Abstract
Evidence of spotted fever group (SFG) rickettsiae was obtained from flea pools and individual ticks collected at three sites in northwestern Peru within the focus of an outbreak of febrile disease in humans attributed, in part, to SFG rickettsia infections. Molecular identification of the etiologic agents from these samples was determined after partial sequencing of the 17-kDa common antigen gene (htrA) as well as pairwise nucleotide sequence homology with one or more of the following genes: gltA, ompA, and ompB. Amplification and sequencing of portions of the htrA and ompA genes in pooled samples (2 of 59) taken from fleas identified the pathogen Rickettsia felis. Four tick samples yielded molecular evidence of SFG rickettsiae. Fragments of the ompA (540-bp) and ompB (2,484-bp) genes were amplified from a single Amblyomma maculatum tick (tick 124) and an Ixodes boliviensis tick (tick 163). The phylogenetic relationships between the rickettsiae in these samples and other rickettsiae were determined after comparison of their ompB sequences by the neighbor-joining method. The dendrograms generated showed that the isolates exhibited close homology (97%) to R. aeschlimannii and R. rhipicephali. Significant bootstrap values supported clustering adjacent to this nodule of the SFG rickettsiae. While the agents identified in the flea and tick samples have not been linked to human cases in the area, these results demonstrate for the first time that at least two SFG rickettsia agents were circulating in northern Peru at the time of the outbreak. Furthermore, molecular analysis of sequences derived from the two separate species of hard ticks identified a possibly novel member of the SFG rickettsiae.
Proteobacteria of the family Rickettsiaceae, order Rickettsiales, are made up of highly specialized obligate intracellular, gram-negative bacteria that survive freely within the cytosol of the host cell. Members of the genus Rickettsia are divided into two genetically similar groups, the spotted fever group (SFG) and the typhus group (TG), on the basis of host specificity, intracellular location, in vitro growth conditions, antigenic characteristics, the molecular sequences of conserved genes, clinical features, and epidemiology (15, 16, 37, 38). Seventeen species of the genus Rickettsia are categorized within the SFG rickettsiae. With the exception of Rickettsia akari (mite-borne) and R. felis (flea-borne), the remaining SFG rickettsia species are recognized as tick-borne rickettsiae that are passed to subsequent generations or stages transovarially and transtadially (21). While the members of the SFG rickettsiae are adapted to existence within specific hosts, they are capable of infecting humans after humans are bitten by infected arthropods. The TG contains two species, R. prowazekii and R. typhi. The former scrub typhus group has been moved into its own genus, Orientia tsutsugamushi, following analysis of the 16S rRNA gene (47).
At least 15 members of SFG rickettsiae have been associated with syndromic diseases that result in diverse clinical presentations, from asymptomatic to severe (25, 49). The illnesses that occur as a result of rickettsial infections are often characterized by an acute onset of fever, accompanied by nonspecific signs and symptoms. Occasionally, a rash follows the occurrence of the systemic symptoms and may be pathognomonic (50). Clinical presentation and manifestations often vary, with the more pathogenic strains causing debilitating disease with rapid onset.
While rickettsioses have been reported on practically every continent, the incidence of rickettsial diseases has been underreported in Central and South America (20) and has generally been undocumented in Peru. Brazilian spotted fever, which is transmitted by the tick Amblyomma cajennense (12), was initially reported in Brazil more than 60 years ago (14), and cases have increasingly been documented over the last 20 years (10, 11, 13, 43, 46). Rickettsiae and rickettsioses have also been recorded in Mexico (52), Costa Rica (19), and Argentina (27, 39). The SFG rickettsia agent, R. felis, has been identified in domestic animals and is thought to infect humans in Peru, although these data remain unpublished (C. Moron, unpublished data). Louse-transmitted typhus infections have previously plagued sierra communities in the department of Cusco in central Peru (33).
Between May and October 2002, a number of febrile cases, including two deaths, were reported in the area around the town of Sapillica in northwestern Peru. A joint investigation with representatives of the Peruvian Ministry of Health documented a high seroprevalence of rickettsial agents and Leptospira species and the molecular identification of an SFG rickettsia (7). As these diseases are often associated with a zoonotic focus, ectoparasites were taken from domestic animals and animals trapped in the wild during the course of this study. Here we provide evidence from the results of PCR, sequencing, and phylogenetic analyses of fragments of the common 17-kDa antigen gene (htrA) (2), the citrate synthase gene (gltA) (24), ompA (18), and ompB (24, 41) that SFG rickettsiae were circulating among arthropods at the time of the disease outbreak. Finally, the 2,484-bp sequence of the ompB gene obtained from two ticks was found to be phylogenetically disparate (≥3%) from those of known SFG rickettsia species, suggesting a putative novel species.
MATERIALS AND METHODS
Ecologic and entomologic samples.
Ecologic and entomologic samples were obtained from the sparsely populated sierras of northwestern Peru in an area that rests roughly 2,700 m above sea level, has a mean average temperature range from −3 to 34°C, and rainfall of 1,000 mm/year. Within the defined study area, which comprised three separate villages 45 km apart, rodents were trapped live by using a combination of Tomahawk (Tomahawk Live Trap Co., Tomahawk, Wis.) and Sherman (H. B. Sherman Traps Inc., Tallahassee, Fla.) traps. Additionally, domesticated dogs, cats, and horses were gathered and screened for fleas and ticks. Ectoparasites were removed with fine forceps and placed individually in vials containing 70% ethanol or snap frozen in liquid nitrogen. DNA was prepared from these as described previously (44).
PCR.
