Abstract
In this study, the enzymes involved in polycyclic aromatic hydrocarbon (PAH) degradation in the chrysene-degrading organism Sphingomonas sp. strain CHY-1 were investigated. [14C]chrysene mineralization experiments showed that PAH-grown bacteria produced high levels of chrysene-catabolic activity. One PAH-induced protein displayed similarity with a ring-hydroxylating dioxygenase beta subunit, and a second PAH-induced protein displayed similarity with an extradiol dioxygenase. The genes encoding these proteins were cloned, and sequence analysis revealed two distinct loci containing clustered catabolic genes with strong similarities to corresponding genes found in Novosphingobium aromaticivorans F199. In the first locus, two genes potentially encoding a terminal dioxygenase component, designated PhnI, were followed by a gene coding for an aryl alcohol dehydrogenase (phnB). The second locus contained five genes encoding an extradiol dioxygenase (phnC), a ferredoxin (phnA3), another oxygenase component (PhnII), and an isomerase (phnD). PhnI was found to be capable of converting several PAHs, including chrysene, to the corresponding dihydrodiols. The activity of PhnI was greatly enhanced upon coexpression of genes encoding a ferredoxin (phnA3) and a reductase (phnA4). Disruption of the phnA1a gene encoding the PhnI alpha subunit resulted in a mutant strain that had lost the ability to grow on PAHs. The recombinant PhnII enzyme overproduced in Escherichia coli functioned as a salicylate 1-hydroxylase. PhnII also used methylsalicylates and anthranilate as substrates. Our results indicated that a single enzyme (PhnI) was responsible for the initial attack of a range of PAHs, including chrysene, in strain CHY-1. Furthermore, the conversion of salicylate to catechol was catalyzed by a three-component oxygenase unrelated to known salicylate hydroxylases.
Contamination of soils and sediments by polycyclic aromatic hydrocarbons (PAHs) is widespread, which raises environmental concerns because many PAHs are cytotoxic and some are mutagenic and/or carcinogenic. A number of microorganisms that are able to degrade PAHs have been isolated (7), and bioremediation strategies based on microbial degradation of these pollutants have been proposed (45). However, while low-molecular-weight PAHs, like naphthalene, are readily degraded by bacteria, high-molecular-weight PAHs are more recalcitrant, and the catabolic pathways leading to their biodegradation are still poorly understood (19). Previous work showed that species belonging to the Burkholderia (18) and Stenotrophomonas (4) genera are able to degrade 4- and 5-ring PAHs. Several Mycobacterium species that are able to degrade phenanthrene, anthracene, and pyrene have been described, and a few of them could also metabolize fluoranthene, benz[a]anthracene, and benzo[a]pyrene (9, 16, 36, 41). The catabolic enzymes involved in the degradation of these PAHs have been investigated, which has led to the identification of dioxygenases that catalyze the initial attack of phenanthrene and pyrene (21, 23).
In recent years, sphingomonad species have been described for their ability to degrade a wide range of aromatic hydrocarbons, including mono- and polycyclic aromatic hydrocarbons (13, 32, 40, 48), naphthalene sulfonate (39), dibenzo-p-dioxin (1), dibenzothiophene, and methylated PAHs (29). The sequences and organization of catabolic genes responsible for PAH degradation were found to be remarkably similar in several sphingomonads (31), but these genes differed from those previously described for pseudomonads (46). In Novosphingobium aromaticivorans F199, sequence analysis of a large plasmid revealed that 79 genes (one-third of all genes) were probably involved in the catabolism of aromatic hydrocarbons (34). The catabolic genes had an unusual arrangement in that genes predicted to participate in the degradation of monoaromatic hydrocarbons were interspersed with genes potentially involved in biphenyl or PAH catabolism. Multiple copies of genes that potentially encoded ring-hydroxylating dioxygenase terminal components were identified, but none of these genes has been assigned a precise function in the catabolic pathway of PAHs. In a phenanthrene-degrading Sphingobium strain carrying catabolic genes very similar to those found in strain F199, it was recently found that three distinct oxygenases had salicylate hydroxylase activity (33). Genetic studies involving other PAH-degrading sphingomonads, as well as mutant strains impaired in the utilization of PAHs, identified a few genes essential for both the xylene and PAH degradation routes, like bphA3 encoding a dioxygenase-associated ferredoxin (22), and revealed the occurrence of convergent points in the catabolic pathways of two- and three-ring PAHs (40). Moreover, genes responsible for the salicylate lower pathway on the one hand and for the protocatechuate lower pathway on the other were shown to be required for PAH degradation (33) and for fluorene degradation (44), respectively. However, little is known about the enzymes involved in the initial steps of PAH degradation. In this respect, although in vivo studies have provided evidence that the initial attack of PAHs is catalyzed by a dioxygenase-type enzyme (15, 48), such an enzyme has not been identified yet in sphingomonads (31).
Chrysene is a four-ring PAH that is highly resistant to biodegradation. A few bacterial isolates that are capable of chrysene mineralization have been described, including Rhodococcus sp. strain UW1 (43) and a Stenotrophomonas maltophilia strain (4). A mutant of Sphingomonas yanoikuyae (B8/36) was found to oxidize chrysene to (+)-cis-3,4-dihydroxy-3,4-dihydrochrysene, a reaction most likely catalyzed by a ring-hydroxylating dioxygenase (6).
In this study, a Sphingomonas strain selected for its ability to grow on chrysene as a sole carbon and energy source was used to identify proteins involved in PAH catabolism. Proteins specifically induced in PAH-grown cells were subjected to peptide analysis. Peptide sequences were used to clone corresponding catabolic genes, and subsequent sequencing revealed two gene clusters that included genes encoding the terminal components of two ring-hydroxylating oxygenases. The physiological functions of these enzymes were investigated.
MATERIALS AND METHODS
Reagents.
PAHs, antibiotics, and most other chemicals were obtained from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Silicone oil (Rhodorsil 47V20), paraffin oil (Merck catalog no. 1.07174), and 2,2,4,4,6,8,8-heptamethylnonane (ICN catalog no. 157322) were purchased from Sodipro (Echirolles, France). [5,6,11,12-14C]chrysene was obtained from Chemsyn Science Laboratories (Lenexa, Kans.). Oligonucleotides, as well as isopropyl-β-d-thiogalactopyranoside (IPTG), were purchased from Eurogentec (Seraing, Belgium). Restriction enzymes were obtained from Promega (Promega France, Charbonnières, France) or Fermentas (Euromedex, Mundolsheim, France).
Bacterial strains, plasmids, and culture conditions.
Sphingomonas sp. strain CHY-1 was isolated from a PAH-contaminated soil by successive enrichment with chrysene as the sole carbon source, as will be described elsewhere (Willison, unpublished results). This bacterium was grown on a mineral salts medium (MSM) (43) supplemented with succinate (10 mM) or with a PAH in biphasic cultures as previously described (23). The PAHs were supplied at a concentration of 0.1 g/liter (naphthalene or phenanthrene) or 0.02 g/liter (chrysene). Sphingomonas sp. strain CHY-1 was also grown on PTYG medium (10 g of glucose per liter, 10 g of yeast extract per liter, 5 g of peptone per liter, 5 g of tryptone per liter, 75 mg of CaCl2 · 2H2O per liter, 0.6 g of MgSO4 per liter) and PTYG20 medium (PTYG medium diluted 20-fold). Growth was carried out at 25°C in Erlenmeyer flasks incubated in a rotary shaker at 150 rpm. Bacterial density was determined by measuring the optical density at 600 nm (OD600).
Escherichia coli and Pseudomonas putida strains and plasmids used in this study are listed in Table 1. Cultures were grown in rich broth (Luria-Bertani medium) containing appropriate antibiotics.
TABLE 1.
