUs 10, 23, and 31, previously reported as highly diverse, seemed “frozen” in Cameroon family strains, whereas MIRU 27 was polymorphic in spite of its very restricted allele distribution in other populations of strains (18). We also found, in contrast with previous studies (6, 24), that MIRU-VNTR typing was less discriminant than spoligotyping. Our study evidences the complexity for interpreting fingerprint data for highly related strains. Interpretation of molecular typing needs to be appropriate to the specific characteristics of the strain population within each setting. To identify strains in epidemiological studies in Cameroon, neither IS6110-RFLP nor MIRU-VNTR typing can be used alone; several markers must be used.
TABLE 3.
Findings of combined analysis of molecular patterns for M. tuberculosis strains with more than five IS6110 copies
| 1st method and pattern | Pattern with the 2nd method
|
|||||
|---|---|---|---|---|---|---|
| Spoligotyping
|
MIRU-VNTR
|
IS6110-RFLP
|
||||
| Other studies | This study | Other studies | This study | Other studies | This study | |
| Spoligotyping | ||||||
| Identical | Identical or different | Identical or different | Identical or different | Identical or different | ||
| Different | Different | Identical or differenta | Different | Identical or differenta | ||
| MIRU-VNTR | ||||||
| Identical | Identical | Identical or differenta | Identical or different | Identical or different | ||
| Different | Identical or different | Identical or different | Different | Identical or differenta | ||
| IS6110-RFLP | ||||||
| Identical | Identical | Identical or differenta | Identical | Identical or differenta | ||
| Different | Identical or different | Identical or different | Identical or different | Identical or different | ||
Cameroon family M. tuberculosis strains could have some selective advantage over other M. tuberculosis genotypes present in Cameroon involving virulence, transmissibility, or the ability to interact with the host immune defense system. However, other possibilities to explain this predominance, like geographic confinement, cannot be excluded. In any case, a comprehensive understanding of this particular genotype with a large impact on TB in the country is valuable to allow greater refinement of TB transmission studies by interpreting data in the context of this geographic area.
Acknowledgments
S.N.N.-E. received a research fellowship from the Agence Universitaire de la Francophonie. This work benefited from the EU Concerted Action Project “New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis” (QLK2-CT-2000-630).
REFERENCES
- 1.Beggs, M. L., K. D. Eisenach, and M. D. Cave. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2923-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho, C. M., M. R. Aires-Barros, and J. M. Cabral. 1999. Cutinase: from molecular level to bioprocess development. Biotechnol. Bioeng. 66:17-34. [DOI] [PubMed] [Google Scholar]
- 4.Choudhary, R. K., S. Mukhopadhyay, P. Chakhaiyar, N. Sharma, K. J. Murthy, V. M. Katoch, S. E. Hasnain, P. W. Hermans, D. van Soolingen, E. M. Bik, P. E. de Haas, J. W. Dale, and J. D. van Embden. 2003. PPE antigen Rv2430c of Mycobacterium tuberculosis induces a strong B-cell response. Infect. Immun. 71:6338-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, Z., C. Doig, D. T. Kenna, N. Smittipat, P. Palittapongarnpim, B. Watt, and K. J. Forbes. 1999. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J. Bacteriol. 181:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang, Z., N. Morrison, B. Watt, C. Doig, and K. J. Forbes. 1998. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J. Bacteriol. 180:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman, C. R., M. Y. Stoeckle, W. D. Johnson, Jr., L. W. Riley, J. Magdalena, A. Vachee, P. Supply, C. Locht, P. W. Hermans, D. van Soolingen, E. M. Bik, P. E. de Haas, J. W. Dale, and J. D. van Embden. 1995. Double-repetitive-element PCR method for subtyping Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 33:1383-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermans, P. W., F. Messadi, H. Guebrexabher, D. van Soolingen, P. E. de Haas, H. Heersma, H. de Neeling, A. Ayoub, F. Portaels, D. Frommel, et al. 1995. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J. Infect. Dis. 171:1504-1513. [DOI] [PubMed] [Google Scholar]
- 13.Hermans, P. W., D. van Soolingen, E. M. Bik, P. E. de Haas, J. W. Dale, and J. D. van Embden. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurepina, N. E., S. Sreevatsan, B. B. Plikaytis, P. J. Bifani, N. D. Connell, R. J. Donnelly, D. van Soolingen, J. M. Musser, and B. N. Kreiswirth. 1998. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber. Lung Dis. 79:31-42. [DOI] [PubMed] [Google Scholar]
- 17.Magdalena, J., A. Vachee, P. Supply, C. Locht, P. W. Hermans, D. van Soolingen, E. M. Bik, P. E. de Haas, J. W. Dale, and J. D. van Embden. 1998. Identification of a new DNA region specific for members of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 36:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mokrousov, I., O. Narvskaya, E. Limeschenko, T. Otten, and B. Vyshnevskiy. 2002. Novel IS6110 insertion sites in the direct repeat locus of Mycobacterium tuberculosis clinical strains from the St. Petersburg area of Russia and evolutionary and epidemiological considerations. J. Clin. Microbiol. 40:1504-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niobe-Eyangoh, S. N., C. Kuaban, P. Sorlin, P. Cunin, J. Thonnon, C. Sola, N. Rastogi, V. Vincent, and M. C. Gutierrez. 2003. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 41:2547-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prod'hom, G., C. Guilhot, M. C. Gutierrez, A. Varnerot, B. Gicquel, and V. Vincent. 1997. Rapid discrimination of Mycobacterium tuberculosis complex strains by ligation-mediated PCR fingerprint analysis. J. Clin. Microbiol. 35:3331-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sampson, S., R. Warren, M. Richardson, G. van der Spuy, P. van Helden, N. Dunlap, and W. H. Benjamin, Jr. 2001. IS6110 insertions in Mycobacterium tuberculosis: predominantly into coding regions. J. Clin. Microbiol. 39:3423-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, C. Locht, I. A. van Gemeren, A. Beijersbergen, C. A. van den Hondel, and C. T. Verrips. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thierry, D., P. Matsiota-Bernard, E. Pitsouni, C. Costopoulos, and J. L. Guesdon. 1993. Use of the insertion element IS6110 for DNA fingerprinting of Mycobacterium tuberculosis isolates presenting various profiles of drug susceptibility. FEMS Immunol. Med. Microbiol. 6:287-297. [DOI] [PubMed] [Google Scholar]
- 26.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Gemeren, I. A., A. Beijersbergen, C. A. van den Hondel, and C. T. Verrips. 1998. Expression and secretion of defined cutinase variants by Aspergillus awamori. Appl. Environ. Microbiol. 64:2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Soolingen, D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1-26. [DOI] [PubMed] [Google Scholar]
- 29.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vera-Cabrera, L., M. A. Hernandez-Vera, O. Welsh, W. M. Johnson, and J. Castro-Garza. 2001. Phospholipase region of Mycobacterium tuberculosis is a preferential locus for IS6110 transposition. J. Clin. Microbiol. 39:3499-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Victor, T. C., P. E. de Haas, A. M. Jordaan, G. D. van der Spuy, M. Richardson, D. van Soolingen, P. D. van Helden, and R. Warren. 2004. Molecular characteristics and global spread of Mycobacterium tuberculosis with a Western Cape F11 genotype. J. Clin. Microbiol. 42:769-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van Der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, K. 1990. Preparation of genomic DNA from bacteria, vol. 1. Wiley Interscience, New York, N.Y.