Nested PCR for the detection of fragments of the Rickettsia genus-specific 17-kDa protein gene (htrA) was performed to discern R. typhus and R. rickettsii members by using the sequences published previously (48). Characterization of SFG rickettsiae was determined after single-stage or nested PCR of specific conserved genes, including ompA (40) and the citrate synthase (gltA) genes (42). The PCR primers and the sizes of the amplicons obtained following amplification are shown in Table 1. Reactions were performed with Ready-To-Go PCR Beads (Amersham Pharmacia Biotech) with Taq Platinum (Invitrogen, Inc., Carlsbad, Calif.). Primers were used at 0.5 to 1.0 μmol/liter, with MgCl2 concentrations held at 1.5 mmol/liter. Following electrophoresis on ethidium bromide-stained 2% low-melting-temperature agarose gels, the PCR products were purified and sequenced on either an automated ABI 373A or an ABI Prism 3100 gene sequencer (Applied Biosystems, Foster City, Calif.) by standard procedures.
TABLE 1.
Description of primers used to amplify gene fragments
| Target gene or species | Primer or probe | Sequence (5′-3′)a | Amplicon size (bp) |
|---|---|---|---|
| htrA | R17-122 (first round) | CAGAGTGCTATGAACAAACAAGG | |
| R17-500 (first round) | CTTGCCATTGCCCATCAGGTT | 378 | |
| TZ15 SFG F | TTCTCAATTCGGTAAGGGC | ||
| TZ16 SFG R | ATATTGACCAGTCGCTATTTC | 247 | |
| RPID TG F | CGGTACACTTCTTGGTGGCGCAGGAGGT | ||
| RP2 TG R | TTCACGGCAATATTGACCTGTACTGTTCC | 286 | |
| htrA 434-bp fragment | Rr1175F | GCTCTTGCAACTTCTATGTT | |
| Rr2608R | CATTGTTCGTCAGGTTGGCG | 434 | |
| ompBc qPCR | RF1396F | ACCCAGAACTCGAACTTTGGTG | |
| RF1524R | CACACCCGCAGTATTACCGTT | ||
| R. felisb | RFQuBP (probe) | CGCGACTTACAGTTCCTGATACTAAGGTTCTT | 129 |
| ACAGGTCGCG-BHQ-2 | |||
| ompB qPCR | RR1595F | GCCGGAGTTGTCCAATTATCA | 128 |
| CCGCCGACAAGAGCAGTTT | |||
| SFG; not R. felis or R. akari | RR1722R | ROX-CCGCGCCGGCATTTCCTAAACGTAACTCGGC | |
| RR1654P | AGCGCGG-DAB | ||
| ompB | ompB2409F | CCGTAACATTAAACAAACAAGCTG | 2,478 |
| ompB4887R | AGAGTACCTTGATGTGCRGTATAYT | ||
| ompA | RR 190-70 (first round) | ATGGCGAATATTTCTCCAAAA | |
| RR 190-701 (first round) | GTTCCGTTAATGGCAGCATCT | 590 | |
| 190-FN1 (nested) | AAGCAATACAACAAGGTC | ||
| 190-RN1 (nested) | TGACAGTTATTATACCTC | 540 | |
| gltA | RpCS.877p | GGGGGCCTGCTCACGGCGG | |
| RpCS. 1258n | ATTGCAAAAAGTACAGTGAACA | 381 |
Abbreviations: ROX, passive reference dye; DAB, diaminobenzidine; Y, pyrimidine.
Primers and probe specific for amplication of ompB gene fragment of R. felis (J. Jiang, unpublished).
Primers specific for amplication of ompB gene fragment of SFG and not R. felis (J. Jiang, unpublished).
For the extended ompB amplifications (41), PCR primers with specificities for conserved regions were designed after alignment of the sequences of 23 SFG rickettsiae (J. Jiang, unpublished data). A nested PCR was performed with one reverse primer and two different forward primers (Table 1). One microliter of the DNA template was added to the reaction mixture, which contained SuperMix High Fidelity (Invitrogen) and 0.5 μM primers. Each PCR was conducted in a T-Gradient thermocycler (Biometra, Goettingen, Germany). Following initial denaturation for 1 min at 94°C, 35 cycles of denaturation for 30 s at 94°C, annealing for 1 min at 50°C, and extension for 4 min at 68°C were performed. A final extension step was done for 20 min at 72°C. The PCR products were purified and cloned with a TOPO XL PCR cloning kit (Invitrogen), following the instructions of the manufacturer. Plasmid DNA was isolated (Miniprep; Qiagen) from one colony each for tick 124 and tick 163. The sequence was obtained with both forward and reverse sequencing primers, as indicated below, with an ABI Prism 3100 genetic analyzer (Applied Biosystems). The sequences were assembled with Sequencher (version 4.0) (Gene Codes Corporation Inc., Ann Arbor, Mich.).
Sequencing primers.
The following M13 forward and reverse primers from the Topo XL PCR cloning kit (Invitrogen) were used: primer *120-2788F (AAACAATAATCAAGGTACTGT), primer R3521F (GATAATGCCAATGCAAATTTCAG), primer R4224F (ACCAAGATTATAAGAAAGGTGATAA), primer R3008R (CGCCTGTAGTAACAGTTACAC), primer R3637R (GAAACGATTACTTCCGGTTACA), and primer *120-4346R (CGAAGAAGTAACGCTGACTT).
qPCR.