Bacterial strains and plasmids used
| Bacterial strain or plasmid | Relevant genotype and/or properties | Source or reference |
|---|---|---|
| Strains | ||
| Sphingomonas sp. strain CHY-1 | Wild type; Phe+ Chry+ | This study |
| Sphingomonas sp. strain M10-1 | Mutant of CHY-1 carrying a phnA1a::Gmr deletion-insertion; Phe− Chry− | This study |
| Pseudomonas putida KT2442 | Rifr | 10 |
| Escherichia coli DH5α | F−endA1 hsdR17 (rk− mk+) supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 φ80d lacZΔM15 | Invitrogen |
| Escherichia coli XL1-Blue MRA | Δ(mcrA) 183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 gyrA96 relA1 lac | Stratagene |
| Escherichia coli BL21(DE3) | F−dcm ompT hsdS (rB− mB−) gal λ (DE3) | Novagen |
| Escherichia coli BL21 AI | F−dcm ompT hsdS (rB− mB−) gal araB::T7RNAP-tetA | Invitrogen |
| Plasmids | ||
| pDrive | Kanr, cloning vector | QIAGEN |
| pGEM-T Easy | Apr, T-cloning vector | Promega |
| pCR-Blunt II-TOPO | Kanr, cloning vector | Invitrogen |
| pET15b | Ampr, expression vector | Invitrogen |
| pVLT31 | Tetr, expression vector | 10 |
| pSD1G3 | Supercos I carrying ca. 40-kb fragment of CHY-1 DNA including phnC | This study |
| pSD2C4 | Supercos I carrying ca. 40-kb fragment of CHY-1 DNA including phnA1aA2a-phnA2a | This study |
| pSD6 | pDrive containing phnA1a-phnA2a | This study |
| pSD8 | pET15b containing phnA1a-phnA2a | This study |
| pSD9 | pVLT31 containing phnA1a-phnA2a | This study |
| pDA11 | pDrive containing phnA1b | This study |
| pDB11 | pDrive containing phnA2b | This study |
| pDAB11 | pDrive containing phnA1b-phnA2b | This study |
| pED11 | pET9a carrying phnA1b-phnA2b | This study |
| pVED11 | pVLT31 containing phnA1b-phnA2b | This study |
| pSD11 | pDrive containing phnA3 | This study |
| pEBA3 | pET15b carrying phnA3 | This study |
| pDRA4 | pDrive containing phnA4 | This study |
| pEBA4 | pET15b carrying phnA4 | This study |
| pEB431 | pET15b carrying phnA4 and phnA3 | This study |
| pEB432 | pET15b carrying phnA4 and phnA3 in opposite orientation | This study |
| pSD13 | pCR-Blunt II-TOPO carrying a 3.06-kb fragment including phnA1a-phnA2a | This study |
| pSD14 | pSD13 carrying a phnA1a::Gmr deletion-insertion | This study |
[14C]chrysene mineralization experiments.
[14C]chrysene mineralization experiments were carried out in 33-ml Warburg flasks closed with rubber stoppers at the two outlets. A mixture of 0.4 μCi of [5,6,11,12-14C]chrysene and 100 nmol of unlabeled chrysene dissolved in acetone was introduced into empty flasks, and the solvent was allowed to evaporate. Next, 2 ml of a strain CHY-1 suspension with an OD600 around 3.0 was placed in the main chamber of each flask, and 0.4 ml of 1 N NaOH was added to the central well. The flasks were incubated at room temperature on a reciprocal shaker. The 14CO2 produced by chrysene mineralization and trapped in NaOH was estimated by scintillation counting. The NaOH solution was renewed and counted every hour for 6 h. The results from four replicate flasks were averaged and expressed in nanomoles of [14C]chrysene mineralized into CO2 normalized to the protein content of the bacterial samples. The initial rates were calculated from data obtained during the first 4 h of incubation.
Analysis of bacterial extracts by 1D and 2D PAGE.
Strain CHY-1 was grown to the late log phase on minimum medium with one of the carbon sources. Protein extracts were prepared and then subjected to one-dimensional (1D) or two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) as previously described (23). The proteins were visualized by silver staining or Coomassie blue staining.
Determination of peptide sequences.
Proteins of interest were cut out from Coomassie blue-stained 1D or 2D PAGE gels and then subjected to in-gel trypsin digestion, as previously described (12). Peptides were extracted and subjected to mass determination analysis and mass spectrometry (MS)-MS sequence analysis as previously reported (23).
In vivo 35S labeling of proteins.
Succinate-grown cultures of Sphingomonas were harvested in the mid-log phase and resuspended to an OD600 of 2.0 in MSM. Portions (25 ml) of the suspension were transferred to 100-ml flasks containing no carbon source or one of the following carbon sources: chrysene (5 mg), phenanthrene (25 mg), naphthalene (25 mg), or succinate (50 mM). PAHs were supplied as 5-ml solutions in paraffin oil. After 1 h of incubation at 25°C, the bacterial suspensions were supplemented with 0.14 mCi (5.18 MBq) of a 35S-labeled mixture of methionine and cysteine (Easytag express protein labeling mixture; NEN Life Science Products) combined with a mixture of two unlabeled amino acids as carriers (12 μM Met plus 6 μM Cys). The bacteria were then incubated for different times depending on the carbon source used (6 h for naphthalene or phenanthrene, 24 h for succinate, or 48 h for chrysene or no carbon source). Cells were then harvested by centrifugation, washed once in MSM, and resuspended in 0.25 ml of 0.1 M Tris-HCl-10 mM EDTA (pH 8.0). Bacterial extracts were prepared as described above and were analyzed by 1D and 2D PAGE. Gels were stained with Coomassie blue, dried, and exposed to X-ray film (Hyperfilm-βmax; Amersham Biosciences) for 4 days.
General DNA manipulations.
Restriction enzyme digestion, ligation, and agarose gel electrophoresis were performed by using standard procedures (35). DNA fragments were purified from agarose gels with a QIAEX II extraction spin kit (QIAGEN S.A., Courtaboeuf, France). Plasmids were prepared by using Nucleospin or Nucleobond AX purification kits from Macherey-Nagel SA (Hoerdt, France). Routine PCRs were carried out by using Taq polymerase from Promega. Sphingomonas DNA was isolated from PTYG medium-grown cells harvested in the stationary phase by using a procedure described previously (23).
Construction of a genomic library.
Genomic DNA from Sphingomonas sp. strain CHY-1 was partially digested with Sau3AI, and approximately 40-kb fragments were purified by agarose gel electrophoresis and cloned into BamHI-predigested Supercos I vector according to the instructions of the supplier (Stratagene Europe, Amsterdam, The Netherlands). The ligated DNA was packaged by using the Gigapack III gold packaging extract (Stratagene) and was propagated as recombinant phages in E. coli XL1-Blue MRA. Clones of E. coli containing corresponding cosmids were transferred to 96-well microtiter plates to facilitate screening of the library.
Cloning of PAH-catabolic genes from strain CHY-1 and DNA sequence analysis.
A set of degenerate primers (5′-TAYGCNGCNGARGTNGCNGGNATG-3′ and 5′-TCRTTNCCRTGNCCRAADATRTC-3′) was designed based on the internal sequences YAAEVAGM and DIFGHGNE of protein P1. By using these primers, a 0.8-kb DNA fragment was amplified by PCR with Sphingomonas DNA as the template. The nucleotide sequence of this fragment was similar to the sequence of an internal region of the bphC gene found on plasmid pNL1 in N. aromaticivorans. The same primers were then used to screen the genomic library by serial rounds of PCR, as previously described (23). One positive cosmid, designated pSD1G3, was selected and subjected to DNA sequencing by gene walking, starting with the primers described above. A sequence analysis was performed for a 3,900-bp DNA region that included the phnC gene encoding the P1 protein. A second pair of degenerate primers, P6-RV (5′-GGRTCYTCCATCCANAC-3′; reverse primer), based on the available internal sequence of protein P6 (VWMEDP), and Diox-1 (5′-CGNTGCCCCTAYCAYGGNTGG-3′; forward primer), based on the internal consensus sequence CPYHGW found in most dioxygenase α-subunit sequences, was used to amplify a 1.4-kb fragment from the genomic DNA of strain CHY-1. After sequence analysis of this fragment, PCR-based screening of the genomic library was performed as described above with primers P6-RV and Diox-1, which resulted in the selection of one cosmid, designated pSD2C4, which was subjected to further sequence analysis by gene walking. The sequence of a 4,432-bp fragment was determined on both strands. DNA sequence analyses, as well as protein sequence comparison and alignment, were performed by using BLAST or programs available on the Infobiogen server (http://www.infobiogen.fr/services/deambulum/fr/index.html.)
Construction of plasmids for protein overexpression.
Construction of the plasmids used in this study (Table 1) involved multiple PCR amplifications and intermediate steps which, for the sake of clarity, are only outlined below (a detailed description is available upon request). The phnA1a and phnA2a genes were amplified by PCR by using cosmid pSD2C4 as the template and were cloned into pET15b, producing pSD8. The phnA1a and phnA2a genes were also transferred into pVLT31 as a 2.4-kb XbaI-HindIII fragment from pSD8, yielding pSD9.
The phnA1b and phnA2b genes were separately amplified from cosmid pSD1G3 and cloned into pDrive to obtain pDA11 and pDB11, respectively. The phnA2b coding sequence, including a suitable ribosome-binding site at the 5′ end, was subcloned downstream from phnA1b in pDA11 to obtain pDAB11. The phnA1b-phnA2b sequence was then cloned into pET9a, producing plasmid pED11. A derivative of pVLT31 carrying phnA1b-phnA2b in the same orientation as the Ptac promoter was also constructed and designated pVED11.
The phnA3 and phnA4 genes were also amplified with cosmid pSD1G3 as the template and were separately cloned into pDrive to obtain pSD11 and pDRA4, respectively. The two genes were then introduced into pET15b, giving pEB431 and pEB432. In pEB431, phnA3 is oriented like phnA4 and both genes are under the control of the T7 promoter, whereas in pEB432, phnA3 is in opposite direction. Plasmids pSD8, pSD9, pVED11 pEB431, and pEB432 were introduced into E. coli strain BL21(DE3) by transformation. Plasmid pSD9 was transferred to P. putida strain KT2442 by conjugation.
In vivo assays of recombinant PhnI and PhnII activity.
The activity of recombinant PhnI expressed in E. coli was assayed as follows. Strain BL21(DE3)(pSD9)(pEB431) was grown in Luria-Bertani medium to an OD600 of 1.0 and then incubated overnight at 25°C with 0.1 mM IPTG. Strains in which pVLT31 replaced pSD9 or pEB432 replaced pEB431 were also tested. Cells were harvested, resuspended to an OD600 of 2.0 in M9 minimal medium containing 0.2% glucose, and distributed as 10-ml aliquots into 50-ml Falcon tubes containing 100 μg of PAH dissolved in 2 ml of silicone oil. The tubes were incubated at 200 rpm for 6 to 24 h at 25°C. In some experiments, PhnI activity was assayed in P. putida KT2442 carrying pSD9. The procedure was similar to that described above, except that 0.2 mM IPTG was used to induce PhnI gene expression. Hydrosoluble products were analyzed for samples of the aqueous phase, which were centrifuged and then passed through solid-phase extraction columns (Upti-clean C18U; 0.5 g; Interchim, Montluçon, France). The columns were washed with 10 ml of water and then eluted with 2 ml of methanol. Extracts were evaporated under a vacuum and derivatized with n-butylboronate (NBB). Gas chromatography (GC)-MS analysis was carried out as previously described (23), except that dihydrodiols were detected by selected ion monitoring by using the calculated mass of the NBB derivative. To determine the mass spectra of the putative dihydrodiols formed from chrysene and benz[a]anthracene, cells were incubated for 24 h in 500 ml of growth medium containing the PAH dissolved in 2,2,4,4,6,8,8-heptamethylnonane. Extracts were then prepared from these cultures as described above, derivatized with NBB, and analyzed by GC-MS in the total ion current mode.
PhnII activity was assayed by using strain BL21(DE3) carrying pVED11 and pEB431 or other pET15b derivatives as controls. Bacteria were grown in rich medium, subjected to IPTG induction, and resuspended in M9 minimum medium as described above for PhnI. Aliquots (10 ml) of a cell suspension were incubated with either 1 mM salicylate, 1 mM (methyl)salicylate, or 1 mM anthranilate. Other substrates, including biphenyl, naphthalene, phenanthrene, were tested as 2-ml solutions (0.05 g/liter) in silicone oil. After incubation at 25°C with agitation for various times (see below), products released into the aqueous phase were either directly analyzed by high-performance liquid chromatography (HPLC) or extracted with ethyl acetate. After solvent evaporation under a stream of nitrogen gas, the extracts were derivatized with NBB and subjected to GC-MS analysis in the total ion current mode as described above.
HPLC analysis of PhnII substrates and products.
Samples to be analyzed were acidified by adding 0.1% acetic acid and were filtered through 0.22-μm-pore-size filter units. HPLC was performed with a Kontron apparatus and a Zorbax C8 column (4.6 by 150 mm; 5 μm; Agilent Technologies). The column was eluted at a rate of 1 ml/min by using a linear 0 to 50% methanol gradient in 0.1% acetic acid for 15 min, and compounds were detected by UV absorption at 276 and 296 nm with a Kontron 230 detector. Peaks were identified and quantified by using catechol and salicylate or their methyl derivatives as standards.
Other analytical methods.
The whole-cell protein content was determined by the method of Lowry et al. (26), after bacterial samples were treated with 0.2 M NaOH for 1 h at 90°C. Routine protein assays were done with a bicinchoninic acid reagent kit (Pierce) by using bovine serum albumin as a standard. Sodium dodecyl sulfate (SDS)-PAGE on mini slab gels was performed as previously described (17).
Construction of phnA1a null mutant strain.
A 3.06-kb DNA region encompassing phnA1a and phnA2a and part of phnB was amplified by PCR with Expand High Fidelity DNA polymerase by using pSD2C4 as the template and the following primers: 5′-CTCTAGATCTCTCCCAACAGC-3′ and 5′-GTCTAGACGGTGTGACACATG-3′ (underlined sequences are XbaI sites). The resulting product was cloned into pCR-Blunt II-TOPO to obtain plasmid pSD13. This plasmid was digested with ClaI and NheI, creating a 1,224-bp deletion in the phnA1a coding sequence. A gentamicin resistance (Gmr) cassette was extracted from plasmid pFRKII (11) as a 1.56-kb PstI-HindIII fragment and ligated into pBluescript (Stratagene). The Gmr cassette was reisolated as a ClaI-SpeI fragment, which was cloned into pSD13 deleted from phnA1a, creating pSD14, which carried a 3.3-kb insert with a phnA1a::Gmr deletion-insertion. This insert was isolated from pSD14 as an XbaI fragment of linear DNA, which was directly introduced into Sphingomonas sp. strain CHY-1 by electroporation. For this purpose, cells were grown on 0.5 liter of PTYG medium to the early stationary phase and were made electrocompetent by double washing in ice-cold water and suspension in 2 ml of 20% glycerol. Aliquots (50 μl) of the suspension were subjected to a 1.25-kV pulse in the presence of 250 ng of DNA. After growth on solid PTYG20 medium for 3 days at 30°C, gentamicin-resistant strains were selected at a rate of approximately 3 × 10−6. Four clones were analyzed by PCR amplification of genomic DNA with the primers described above, all of which gave a 3.3-kb fragment instead of a 3.06-kb fragment for the wild-type strain, which is consistent with the hypothesis that the phnA1a::Gmr deletion-insertion was incorporated into the genome by double recombination. This was verified by Southern hybridization in which a 525-bp fragment spanning a region between the 3′ end of phnA1a and the 5′ end of phnA2a was used as a probe. The probe was obtained by PCR amplification by using pSD2C4 as the template and primers 5′-CCGATTGGGCGAGCGTGAAGG-3′ and 5′-CTGGCGGCGGTTGCGGTAGAC-3′ and was subsequently labeled with digoxigenin by using a DIG-DNA labeling kit (Roche Diagnostics). Genomic DNA from CHY-1 and two mutant strains were digested with either SalI or EcoRI, separated by agarose gel electrophoresis, and hybridized with the probe as recommended by the manufacturer. Two hybridizing bands at 3.0 and 1.5 kb (CHY-1) and at 3.0 and 1.8 kb (mutants) were obtained for the SalI digests, which was consistent with the presence of a SalI site at the 5′ end of phnA2a. The EcoRI digests yielded a single band at 0.66 kb for CHY-1 and at 2.3 kb for the two mutants.