A Rickettsia genus-specific quantitative real-time PCR (qPCR) (26) and an R. felis-specific qPCR were performed to detect the presence of the R. felis ompB gene. The R. felis qPCR assay was specifically designed to detect a portion of the R. felis ompB gene (in the open reading frame between positions 1179 and 1307). Preparations of DNA from R. felis-infected cat fleas (Ctenocephalides felis; FleaData, Inc. Freeville, N.Y.) (8) were positive by this assay; but those of DNA from R. typhi Wilmington or Museibov strains, R. prowazekii, 11 species of SFG rickettsiae, Orientia, Ehrlichia, Bartonella, and 12 other bacteria were not (J. Jiang, unpublished). The host cell DNA included in the bacterial cultures did not produce false-positive reactions. For the R. felis qPCRs, 2 μl of template was reacted with 0.5 μM both forward and reverse primers and 0.4 μM Quasar 670 probe (Biosearch Technologies, Inc., Novato, Calif.). MgCl2 (5 mM) and premixed OmniMix HS beads (Cepheid, Sunnyvale, Calif.) were added to the reaction mixture. Amplification for the qPCR assay was conducted with a SmartCycler thermocycler (Cepheid) with the following temperatures and cycle parameters: an initial denaturation of 3 min at 94°C and then 50 cycles of denaturation (94°C for 5 s) and annealing and elongation (60°C for 30 s). The threshold cycle value was determined when experimental samples produced fluorescence greater than the calculated threshold cycle value on the basis of the background fluorescence measured during amplification. No-template controls (which contained 2 μl of double-distilled H2O instead of DNA), which were produced at the same time and under the same conditions as the experimental positive control samples, were consistently negative (i.e., the fluorescence did not cross the threshold of background fluorescence).
Phylogenetic analysis.
The sequences were compared to those downloaded from GenBank by using MacVector (version 7.22) software (Accelrys, Inc., San Diego, Calif.), and alignments were created with Sequencher (version 3.0) software (Gene Codes Corporation Inc.). Phylogenetic analyses were performed with the PAUP (version 5.0) program (Sinauer Associates, Inc., Sunderland, Mass.) with the criterion of maximum parsimony and neighbor joining. Reference sequences from GenBank and two voucher specimens (R. akari and R. rickettsii) were used to confirm the results for the experimental samples. For the maximum-parsimony analysis, all characters were weighted equally, and a heuristic search with 100 replicates of a random taxon was performed to facilitate branch swapping. Confidence in nodes was assessed by the bootstrap procedure with 500 resampling replicates. Bootstrap values >70% were considered well supported.
Nucleotide sequence accession numbers.
The sequences obtained from the DNA extracted from tick 163 were assigned GenBank accession numbers AY590796 (540 bp, ompA), AY590797 (381 bp, gltA), and AY652981 (2,484 bp, ompB).
RESULTS
Sample collection.
Sampling was conducted at three sites during an outbreak of febrile illness that began in June 2002 and that ended in November 2002. These sites, Coletas, Naranjo, and Sapillica, are located in the department of Piura, northeast of the city of Sullana, in northwestern Peru. A total of 16 ticks and 59 pools of fleas were collected and categorized (Table 2). Ectoparasites (fleas and ticks) were taken from individual domestic animals and animals trapped in the wild and sorted by species. Ticks were identified by using taxonomic keys for the tick species found in Peru (28). A total of six tick species and six flea species were collected from dogs, horses, cats, and rodents. No ticks or fleas were taken from human subjects in the area.
TABLE 2.
Ectoparasite collected and percentage of parasites positive for an SFG agent
| Ectoparasite | No. (% of totala) | No. SFG positive (% of totala) |
|---|---|---|
| Tick species | ||
| Amblyomma tigrinum | 2 (12.5) | 0 |
| Amblyommma maculatum | 3 (18.7) | 2 (12.5) |
| Anocenter nitens | 1 (6.3) | 1 (6.2) |
| Boophilus sp. | 5 (31.3) | 0 |
| Ixodes boliviensis | 3 (18.7) | 1 (6.2) |
| Ixodes pararicinus | 2 (12.5) | 0 |
| Flea speciesb | ||
| Adoratopsilla intermedia | 2 (3.4) | 0 |
| Ctenocephalides felis | 33 (55.9) | 2 (3.4) |
| Ctenocephalides canis | 6 (10.2) | 0 |
| Pulex irritan | 11 (18.6) | 0 |
| Neotyphloceras crassispina | 2 (3.4) | 0 |
| Xenopsylla cheopis | 5 (8.5) | 0 |
The percentage based on the total number of either ticks, or fleas.
Fleas of the same species were placed into sorted pools, based on individual hosts.
PCR and qPCR analysis of R. felis from flea samples.
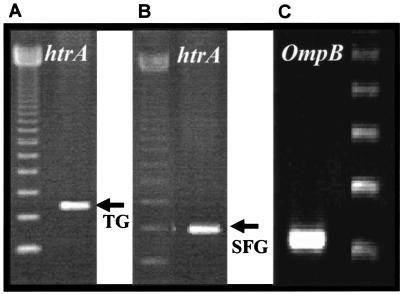
Once the fleas were keyed and sorted, DNA was extracted from pools removed from a single domestic animal or an animal trapped in the wild. Initially, all samples were tested by PCR for detection of the 17-kDa common antigen gene (htrA) found in both TG and SFG members of the rickettsiae (2, 48). In the initial reaction, genus-specific primers were used to amplify a PCR product, and then a nested reaction was conducted to differentiate the TG from the SFG members of the rickettsiae. Nested reactions for the TG rickettsiae yielded a product of 286 bp, while reactions for the SFG rickettsiae gave a product of 247 bp. The results of the analyses was confirmed by PCR and sequencing of a 434-bp amplicon (51). After screening, 2 of 59 pools of fleas were found to contain PCR bands and sequences that signified an infection with an SFG agent. Both samples were extracted from pools of C. felis fleas removed from domestic canines in Sapillica. In flea sample FL105, an SFG-specific band and a TG-specific band were both evident (Fig. 1A and B). In order to rule out possible dual infection, a qPCR assay specific for R. typhi (J. Jiang, unpublished) was conducted, and the results were found to be negative (data not shown). Sequence analysis of the htrA PCR product demonstrated 100% similarity with that of R. felis (GenBank accession number AF195118.1). To differentiate the possible SFG agent in the flea samples, ompB qPCR assays specific for either R. felis or SFG rickettsiae but not R. felis were conducted. Flea samples FL103 and FL105 were positive by the R. felis qPCR. The 129-bp R. felis PCR amplicon produced by FL105 is shown in Fig. 1. Additional characterization of the nucleic acid preparations from the flea samples was attempted by using a PCR assay specific for a portion of the ompA gene. A 540-bp PCR product for ompA was obtained from sample FL105 and sequenced. This product also demonstrated 100% homology with the R. felis sequence. Collectively, these data confirm the presence of R. felis within C. felis fleas collected in Peru.
FIG. 1.
Representative example of htrA gene PCR denoting a positive sample from FL105. Evident are both a TG Rickettsia-specific (A) and SFG Rickettsia-specific band (B). The 129-bp product of the ompB gene amplified from DNA from FL105 with R. felis-specific primers is also shown (C). A 123-bp ladder (Gibco Life Sciences, Gaithersburg, Md.) was used as a guide for molecular sizes in all three gel slices.
PCR and phylogentic analysis of a potentially novel SFG agent from ticks.
DNA from a total of 16 ticks was screened by the htrA PCR. Four ticks were positive for SFG DNA. Of these, two were Amblyomma maculatum (tick 047, from a dog, and tick 124, from a horse), one was Anocenter nitens (tick 127, from a horse), and the last one was Ixodes boliviensis (tick 163, from a horse). Sequence data for the tick 127 htrA amplicon indicated that this agent was an SFG rickettsia (99.7% homology to the htrA sequences of R. conorii, R. rickettsii, R. sibirica, and R. peacockki). No additional data were obtained from this sample.
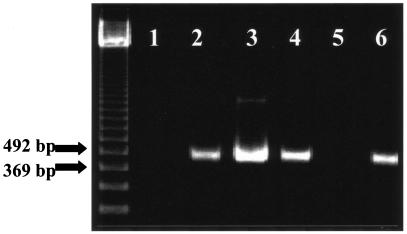
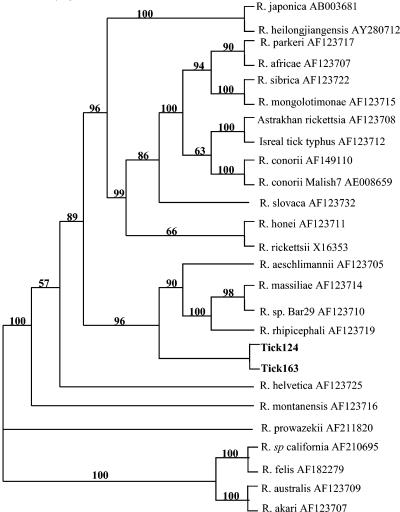
Samples tick 124 and tick 163 yielded positive PCR products for ompA (Fig. 2). These samples, as well as tick 047, were also positive by PCR for the citrate synthase gene, gltA. Sequencing of the 540-bp ompA and the 381-bp gltA fragments indicated that they were most closely aligned with the gene sequences of R. massiliae (6) and with those of Rickettsia sp. strain RPA4 and Rickettsia sp. strain DNS14 (isolates from Siberia [44]). To more closely align the experimental sequences with those of known species, a strategy was developed to sequence 2,484 bp of the ompB genome with broad-range primers. By this technique, 2,484-bp fragments from tick 124 and tick 163 samples were amplified and sequenced, and then these sequences were compared to those of known Rickettsia sp. members (Fig. 3). With the criterion set to distance, a neighbor-joining search on 100 bootstrap replicates provided node values that bracketed the northern Peruvian tick samples in a phylogenetic tree with described SFG members. Of note, while the sequences from the experimental tick samples were 100% similar, they differed by 3% from the most closely aligned sequences representing the R. massiliae group (Table 3).
FIG. 2.
PCR of the 540-bp ompA gene from tick samples. Lane 1, negative extraction control; lanes 2 and 3, positive controls (R. akari and R. rickettsii, respectively); lanes 4 to 6, ticks 124, 127, and 163, respectively. Samples were electrophoresed in 2% agarose gels.
FIG. 3.
Dendrogram of the sequence alignment of rickettsia species based on the nucleotide sequences of a 2,487-bp segment of the ompB gene. The analysis, including maximum parsimony and neighbor joining, was performed with the PAUP program. Bootstrap values are indicated at the nodes.
TABLE 3.
Taxonomy comparison of 2,484-bp ompB fragment from tick 163
| Rickettsia species | GenBank accession no. | No. of identical nucleotides/total no. tested | % Identity |
|---|---|---|---|
| R. aeschlimannii | AF123705 | 2,410/2,479 | 97 |
| R. rhipicephali | AF123719 | 2,406/2,479 | 97 |
| Rickettsia sp. strain Bar 29 | AF123710 | 2,402/2,482 | 96 |
| R. massiliae | AF123714 | 2,397/2,479 | 96 |
| R. slovaca | AF123723 | 2,394/2,479 | 96 |
| R. conorii Malish | AE008659 | 2,393/2,479 | 96 |
| R. conorii | AF149110 | 2,391/2,479 | 96 |
| R. parkeri | AF123717 | 2,390/2,479 | 96 |
| R. sibirica | AF123722 | 2,387/2,479 | 96 |
| R. africae | AF123706 | 2,387/2,479 | 96 |
| R. mongolotimonae | AF123715 | 2,386/2,479 | 96 |
| R. honei | AF123711 | 2,385/2,478 | 96 |
| R. honei strain Thai tick typhus | AF123724 | 2,383/2,478 | 96 |
| Isreali tick typhus rickettsia | AF123712 | 2,379/2,477 | 96 |
| R. japonica | AB003681 | 1,809/1,879 | 96 |
| R. heilongjiangensis | AY280712 | 2,376/2,478 | 95 |
| Astrakhan rickettsia | AF123708 | 2,379/2,479 | 95 |
| R. rickettsii | X16353 | 2,377/2,479 | 95 |
DISCUSSION
Pathogenic rickettsiae are transmitted to vertebrate hosts by arthropod vectors such as fleas, ticks, lice, and mites. Many of these diseases have resulted in marked morbidity, with occasional fatalities. In the last 10 years, the number of human rickettsioses has dramatically increased, with 15 species recognized throughout the world (25, 31, 37). The incidence of rickettsial pathogens has been underdocumented in Peru, and the species remain undercharacterized. Here we report evidence of SFG rickettsiae among ectoparasites collected during an investigation to determine the zoonotic factors possibly related to an increase in the rate of febrile illnesses in the northwestern region of the country (Table 4).