Nucleotide sequence accession numbers.
The nucleotide sequences described in this report have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers AJ633551 and AJ633552.
RESULTS
Induction of chrysene mineralization in Sphingomonas sp. strain CHY-1.
A bacterium capable of growing on chrysene as the sole carbon source was isolated from a PAH-polluted soil after several rounds of selective enrichment on mineral salts medium supplemented with chrysene (Willison, unpublished). This bacterium is related to the sphingomonads based on its 16S rRNA gene sequence, and the closest relative in the databases is Sphingomonas xenophaga (96% sequence identity). It was therefore provisionally designated Sphingomonas sp. strain CHY-1. In addition to chrysene, strain CHY-1 can grow on naphthalene, phenanthrene, and anthracene (Fig. 1), but it is unable to grow on, or to metabolize at significant rates, acenaphthene, fluorene, fluoranthene, pyrene, benz[a]anthracene, and benzo[a]pyrene when these compounds are provided as sole carbon substrates. Bacterial cells grown on PAH substrates or on glucose or succinate as alternate carbon sources were compared with respect to the ability to mineralize [14C]chrysene (Table 2). Chrysene-grown cells showed the highest activity, with initial mineralization rates of 4.94 ± 0.20 nmol/h/mg of protein. Naphthalene- and phenanthrene-grown cells displayed intermediate levels of activity, whereas succinate-grown cells were essentially inactive. These results suggested that the enzymes responsible for chrysene degradation were not expressed in succinate-grown cells, whereas intermediate levels of these enzymes might be expected in cells grown on PAHs other than chrysene.
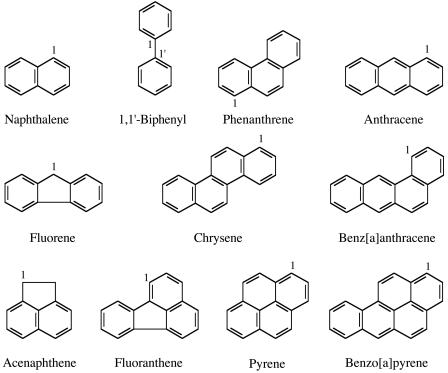
FIG. 1.
Chemical structures of 1,1′-biphenyl and the 10 PAHs tested in this study. The positions of substitutions in the aromatic rings are numbered clockwise from the carbon atoms shown. Sphingomonas sp. strain CHY-1 was isolated for its ability to grow on chrysene, but it is also able to utilize naphthalene, phenanthrene, and anthracene as growth substrates. These compounds, as well as 1,1′-biphenyl, fluorene, and benz[a]anthracene, were converted to the corresponding dihydrodiols by recombinant E. coli strains overproducing PhnI dioxygenase, whereas acenaphthene, fluoranthene, pyrene, and benzo[a]pyrene were not oxidized at detectable rates by these strains.
TABLE 2.
Initial rates of chrysene mineralization by strain CHY-1 grown on various carbon sources
| Carbon source(s)a | Chrysene mineralization rate (nmol/h/mg of protein)b |
|---|---|
| Succinate | 0.16 ± 0.025 |
| Glucose | 0.43 ± 0.078 |
| Naphthalene | 2.56 ± 0.27 |
| Phenanthrene | 3.99 ± 0.39 |
| Naphthalene + chrysene | 2.29 ± 0.12 |
| Phenanthrene + chrysene | 2.70 ± 0.18 |
| Chrysene | 4.94 ± 0.20 |
The PAHs used as growth substrates were provided as solutions in silicone oil.
The values are means ± standard deviations for four separate determinations.
Identification of proteins involved in PAH catabolism.
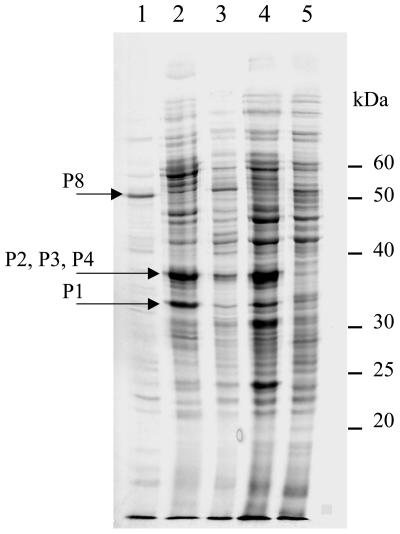
When the protein patterns of PAH-grown and succinate-grown cells were compared by SDS-PAGE, a few prominent polypeptides appeared to be synthesized only in the presence of PAH. In vivo 35S labeling performed as described in Material and Methods highlighted such PAH-induced polypeptides (Fig. 2). Three protein bands at Mr of ca. 33,000, 37,000, and 50,000, which were highly labeled and/or dominant on SDS-PAGE gels prepared from PAH-grown cells, were subjected to peptide sequence analysis (Table 3). The 33,000-Mr band produced a set of six tryptic peptides, all of which perfectly matched internal sequences of the bphC gene product from N. aromaticivorans encoded by the pNL1 catabolic plasmid (34). Protein P1 is therefore analogous to BphC, which was described as an extradiol ring cleavage dioxygenase. The 37,000-Mr band yielded several peptides which could be grouped into three sets, each of which likely corresponded to a separate protein (Table 3). The three polypeptides identified showed remarkable similarities to the products of nahE, xylQ, and a gene with an unknown function (orf064) found in a large aromatic hydrocarbon catabolic gene cluster in three sphingomonads (22, 33, 34). Peptide analysis of a 50,000-Mr protein band from an extract of chrysene-grown cells resulted in identification of a protein resembling an S-adenosylhomocysteine hydrolase (protein P8), which is probably not related to aromatic hydrocarbon degradation but comigrated with the protein of interest (50-kDa labeled band from chrysene-induced cells in Fig. 2).
FIG. 2.
SDS-PAGE analysis and autoradiography of 35S-labeled protein extracts from Sphingomonas sp. strain CHY-1. Protein extracts were prepared as described in Materials and Methods from bacteria subjected to in vivo labeling in the presence of the following carbon sources: chrysene (lane 1), naphthalene and chrysene (lane 2), phenanthrene (lane 3), naphthalene (lane 4), and succinate (lane 5). The arrows indicate major labeled bands which were subjected to peptide analysis. For this, extracts from unlabeled cells grown on naphthalene plus chrysene or on chrysene alone were analyzed on a similar SDS-PAGE gel, and protein bands with relative mobilities identical to those of the labeled bands of interest were analyzed. P1, P2, P3, P4, and P8 are the proteins described in Table 3.
TABLE 3.
Sequence analysis of PAH-induced polypeptides from strain CHY-1 and possible functions based on sequence similarities
| Polypeptide | Mr | Internal peptide sequencesa | Best database matchb | % Amino acid identityc | Possible function |
|---|---|---|---|---|---|
| P1 (1D) | 33,000 | SYAAEVAGMEIVDEGEGDR | BphC (AF079317) 22-40 | 100 | Extradiol ring cleavage |
| DIFGHGNEAAGYGMDIPLG | 281-299 | dioxygenase | |||
| [LI]V[LI]HASDSDD[LI]AY[LI]GWR | 52-68 | ||||
| [LI]TAAG[LI]S[LI]TVTASEAEAR | 82-98 | ||||
| QDVPAAAAFYGLR | 156-166 | ||||
| XXTE[LI]GY[LI]G[LI]TVTN[LI]DAWR | 5-21 | ||||
| P2 (1D) | 37,000 | [KR]XXSTYN[LI]A[LI]EK | NahE (AB091692) 270-281 | 95 | 2′-Hydroxybenzal- |
| [KR]VPV[FM]VGTTC[LI]NTR | 86-99 | pyruvate aldolase | |||
| [KR]([LI][LI])P[LI]D[FM]DYYGAAR | 197-210 | ||||
| P3 (1D) | RTPVSGTVLSWDLMR | ORF064 (AF079317) 148-162 | 100 | Unknown | |
| WTIVLDPSVYTAIEK | 88-102 | ||||
| TATYASALANVK | 271-282 | ||||
| AGPILPPVADVTPEK | 292-306 | ||||
| LADIDTVIGVK | 164-174 | ||||
| P4 (1D) | [KR]XX[LI][FM]T[LI]TDQVDEDA[LI]R | XylQ (AB091692) 205-221 | 100 | Acetaldehyde dehydrogenase | |
| P5 (2D) | 21,000 | AFVPLFAPATSDEAK | GST (AF001779) 108-122 | 100 | Glutathione S- |
| TEAGEDFLTVNPSGK | 37-51 | transferase | |||
| P6 (2D) | 21,000 | [KR][LI]YSG[QK]VWMEDPRR | BphA2f (AF079317) 84-95 | 83 | β Subunit of dioxygenase |
| P7 (2D) | 52,000 | IGLFGGAGVGK(T) | AtpD (AJ294407) 46-57 | 100 | β Subunit of ATP |
| VALVYGQMNEPPGAR | 31-45 | synthase | |||
| P8 (1D) | 50,000 | LPFPAINVNDSVTK | Saro2122(ZP_00095096) 211-224 | 74 | S-Adenosylhomocysteine hydrolase |
| [KR]XXS[LI][QK]EYWDY[FM]VR | 110-117 | ||||
| [KR]VAACVAG[FM]GDVGK | 254-264 |
Boldface letters indicate identical residues in relevant peptides and best database matches. Letters in parentheses indicate uncertain residues, and letters in brackets indicate residues that are equally possible at the position. X, undetermined residue.
Accession numbers are indicated in parentheses. The numbers not in parentheses indicate the positions in the sequence of the protein homologue which best match relevant peptide sequences.
Calculated from available peptide sequences.
Further analysis of strain CHY-1 protein extracts by 2D PAGE revealed 15 additional PAH-induced proteins, 5 of which were chosen for peptide sequence analysis. A 21-kDa protein (protein P5) was identified as a glutathione S-transferase (GST) (Table 3) similar to the GST found in several sphingomonads, including Sphingomonas paucimobilis EPA505 (25) and N. aromaticivorans (BphK) (34). GST-encoding genes are often associated with catabolic genes in PAH-degrading bacteria (25), but their exact function is unknown (42). Protein P6 yielded a 14-residue tryptic peptide that closely matched an internal sequence of the bphA2f gene product from N. aromaticivorans. The latter protein was identified as a ring-hydroxylating dioxygenase beta subunit (34). Lastly, the 52,000-Mr protein found in chrysene-grown cells (protein P7) was identified as a putative ATP synthase β subunit. Its role, if any, in PAH catabolism is not yet clear.
Cloning and sequence analysis of two catabolic gene clusters.
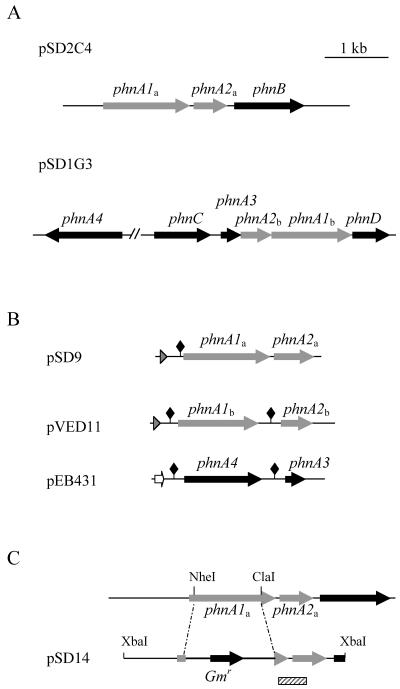
Based on the peptide sequence analysis described above, proteins P1 and P6 might be expected to participate in key steps of PAH degradation in strain CHY-1. The genes encoding these proteins were therefore chosen as targets of a cloning strategy to identify relevant catabolic genes. To this end, a cosmid library consisting of ∼40-kb fragments of genomic DNA was constructed and screened by using probes specific for P1 or P6. Two cosmids, designated pSD1G3 and pSD2C4, which carried the genes of interest, were selected and subjected to DNA sequence analysis. The results of this analysis revealed two clusters of putative catabolic genes (Fig. 2). In the first locus, the phnC gene encoding protein P1, a meta-cleavage dioxygenase, was followed by genes coding for a ferredoxin (phnA3), beta and alpha subunits of a putative ring-hydroxylating oxygenase (phnA2b, phnA1b), and an isomerase (phnD) similar to NahD involved in naphthalene catabolism. In the second locus, two genes encoding the subunits of another oxygenase component (phnA1a, phnA2a) were followed by a gene coding for a putative aryl alcohol dehydrogenase (phnB). The two defined loci were very similar in both sequence and gene arrangement to DNA regions found previously on the catabolic plasmid pNL1 from N. aromaticivorans (34) and in related sphingomonads (31). Since none of these gene products has been assigned a specific function in PAH catabolism, we studied the properties of the two oxygenase components found in strain CHY-1, which are referred to in this paper as PhnI (the product of phnA1a-phnA2a) and PhnII (the product of phnA1b-phnA2b).
Overproduction of PhnI and auxiliary protein requirement for in vivo activity.
When the phnA1a-phnA2a coding sequence was overexpressed in E. coli BL21AI by using the T7 promoter carried on plasmid pSD8, two polypeptides of the expected sizes were overproduced in recombinant cells, as detected by SDS-PAGE. However, purification attempts led to low yields of a colorless protein, which probably corresponded to a misfolded or apoform of the enzyme. Moreover, recombinant cells carrying pSD8 did not show any hydroxylase activity with naphthalene, phenanthrene, or biphenyl. The phnA1a-phnA2a sequence was then subcloned as a His-tagged fusion behind the Ptac promoter in the broad-host-range plasmid pVLT31, and the resulting plasmid, designated pSD9, was introduced into either E. coli BL21 or P. putida KT2442. When incubated with IPTG as an inducer, both types of recipient cells were found to overproduce high levels of 20- and 50-kDa recombinant proteins. In addition, a brown protein was recovered upon purification by Ni-immobilized metal affinity chromatography, which produced an absorbance spectrum typical for a metalloprotein containing a Rieske type of cluster (data not shown). When induced recombinant cells harboring pSD9 were incubated in the presence of phenanthrene, an oxidation product was generated, which was identified as cis-3,4-dihydroxy-3,4-dihydrophenanthrene since it had GC-MS characteristics identical to those of a product identified as this compound in previous work (23). However, the low rate of conversion observed suggested that the host cells did not provide PhnI with the appropriate electron carriers. The phnA3 gene product described above as a putative ferredoxin (Fig. 3) might be expected to serve as an electron donor to PhnI. Additionally, a gene possibly encoding a reductase was detected by PCR on cosmid pSD1G3 carrying phnA3 and other phn genes (Fig. 3). This gene, designated phnA4, was 97.8% identical to bphA4 found on plasmid pNL1 in N. aromaticivorans (34). The phnA4 gene was cloned together with phnA3 into pET15b, and the resulting plasmid, designated pEB431 (Fig. 3B), was introduced together with plasmid pSD9 into E. coli by transformation. Under appropriate expression conditions, strain BL21(DE3)(pSD9)(pEB431) was found to convert 35 times as much phenanthrene to the corresponding dihydrodiol as the strain lacking pEB431 converted (Table 4). Moreover, strain BL21(DE3)(pSD9)(pEB342) carrying a variant of pEB431 in which the phnA3 gene was cloned in the orientation opposite the direction of transcription displayed an intermediate level of activity that was equivalent to about 15% of the maximum rate. Analysis of controls of protein expression by SDS-PAGE showed that cell extracts from strain BL21(DE3)(pSD9) carrying pEB431 or pEB432 produced an equivalent amount of the PhnI α and β subunits, as well as an additional 45-kDa protein whose size corresponded to the polypeptide size of PhnA4 (data not shown). The PhnA3 product was not detected, perhaps because the standard SDS-PAGE method employed was not appropriate for resolving polypeptides in the 10,000- to 15,000-Mr range. These results indicated that the phnA3 and phnA4 gene products were correctly synthesized in E. coli host cells and supported phenanthrene dihydroxylation activity in vivo by supplying electrons to the PhnI dioxygenase component.