TABLE 4.
Summary for samples found to be positive for SFG
| Sample | Site | Sample description | Host | Gene(s) sequenced | Closest match (%) |
|---|---|---|---|---|---|
| FL103 | Sapillica | C. felis | Dog | htrA | R. felis (100) |
| FL105 | Sapillica | C. felis | Dog | htrA | R. felis (100) |
| ompA | R. felis (100) | ||||
| Tick 047 | Coletas | A. maculatum | Dog | htrA | SFG (99.7)a |
| gltA | R. heilongjiangensis (98) | ||||
| Rickettsia sp. strain RpA4 (98) | |||||
| Tick 124 | Coletas | A. maculatum | Horse | htrA | SFG (98.4) |
| ompA | R. heilongjiangensis (98) | ||||
| gltA | R. heilongjiangensis (98) | ||||
| ompB (2,484 bp) | R. aeschlimannii (97) | ||||
| R. rhipicephali (97) | |||||
| Tick 127 | Coletas | A. nitens | Horse | htrA | SFG (98.1) |
| Tick 163 | Naranjo | I. boliviensis | Horse | htrA | SFG (99.7) |
| ompA | Rickettsia sp. strain RpA4 (97.5) | ||||
| gltA | Rickettsia sp. strain RpA4 (98) | ||||
| ompB (2,484-bp) | R. aeschlimannii (97) | ||||
| R. rhipicephali (97) |
Denotation of SFG represents similar sequence homology to the sequences of four or more SFG members.
Toward this end, we used gene sequence-based criteria for the taxonomic positioning of the samples obtained to characterize suspected rickettsiae. Initially, all samples were tested for expression of the genus-specific gene htrA and were then segregated into either the TG or the SFG. Samples were also screened for expression of the citrate synthase gene (gltA). The nucleotide sequence of the gltA gene serves as a better discriminator of closely related rickettsiae than the 16S rRNA gene (42). To better characterize the rickettsial agents in our samples, ompA and ompB PCRs were conducted. These genes encode a highly antigenic, large-molecular-mass membrane protein specific for SFG members and have proven useful in delineating relationships (40, 41). Ultimately, phylogenetic relationships among SFG members and our unique sequences were determined by comparison of a large 2,484-bp ompB fragment.
The prevalences obtained by these tests demonstrated that 3.3% of flea pools (2 of 59) and 25% of ticks (4 of 16) were positive for SFG agents. Following the htrA PCR, no sample yielded solely a TG-specific band, although such a band was evident from two flea samples in combination with an SFG-specific band. The R. typhi-specific qPCR assay was negative with this sample, providing evidence that the TG-specific band did not result from a dual R. felis-R. typhi infection. The sequence generated after both htrA and ompA partial gene amplification demonstrated 100% homology to the R. felis sequence, which is in a clade closely aligned with but distinct from that for R. australis and R. akari. These samples were disparate from the flea-borne rickettsiae, R. typhi, and the louse-borne agent, R. prowazekii, implying genetic distance.
R. felis is an emerging pathogen that was first isolated in a commercial colony of cat fleas (1). Upon isolation, R. felis was designated a new species (22) and on the basis of molecular data was placed in the SFG rather than the TG (9). R. felis is transmitted by fleas horizontally and vertically (4). It was recognized as a pathogenic rickettsia after identification in samples from the Americas (35). Infected fleas, particularly C. felis, were found to be the likely cause of outbreaks in humans in Mexico (53) and Brazil (29). These examples provided evidence that R. felis exists in Central and South America. Our samples were taken from C. felis, a parasite commonly found on both canines and felines in Peru. We did not find evidence of R. typhi among Xenopsylla cheopis fleas (3) removed from rats (Rattus norvegicus) trapped during our investigation, nor did we identify R. prowazekii, the etiologic agent of epidemic typhus (data not shown). However, we did note the presence of nucleic acid extracted from the blood of rodents collected in Sapillica that was similar to that of R. felis (data not shown). C. felis is known to feed on mice and rodents and may represent an important reservoir for R. felis in northern Peru.
Similar molecular analyses were conducted with 16 ticks removed from domestic animals and animals trapped in the wild in the area of the fever outbreak. Following htrA PCR, positive reactions were found for 4 of 16 nucleic acid preparations from ticks, and sequence data obtained from the ompA and gltA amplicons supported the inclusion of the isolates from these samples in the SFG. To better align our sequences with those established previously, we compared the ompB amplicons generated from two ticks (tick 124 and tick 163) with those of other SFG species. The sequences from the Peruvian ticks aligned with sequences from strains just outside a nodule containing the R. massiliae group, including R. rhipicephali (97%; GenBank accession number AF123719), an isolate from the Astrakhan region of the former Soviet Union (44), R. aeschlimannii (97%; GenBank accession number AF123705) from southern Croatia (5, 32), Rickettsia sp. strain Bar 29 (GenBank accession number AF123710), and R. massiliae (GenBank accession number AF1213714) (6) from the south of France. The tick R. aeschlimannii isolate was identified in a patient returning from Morocco. R. aeschlimannii is pathogenic for humans; and infection with this rickettsia results in an infection characterized by an eschar, fever, and a generalized maculopapular skin rash (34). R. rhipicephali has not been shown to infect humans. Our samples differed from those of the aforementioned SFG members by 3% or more (Table 3). These results infer considerable divergence as well as geographic disparity from the most closely linked matches.