FIG. 3.
Physical maps of two clusters of PAH-catabolic genes from strain CHY-1 and relevant plasmid constructions. (A) Maps of the phn gene clusters found on cosmids pSD1G3 and pSD2C4. The position and orientation of the phnA4 gene with respect to the other phn genes carried on cosmid pSD1G3 are uncertain. (B) Schematic representation of plasmid constructions used for recombinant overproduction of the PhnI and PhnII enzymes, as well as accessory electron carriers. In plasmids pSD9 and pVED11, genes are under the control of the Ptac promoter (grey arrowhead), whereas in pEB431 expression is directed by a T7 promoter (open arrow). The solid diamonds indicate Shine-Dalgarno sequences introduced in front of gene coding sequences to improve recombinant expression. (C) Genetic construction used for phnA1a inactivation. Plasmid pSD14 carried a 3.4-kb insert in which an NheI-ClaI internal fragment of phnA1a was replaced by a gentamicin resistance cassette. The hatched bar shows the position of the DNA probe used for Southern hybridization analysis of mutant Sphingomonas strains.
TABLE 4.
Relative activities of PhnI and PhnII in recombinant cells depending on coexpression of PhnA3 and PhnA4
| Expression system | Gene product(s) | Hydroxylase activity with:
|
|
|---|---|---|---|
| Phenanthrenea | Salicylateb | ||
| BL21(DE3)(pSD9) | PhnI | 2.8 | —c |
| BL21(DE3)(pSD9)(pEB432) | PhnI + PhnA4 | 14.8 | — |
| BL21(DE3)(pSD9)(pEB431) | PhnI + PhnA4 + PhnA3 | 100 | — |
| KT2442(pSD9) | PhnI | 28.7 | — |
| BL21(DE3)(pVDE11) | PhnII | — | 4.6 |
| BL21(DE3)(pVDE11)(pEB432) | PhnII + PhnA4 | — | 8.5 |
| BL21(DE3)(pVDE11)(pEBA3) | PhnII + PhnA3 | — | 34.8 |
| BL21(DE3)(pVDE11)(pEB431) | PhnII + PhnA4 + PhnA3 | NDd | 100 |
Determined by GC-MS analysis of the diol product formed after 6 h of incubation. The values are the relative GC-MS peak areas normalized to 100% maximal activity.
Calculated from HPLC measurements and expressed as percentages of the maximal activity (100% was 0.24 μmol of catechol/h/mg of protein).
—, not tested.
ND, no product detected.
Substrate range of PhnI.
The recombinant E. coli strain producing PhnI, PhnA3, and PhnA4 was incubated separately with 10 representative PAHs, as well as biphenyl (Fig. 1), and the hydrosoluble products released into the culture medium were extracted and analyzed by GC-MS. Six PAHs gave rise to the formation of at least one detectable product identified as a dihydrodiol derivative based on mass spectroscopic analysis (Table 5). When strain BL21(DE3)(pVLT31)(pEB431), which lacked PhnI, was incubated with PAHs under identical conditions, no diol product could be detected, demonstrating that PhnI was responsible for PAH transformation. Naphthalene was the preferred substrate, and phenanthrene, anthracene, and biphenyl were converted at significant but lower rates to the corresponding diols. Interestingly, PhnI also oxidized fluorene, chrysene, and benz[a]anthracene, and the latter substrate gave rise to three compounds which probably corresponded to isomeric dihydrodiol derivatives. The putative chrysene dihydrodiol had a GC retention time of 27.1 min for the NBB derivative, and the mass spectrum was characterized by the following principal components: m/z = 329 (relative abundance 23%; M++1); 328 (100%; M+); 327 (49%; M+−1); 271 (15%; M+−C4H9); 270 (17%); 244 (18%; M+−C4H9,−BO); 229 (14%); 228 (48%; M+−C4H9,−BO,−O); 227 (26%); 226 (47%; M+− M+−C4H9,−BO,−OH2,); 216 (47%; M+−C4H9,−BO,−CO); 215 (58%); 213 (14%); 113 (12%; M+− C4H9,−BO,−CO,−C8H7). In concentrated extracts, three putative benz[a]anthracene hydrodiols were detected after derivatization, and they had retention times of 26.6, 27.1, and 27.6 min. The major product, the product with the shortest retention time, showed the following mass spectral characteristics: m/z = 329 (relative abundance 18%); 328 (100%); 327 (67%); 271 (17%); 244 (17%); 229 (19%); 228 (58%); 227 (16%); 226 (44%); 216 (32%); 215 (36%); 213 (19%). The amounts of the other two products were not sufficient to allow accurate quantification of their mass spectra, but both NBB derivatives showed characteristic mass spectral peaks at m/z = 328, 271, 228, and 216.
TABLE 5.
PAH selectivity of PhnI as expressed in recombinant form in E. coli
| Substratea | Product | Molecular mass of NBB derivative (Da) | Retention time (min) | Relative amt (%)b |
|---|---|---|---|---|
| Naphthalene | 1,2-Dihydroxy-1,2-dihydronaphthalene | 228 | 15.2 | 100 |
| Biphenyl | 2,3-Dihydroxy-2,3-dihydrobiphenyl | 254 | 17.1 | 20.5 |
| Phenanthrene | 3,4-Dihydroxy-3,4-dihydrophenanthrene | 278 | 20.5 | 14.4 |
| Anthracene | Dihydrodiol | 278 | 20.9 | 10.2 |
| Fluorene | Dihydrodiol | 266 | 18.8 | 1.03 |
| Chrysene | Dihydrodiol | 328 | 27.1 | 1.5 |
| Benz[a]anthracene | Dihydrodiol 1 | 328 | 26.5 | 0.10 |
| Dihydrodiol 2 | 27.1 | 0.13 | ||
| Dihydrodiol 3 | 27.6 | NDc |
Acenaphthene, pyrene, fluoranthene, and benzo[a]pyrene did not give any detectable product.
Calculated from the GC-MS-selected ion monitoring peak areas of the NBB derivatives of the products formed after 6 h of incubation and expressed as percentages of the maximum peak area. The values are averages of at least two separate determinations.
ND, not detected.
Using [14C]chrysene as the substrate, we observed that strain BL21(DE3)(pSD9)(pEB431) converted it to a single radioactive product, as judged by thin-layer chromatography analysis and autoradiography, whereas control cells lacking PhnI did not release any hydrosoluble radioactive product. Assuming complete recovery of the labeled diol upon extraction from the culture medium, we estimated that recombinant cells converted chrysene at a rate of 0.0064 nmol/h/mg of protein at 25°C.
Overproduction of the PhnII oxygenase component and catalytic activity.
The structural genes for PhnII are unusually arranged in the genome of CHY-1 in that the phnA2b gene encoding the small subunit precedes phnA1b encoding the large subunit. Attempts to overexpress the two genes in this order in E. coli led to virtually no synthesis of the large subunit, most likely because the phnA1b gene was not preceded by a ribosome-binding site suitable for expression in enteric bacteria. Plasmid pED11 bearing phnA1b and phnA2b with appropriate translation signals was therefore constructed and transferred into strain BL21(DE3). Cells carrying pED11 produced two recombinant polypeptides with Mrs consistent with the size of the PhnA1b product (Mr, 47,000) and the PhnA2b product (Mr, 18,000) but did not show any detectable hydroxylase activity. On the other hand, salicylate hydroxylase activity was detected in strain BL21(DE3) carrying pVED11, a plasmid resulting from the transfer of phnA1b-phnA2b into pVLT31. The hydroxylase activity was dramatically stimulated in cells coexpressing the phnA3 and phnA4 genes, whereas intermediate levels were observed in cells expressing either phnA3 or phnA4 (Table 4). These results suggested that maximal PhnII activity required specific reductase and ferredoxin components which were only partially replaced by endogenous E. coli electron carriers. In this respect, the activity was much higher in cells coexpressing phnA3 alone than in cells coexpressing only phnA4, indicating that the requirement for the ferredoxin was greater than the requirement for the reductase. Kinetic experiments performed over a 4-h period showed that cells expressing the three enzyme components converted salicylate into catechol at a fairly linear rate, 0.240 μmol · h−1 · mg of protein−1, although lower conversion rates were occasionally observed, probably depending on the level of expression of the enzyme components in recombinant cells. Quantitative measurements showed that salicylate was stoichiometrically transformed into catechol. When compounds analogous to salicylate were tested as alternative substrates, it was found that PhnII catalyzed the hydroxylation of 3-, 4-, and 5-methylsalicylates to the corresponding catechols and that there was a preference for the latter substrate (Table 6). In addition, anthranilate was converted to a product which was identified as 2-aminophenol by HPLC and GC-MS analyses. Recombinant cells overproducing PhnII, PhnA3, and PhnA4 were unable to oxidize biphenyl, naphthalene, or phenanthrene at significant rates.
TABLE 6.
Catalytic activity of PhnII with methylsalicylates and anthranilate
| Substrate | Product | Activitya
|
|
|---|---|---|---|
| μmol · h−1 · mg of protein−1 | % | ||
| Salicylate | Catechol | 0.239 | 100 |
| 3-Methylsalicylate | 3-Methylcatechol | 0.127 | 53 |
| 4-Methylsalicylate | 4-Methylcatechol | 0.203 | 85 |
| 5-Methylsalicylate | 4-Methylcatechol | 0.241 | 101 |
| Anthranilate | 2-Aminophenol | 0.075 | 31 |
Determined from HPLC measurements performed after 2 h of incubation of the substrates with BL21(DE3)(pVED11)(pEB431) recombinant cells.
Construction and functional analysis of a mutant carrying a disrupted phnA1 gene.
As described above, the PhnI dioxygenase component, but not PhnII, could convert various PAHs to the corresponding dihydrodiols, suggesting that the former enzyme was responsible for the initial oxidative attack of PAHs in strain CHY-1. In order to assess the physiological function of PhnI, a mutant strain carrying a deletion-insertion in phnA1a was constructed. For this purpose, a phnA1a::Gmr deletion-insertion was constructed (Fig. 2) and introduced as a linear DNA fragment into strain CHY-1 by electroporation. Gmr strains were selected at a rate of about 3 × 10−6. Two clones were found to contain the desired insertion integrated into the genome by double recombination, as verified by Southern blot hybridization (data not shown). One mutant strain, designated M10-1, was chosen for further study. This mutant showed normal growth on glucose or succinate but was unable to grow on naphthalene, phenanthrene, chrysene, and anthracene. Compared to the wild-type strain, the M10-1 mutant had totally lost the ability to mineralize [14C]chrysene. In M10-1, the orientation of the Gmr cassette was the same as the orientation of the phnA1a-phnA2a-phnB cluster, so that the genes located downstream from phnA1a should have been transcribed from the promoter carried on the Gmr cassette. It therefore seems plausible that the observed phenotype of the M10-1 mutant can be attributed to inactivation of phnA1a, leading to the loss of PhnI, rather than to a polar effect on transcription of a catabolic gene located downstream. These results suggest that PhnI is absolutely required for PAH catabolism in strain CHY-1.
DISCUSSION
Sphingomonads have attracted growing interest since several members of this bacterial group show remarkable metabolic capabilities that allow them to degrade a wide range of organic pollutants, such as PAHs (3, 13, 28, 37, 38, 48). The phylogenetic diversity of Sphingomonas species in PAH-polluted soils has recently been investigated (24). Most of the species described were able to grow on PAHs having up to three rings and to occasionally degrade four- or five-ring PAHs almost exclusively by cooxidation. Here, a Sphingomonas strain that had the rare ability to grow on chrysene as a sole carbon source was used to investigate the enzymes responsible for the catabolism of PAHs and, in particular, the enzyme that catalyzes the initial step in chrysene degradation. The strategy employed in this study involved direct peptide analysis of most prominent PAH-induced proteins and subsequent cloning and functional analysis of genes encoding selected target proteins. This approach led to the identification of two oxygenases, one of which was shown to catalyze ring hydroxylation of several PAHs. A dioxin dioxygenase that catalyzes angular dioxygenation in Sphingomonas wittichii RW1 has been described previously (1), and in this report we describe for the first time an enzyme responsible for PAH dihydroxylation in a Sphingomonas strain.
Results of chrysene mineralization experiments (Table 2) indicated that specific catabolic enzymes were induced in PAH-grown cells. However, the mineralization rates varied depending on the PAH used as the substrate; the lowest rates were observed with naphthalene either alone or in combination with chrysene. Also, phenanthrene alone promoted a high level of activity, but curiously, cells grown on both chrysene and phenanthrene showed a lower mineralization rate. These data might reflect inhibitory or antagonistic phenomena, as previously observed in other PAH-degrading strains (5). Toxic effects of more water-soluble PAHs, such as naphthalene, and accumulation of metabolites have been proposed as possible causes to explain the lower degradation rates in bacteria exposed to two or more PAHs. In addition, the level of induction of the catabolic enzymes may differ in PAH-grown cells depending on the substrate. This is illustrated in Fig. 2, in which the pattern of protein induction in strain CHY-1 appears to vary as a function of the nature of the PAH supplied. Such observations suggest that complex regulation controls the expression of the catabolic genes involved in PAH degradation in strain CHY-1.
Previous studies on PAH-degrading strains revealed that catabolic genes responsible for biphenyl and PAH biodegradation were scattered over a DNA region that was more than 30 kb long (22, 33, 34). Sequence analysis indicated that genes resembling xyl genes found in monoaromatic compound-degrading pseudomonads were interspersed with genes probably involved in PAH degradation. This gene arrangement appears to be highly conserved in strains exhibiting widely diverse origins and might be typical for PAH-degrading sphingomonads (31). In N. aromaticivorans, the xyl and bph catabolic genes are borne on the pNL1 megaplasmid (34). Although limited DNA sequence analysis was performed in this study, it is likely that the same gene organization also occurs in strain CHY-1. In addition to the striking sequence similarities observed between strain CHY-1 phn genes and corresponding bph genes found on plasmid pNL1, the gene arrangement was the same. Moreover, several PAH-induced proteins found in CHY-1 cell extracts showed strong sequence similarity to the products of genes found on the pNL1 plasmid, including, xylQ, nahE, and bphK. Preliminary experiments indicated that strain CHY-1 contains at least three plasmids, none of which carries the phn genes (data not shown). We inferred that the PAH-catabolic genes are probably located on the chromosome in strain CHY-1, although we cannot rule out the possibility that they are borne on a very large plasmid which could not be detected. In this respect, a recent survey of plasmids in several xenobiotic compound-degrading Sphingomonas strains detected at least two large plasmids that were 120 to 500 kb long in almost all strains analyzed and showed that catabolic genes were often plasmid borne (2).
Five sets of genes potentially encoding terminal oxygenase components were found in the ca. 30-kb DNA region containing most catabolic genes in three Sphingomonas strains (22, 33, 34). Genetic analysis indicated that none of these enzymes was required for growth on PAHs, suggesting that other genes mapping outside this region might encode a PAH-hydroxylating dioxygenase (22). Indeed, Romine et al. found a sixth pair of genes designated bphA1f and bphA2f that were about 30 kb from the main cluster of catabolic genes on plasmid pNL1 (34). Based on sequence similarity with known enzymes, it was proposed that the bphA1f-bphA2f gene products might constitute the terminal component of a naphthalene dioxygenase. In the present study, we obtained experimental evidence that the closely related phnA1a-phnA2a gene products from strain CHY-1 assemble to form the terminal component of an enzyme catalyzing PAH dihydroxylation.