The hard ticks A. maculatum and I. boliviensis represent species that are common in the Peruvian Andes. Moreover, these are typical vector genera of rickettsial agents and have been shown to transmit disease in regions as far removed as the Pacific coast of the United States (23), Australia (45), and the former Yugoslavian republics (17). Recently, A. maculatum has been shown to be naturally infected with R. parkeri, a human pathogen in the southern United States (30).
A high serologic prevalence of the bacterium Leptospira and SFG rickettsia in the area of the outbreak was determined previously (7), and the SFG rickettsiae from samples from four febrile humans were identified by molecular analyses (7). We have no clear evidence that the SFG rickettsial agent identified by molecular analysis in the four serum samples from febrile humans from northern Peru is the same agent in our tick samples. The patients seen during our study experienced a mild disease typified by fever and headache, often malaise, and less frequently, a stiff neck and joint pain (7). No eschar was reported, and unlike previously reported cases of “spotless” rickettsiosis (for instance, following R. slovaca infection [36]), all our patients exhibited fever. The symptoms resolved upon treatment with doxycycline for 1 week.
We report for the first time the identification of R. felis from C. felis flea samples in Peru. We were also able to amplify conserved rickettsial genes from both a single A. maculatum tick and a single I. bolivensis tick isolated in northwestern Peru. We do not know whether the SFG agent identified in the tick samples is homologous to that seen in the human samples or, for that matter, whether it can even be transmitted to humans. As was the case for the flea samples, analysis of blood samples taken from rodents trapped during the investigation showed that they were infected with the pathogen R. felis (P. J. Blair, unpublished data). However, partial sequencing of the htrA gene from human samples suggested infection with another SFG species (P. J. Blair, unpublished). Certainly, the close proximity of the agrarian Andean societies with domesticated canines, felines, and herd animals elevates the risk of vertical transmission. A clearer epidemiological assessment of rickettsial diseases in northwestern Peru will become apparent following isolation and characterization of the agents from host species. Efforts are continuing toward that end. Finally, on the basis of the sequence and phylogenetic data presented herein, we believe that we have identified a putative novel SFG agent, for which we propose the name “Candidatus Rickettsia andeanae” in recognition of the area where it was first detected.
Acknowledgments
We thank Roxana Lescano for administrative assistance, David Walker (University of Texas Medical Center) for provision of the R. akari DNA control (to C.M.), and Herbert Thompson (Centers for Disease Control and Prevention) for the R. rickettsii DNA control. We are grateful for the help provided by Carmen Flores and Jeffrey Stancil in identifying tick and flea samples. We acknowledge the assistance of David W. Tandy (University of Tennessee) for help in clarifying the Latin roots of the proposed novel agent names.
The U.S. Department of Defense Global Emerging Infection System funded the investigation in northwestern Peru.
The views expressed in this article are those of the authors and do not reflect the official policy or position of the U.S. Department of Defense, the Department of the Navy, or the U.S. Government.
REFERENCES
- 1.Adams, J. R., E. T. Schmidtmann, and A. F. Azad. 1990. Infection of colonized cat fleas, Ctenocephalides felis (Bouche), with a rickettsia-like microorganism. Am. J. Trop. Med. Hyg. 43:400-409. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. E., and T. Tzianabos. 1989. Comparative sequence analysis of a genus-common rickettsial antigen gene. J. Bacteriol. 171:5199-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad, A. F., S. Radulovic, J. A. Higgins, B. H. Noden, and J. M. Troyer. 1997. Flea-borne rickettsioses: ecologic considerations. Emerg. Infect. Dis. 3:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azad, A. F., J. B. Sacci, Jr., W. M. Nelson, G. A. Dasch, E. T. Schmidtmann, and M. Carl. 1992. Genetic characterization and transovarial transmission of a typhus-like rickettsia found in cat fleas. Proc. Natl. Acad. Sci. USA 89:43-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beati, L., M. Meskini, B. Thiers, and D. Raoult. 1997. Rickettsia aeschlimannii sp. nov., a new spotted fever group rickettsia associated with Hyalomma marginatum ticks. Int. J. Syst. Bacteriol. 47:548-554. [DOI] [PubMed] [Google Scholar]
- 6.Beati, L., and D. Raoult. 1993. Rickettsia massiliae sp. nov., a new spotted fever group Rickettsia. Int. J. Syst. Bacteriol. 43:839-840. [DOI] [PubMed] [Google Scholar]
- 7.Blair, P. J., G. B. Schoeler, C. Moron, E. Anaya, R. Caceda, M. Cespedes, C. Cruz, V. Felices, C. Guevara, A. Huaman, R. Luckett, L. Mendoza, A. L. Richards, Z. Rios, J. W. Sumner, P. Villaseca, and J. G. Olson. 2004. Evidence of rickettsial and leptospira infections in Andean northern Peru. Am. J. Trop. Med. Hyg. 70:357-363. [PubMed] [Google Scholar]
- 8.Boostrom, A., M. S. Beier, J. A. Macaluso, K. R. Macaluso, D. Sprenger, J. Hayes, S. Radulovic, and A. F. Azad. 2002. Geographic association of Rickettsia felis-infected opossums with human murine typhus, Texas. Emerg. Infect. Dis. 8:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouyer, D. H., J. Stenos, P. Crocquet-Valdes, C. G. Moron, V. L. Popov, J. E. Zavala-Velazquez, L. D. Foil, D. R. Stothard, A. F. Azad, and D. H. Walker. 2001. Rickettsia felis: molecular characterization of a new member of the spotted fever group. Int. J. Syst. Evol. Microbiol. 51:339-347. [DOI] [PubMed] [Google Scholar]
- 10.de Lemos, E. R., F. B. Alvarenga, M. L. Cintra, M. C. Ramos, C. D. Paddock, T. L. Ferebee, S. R. Zaki, F. C. Ferreira, R. C. Ravagnani, R. D. Machado, M. A. Guimaraes, and J. R. Coura. 2001. Spotted fever in Brazil: a seroepidemiological study and description of clinical cases in an endemic area in the state of Sao Paulo. Am. J. Trop. Med. Hyg. 65:329-334. [DOI] [PubMed] [Google Scholar]
- 11.de Lemos, E. R., R. D. Machado, and J. R. Coura. 1994. Rocky Mountain spotted fever in an endemic area in Minas Gerais, Brazil. Mem. Inst. Oswaldo Cruz 89:497-501. [DOI] [PubMed] [Google Scholar]
- 12.de Lemos, E. R., R. D. Machado, J. R. Coura, M. A. Guimaraes, N. M. Freire, M. Amorim, and G. S. Gazeta. 1997. Epidemiological aspects of the Brazilian spotted fever: seasonal activity of ticks collected in an endemic area in Sao Paulo, Brazil. Rev. Soc. Bras. Med. Trop. 30:181-185. [DOI] [PubMed] [Google Scholar]
- 13.de Lima, V. L., S. S. de Souza, C. E. de Souza, M. F. Vilela, P. M. Papaiordanou, V. M. Del Guercio, and M. M. Rocha. 2003. Spotted fever in Campinas region, State of Sao Paulo, Brazil. Cad. Saude Publica. 19:331-334. [DOI] [PubMed] [Google Scholar]
- 14.Dias, C. G., M. V. Ropke, S. Superti, L. Berquo, and P. d'Azevedo. 2004. Use of a novel selective medium to detect methicillin-resistant Staphylococcus aureus in colonized patients of an intensive care unit. Infect. Control Hosp. Epidemiol. 25:130-132. [DOI] [PubMed] [Google Scholar]
- 15.Drancourt, M., and D. Raoult. 1994. Taxonomic position of the rickettsiae: current knowledge. FEMS Microbiol. Rev. 13:13-24. [DOI] [PubMed] [Google Scholar]
- 16.Eremeeva, M., X. Yu, and D. Raoult. 1994. Differentiation among spotted fever group rickettsiae species by analysis of restriction fragment length polymorphism of PCR-amplified DNA. J. Clin. Microbiol. 32:803-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fournier, P. E., J. P. Durand, J. M. Rolain, J. L. Camicas, H. Tolou, and D. Raoult. 2003. Detection of Astrakhan fever rickettsia from ticks in Kosovo. Ann. N. Y. Acad. Sci. 990:158-161. [DOI] [PubMed] [Google Scholar]
- 18.Fournier, P. E., V. Roux, and D. Raoult. 1998. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int. J. Syst. Bacteriol. 48(Pt 3):839-849. [DOI] [PubMed] [Google Scholar]
- 19.Fuentes, L. 1986. Ecological study of Rocky Mountain spotted fever in Costa Rica. Am. J. Trop. Med. Hyg. 35:192-196. [DOI] [PubMed] [Google Scholar]
- 20.Galvao, M. A., C. L. Mafra, C. Moron, E. Anaya, and D. H. Walker. 2003. Rickettsiosis of the genus Rickettsia in South America. Ann. N. Y. Acad. Sci. 990:57-61. [DOI] [PubMed] [Google Scholar]
- 21.Hackstadt, T. 1996. The biology of rickettsiae. Infect. Agents Dis. 5:127-143. [PubMed] [Google Scholar]
- 22.Higgins, J. A., S. Radulovic, M. E. Schriefer, and A. F. Azad. 1996. Rickettsia felis: a new species of pathogenic rickettsia isolated from cat fleas. J. Clin. Microbiol. 34:671-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, L. E., C. M. Clifford, R. Gresbrink, L. A. Thomas, and J. E. Keirans. 1976. Isolation of a spotted fever group rickettsia from the Pacific Coast tick, Ixodes pacificus, in Oregon. Am. J. Trop. Med. Hyg. 25:513-516. [DOI] [PubMed] [Google Scholar]
- 24.Inokuma, H., P. Brouqui, M. Drancourt, and D. Raoult. 2001. Citrate synthase gene sequence: a new tool for phylogenetic analysis and identification of Ehrlichia. J. Clin. Microbiol. 39:3031-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensenius, M., P. E. Fournier, and D. Raoult. 2004. Tick-borne rickettsioses in international travellers. Int. J. Infect. Dis. 8:139-146. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, J., T. C. Chan, J. J. Temenak, G. A. Dasch, W. M. Ching, and A. L. Richards. 2004. Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am. J. Trop. Med. Hyg. 70:351-356. [PubMed] [Google Scholar]
- 27.Martino, O., T. Orduna, L. Lourtau, P. Scapellato, B. Cernigo, and A. Seijo. 2001. Spotted fever group rickettsial disease in Argentinean travelers. Rev. Soc. Bras. Med. Trop. 34:559-562. (In Portuguese.) [DOI] [PubMed] [Google Scholar]
- 28.Need, J. T., W. E. Dale, J. E. Keirans, and G. A. Dasch. 1991. Annotated list of ticks (Acari: Ixodidae, Argasidae) reported in Peru: distribution, hosts, and bibliography. J. Med. Entomol. 28:590-597. [DOI] [PubMed] [Google Scholar]
- 29.Oliveira, R. P., M. A. Galvao, C. L. Mafra, C. B. Chamone, S. B. Calic, S. U. Silva, and D. H. Walker. 2002. Rickettsia felis in Ctenocephalides spp. fleas, Brazil. Emerg. Infect. Dis. 8:317-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paddock, C. D., J. W. Sumner, J. A. Comer, S. R. Zaki, C. S. Goldsmith, J. Goddard, S. L. McLellan, C. L. Tamminga, and C. A. Ohl. 2004. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin. Infect. Dis. 38:805-811. [DOI] [PubMed] [Google Scholar]
- 31.Parola, P., and D. Raoult. 2001. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin. Infect. Dis. 32:897-928. [DOI] [PubMed] [Google Scholar]
- 32.Punda-Polic, V., M. Petrovec, T. Trilar, D. Duh, N. Bradaric, Z. Klismanic, and T. Avsic-Zupanc. 2002. Detection and identification of spotted fever group rickettsiae in ticks collected in southern Croatia. Exp. Appl. Acarol. 28:169-176. [DOI] [PubMed] [Google Scholar]
- 33.Raoult, D., R. J. Birtles, M. Montoya, E. Perez, H. Tissot-Dupont, V. Roux, and H. Guerra. 1999. Survey of three bacterial louse-associated diseases among rural Andean communities in Peru: prevalence of epidemic typhus, trench fever, and relapsing fever. Clin. Infect. Dis. 29:434-436. [DOI] [PubMed] [Google Scholar]
- 34.Raoult, D., P. E. Fournier, P. Abboud, and F. Caron. 2002. First documented human Rickettsia aeschlimannii infection. Emerg. Infect. Dis. 8:748-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raoult, D., B. La Scola, M. Enea, P. E. Fournier, V. Roux, F. Fenollar, M. A. Galvao, and X. de Lamballerie. 2001. A flea-associated Rickettsia pathogenic for humans. Emerg. Infect. Dis. 7:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raoult, D., A. Lakos, F. Fenollar, J. Beytout, P. Brouqui, and P. E. Fournier. 2002. Spotless rickettsiosis caused by Rickettsia slovaca and associated with Dermacentor ticks. Clin. Infect. Dis. 34:1331-1336. [DOI] [PubMed] [Google Scholar]
- 37.Raoult, D., and V. Roux. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regnery, R. L., C. L. Spruill, and B. D. Plikaytis. 1991. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 173:1576-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ripoll, C. M., C. E. Remondegui, G. Ordonez, R. Arazamendi, H. Fusaro, M. J. Hyman, C. D. Paddock, S. R. Zaki, J. G. Olson, and C. A. Santos-Buch. 1999. Evidence of rickettsial spotted fever and ehrlichial infections in a subtropical territory of Jujuy, Argentina. Am. J. Trop. Med. Hyg. 61:350-354. [DOI] [PubMed] [Google Scholar]
- 40.Roux, V., P. E. Fournier, and D. Raoult. 1996. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J. Clin. Microbiol. 34:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roux, V., and D. Raoult. 2000. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int. J. Syst. Evol. Microbiol. 50(Pt 4):1449-1455. [DOI] [PubMed] [Google Scholar]
- 42.Roux, V., E. Rydkina, M. Eremeeva, and D. Raoult. 1997. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int. J. Syst. Bacteriol. 47:252-261. [DOI] [PubMed] [Google Scholar]
- 43.Rozental, T., M. C. Bustamante, M. Amorim, N. M. Serra-Freire, and E. R. Lemos. 2002. Evidence of spotted fever group rickettsiae in state of Rio de Janeiro, Brazil. Rev. Inst. Med. Trop. Sao Paulo 44:155-158. [DOI] [PubMed] [Google Scholar]
- 44.Rydkina, E., V. Roux, N. Rudakov, M. Gafarova, I. Tarasevich, and D. Raoult. 1999. New rickettsiae in ticks collected in territories of the former Soviet Union. Emerg. Infect. Dis. 5:811-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sexton, D. J., B. Dwyer, R. Kemp, and S. Graves. 1991. Spotted fever group rickettsial infections in Australia. Rev. Infect. Dis. 13:876-886. [DOI] [PubMed] [Google Scholar]
- 46.Sexton, D. J., M. Muniz, G. R. Corey, E. B. Breitschwerdt, B. C. Hegarty, S. Dumler, D. H. Walker, P. M. Pecanha, and R. Dietze. 1993. Brazilian spotted fever in Espirito Santo, Brazil: description of a focus of infection in a new endemic region. Am. J. Trop. Med. Hyg. 49:222-226. [DOI] [PubMed] [Google Scholar]
- 47.Tamura, A., N. Ohashi, H. Urakami, and S. Miyamura. 1995. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int. J. Syst. Bacteriol. 45:589-591. [DOI] [PubMed] [Google Scholar]
- 48.Tzianabos, T., B. E. Anderson, and J. E. McDade. 1989. Detection of Rickettsia rickettsii DNA in clinical specimens by using polymerase chain reaction technology. J. Clin. Microbiol. 27:2866-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker, D. H. 1989. Rickettsioses of the spotted fever group around the world. J. Dermatol. 16:169-177. [DOI] [PubMed] [Google Scholar]
- 50.Walker, D. H., G. A. Valbuena, and J. P. Olano. 2003. Pathogenic mechanisms of diseases caused by Rickettsia. Ann. N. Y. Acad. Sci. 990:1-11. [DOI] [PubMed] [Google Scholar]
- 51.Webb, L., M. Carl, D. C. Malloy, G. A. Dasch, and A. F. Azad. 1990. Detection of murine typhus infection in fleas by using the polymerase chain reaction. J. Clin. Microbiol. 28:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zavala-Velazquez, J. E., J. Ruiz-Sosa, I. Vado-Solis, A. N. Billings, and D. H. Walker. 1999. Serologic study of the prevalence of rickettsiosis in Yucatan: evidence for a prevalent spotted fever group rickettsiosis. Am. J. Trop. Med. Hyg. 61:405-408. [DOI] [PubMed] [Google Scholar]
- 53.Zavala-Velazquez, J. E., J. A. Ruiz-Sosa, R. A. Sanchez-Elias, G. Becerra-Carmona, and D. H. Walker. 2000. Rickettsia felis rickettsiosis in Yucatan. Lancet 356:1079-1080. [DOI] [PubMed] [Google Scholar]