The phnA1a and phnA2a gene products from CHY-1 displayed 77 and 70% sequence identity with the bphA1f and bphA2f gene products from N. aromaticivorans and 61 and 52% identity with the phnA1 and phnA2 gene products from Cycloclasticus sp. strain A5 (20). The latter gene products are subunits of a dioxygenase that is active on naphthalene, (methyl)naphthalene, and phenanthrene. Since strain CHY-1 was unable to grow on biphenyl (see below), it seemed more appropriate to use phn rather than bph to designate the genes coding for the PAH dioxygenase described in this study. Furthermore, according to the dioxygenase classification proposed by Nam et al. (30), the PhnI enzyme belongs to group III, which includes all the naphthalene and PAH dioxygenases, whereas biphenyl dioxygenases are part of a distinct group.
The PAH selectivity of PhnI resembled that of naphthalene dioxygenases found in pseudomonads in that it showed the best activity with naphthalene; the other preferred substrates were biphenyl, anthracene, and phenanthrene. Although biphenyl is oxidized at high rates by PhnI, cell cultures incubated in the presence of biphenyl accumulated a yellow metabolite, suggesting that biphenyl is incompletely metabolized by the cells. The peculiarity of the PhnI enzyme comes from its ability to hydroxylate the four-ring PAHs benz[a]anthracene and chrysene. In this respect, PhnI more closely resembles the PAH dioxygenase from S. yanoikuyae B1. This enzyme has not been isolated yet, but its selectivity was deduced from analysis of a mutant defective in dihydrodiol dehydrogenase catalyzing the second step in PAH degradation (14). Studies with this mutant strain showed that the S. yanoikuyae dioxygenase used a range of PAH substrates very similar to that found for PhnI, including the four-ring PAHs benz[a]anthracene and chrysene, as well as biphenyl, naphthalene, phenanthrene, and anthracene (6, 14, 15). Interestingly, both enzymes generate three cis-dihydrodiols from benz[a]anthracene, which were previously identified as derivatives bearing hydroxyl groups at the 1,2 positions, the 8,9 positions, and the 10,11 positions (14, 15). While it is not known whether all these oxidation reactions are catalyzed by one or multiple dioxygenases in S. yanoikuyae, the results presented here provide evidence that a single enzyme is responsible for PAH oxidation in strain CHY-1. Indeed, the fact that a phnA1a deletion abolishes PAH-dependent growth as well as PAH oxidation in strain M10-1 suggests that no other dioxygenase can substitute for PhnI in the initial attack of PAHs. However, we cannot rule out the possibility that the phenotype of the M10-1 mutant could arise from a polar effect of the cassette insertion on an essential catabolic gene located downstream or that indirect regulatory effects affect the expression of other oxygenase genes. Complementation experiments will be undertaken to definitively assess the physiological role of PhnI.
The catalytic properties of PhnI obviously contribute to the extended capacity of strain CHY-1 for PAH degradation, including the capacity to grow at the expense of chrysene as a sole carbon source. The dioxygenase activity of PhnI with chrysene was estimated to be 1.5% of that with naphthalene and 10% of that with phenanthrene. This relatively low activity might explain why strain CHY-1 grows approximately 10-fold more slowly on chrysene than on the other two PAH substrates. During growth on chrysene, no metabolite accumulated in appreciable amounts (data not shown), suggesting that the initial attack of chrysene is a major rate-limiting step in the degradation pathway. Assuming that chrysene is hydroxylated at the 3,4 positions, as demonstrated for the S. yanoikuyae enzyme (6), subsequent oxidations and ring cleavage should lead to a phenanthrene derivative with substituents (possibly a carboxyl and a hydroxyl) at the 1,2 positions. Phenanthrene degradation generally proceeds through oxidation at the 3,4 positions, although 2-hydroxy 3-phenanthroic acid and 3-hydroxy 2-phenanthroic acid were identified as intermediates in the metabolism of benz[a]anthracene by S. yanoikuyae (27). Further work is needed to elucidate how the chrysene dihydrodiol generated by the PhnI dioxygenase is further oxidized in strain CHY-1 and to identify the enzymes involved.
The second enzyme characterized in this study catalyzed the oxidative decarboxylation of salicylate and methylsalicylates to the corresponding catechols. It appears to be a three-component enzyme in which the PhnII terminal component requires a ferredoxin and a reductase for activity. Interestingly, both PhnI and PhnII recombinant enzymes showed enhanced activity when they were associated with PhnA3 and PhnA4, suggesting that they might share the same electron carriers. In related sphingomonad strains, sharing of electron carriers by several oxygenases is also plausible given that a single copy of genes encoding such proteins was found in the 30-kb catabolic gene region (22, 31) or on the pNL1 catabolic plasmid (34). Accordingly, the genes encoding similar ferredoxin (bphA3) and reductase (bphA4) components in S. yanoikuyae were found to be essential for metabolism of biphenyl and PAH (22).
In naphthalene-degrading pseudomonads, the conversion of salicylate to catechol is catalyzed by the product of nahG, which encodes a flavoprotein monooxygenase (47). The absence of an nahG-like gene associated with the PAH-catabolic genes in three strains of Sphingomonas suggested that other gene products were responsible for this reaction (31, 34, 48). Recently, three sets of genes located in the main catabolic gene region in Sphingobium sp. strain P2 were found to encode salicylate 1-hydroxylases (33). These enzymes likely substitute for NahG monooxygenase in strain P2, as well as in other sphingomonads in which similar genes were identified (31).
PhnII is encoded by the phnA2b-phnA1b sequence, in which the gene for the small subunit precedes the gene for the large subunit, a situation unusual for an enzyme of this family, in which the reverse gene order is generally observed. A similar inversion was observed for one of the five sets of terminal oxygenase genes found in two other strains of Sphingomonas (33, 34). Likewise, the genes encoding a pyrene dioxygenase were found to be in the reverse order in two Mycobacterium strains (21, 23). The physiological significance of such a gene inversion is unknown.
Amino acid sequence comparison indicated that the PhnII α and β subunits were highly similar to the products of ahdA1c (81% identity) and ahdA2c (67% identity) from strain P2. Significant sequence similarities were also observed between PhnII and anthranilate dioxygenase from Burkholderia cepacia DBO1 (8) (52 and 38% identity for the α subunits and the β subunits, respectively). Despite these similarities, the PhnII enzyme from CHY-1 exhibited distinctive catalytic properties. In contrast to the enzyme encoded by ahdA1c-ahdA2c from strain P2, which displayed a preference for salicylate over methylsalicylates and had poor activity with 4-methylsalicylate, PhnII was almost as active with salicylate as it was with its 4-methyl and 5-methyl derivatives and was half as active with 3-methylsalicylate. Also, PhnII converted anthranilate to 2-aminophenol, in contrast to the anthranilate dioxygenase from B. cepacia DBO1, which converted anthranilate to catechol. The latter enzyme showed a narrow substrate specificity, with some activity with salicylate but no activity with its methyl derivatives (8). Our results indicate that PhnII functions essentially as a monooxygenase, with broad specificity for methyl-substituted salicylates. Preliminary experiments with a mixture of PAHs extracted from a contaminated sediment have shown that CHY-1 is able to degrade a wide range of alkylated PAH derivatives. The results suggest that PhnI might be capable of oxidizing methylnaphthalene, like the related enzyme from Cycloclasticus (20). In future work we will address both the selectivity and the catalytic properties of the PhnI and PhnII enzymes described in this study.
Acknowledgments
This work was supported by grants from the Centre National de la Recherche Scientifique and the Commissariat à l'Energie Atomique.
REFERENCES
- 1.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basta, T., A. Keck, J. Klein, and A. Stolz. 2004. Detection and characterization of conjugative degradative plasmids in xenobiotic-degrading Sphingomonas strains. J. Bacteriol. 186:3862-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastiaens, L., D. Springael, P. Wattiau, H. Harms, R. deWachter, H. Verachtert, and L. Diels. 2000. Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl. Environ. Microbiol. 66:1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boonchan, S., M. L. Britz, and G. A. Stanley. 1998. Surfactant-enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia. Biotechnol. Bioeng. 59:482-494. [DOI] [PubMed] [Google Scholar]
- 5.Bouchez, M., D. Blanchet, and J. P. Vandecasteele. 1995. Degradation of polycyclic aromatic hydrocarbons by pure strains and by defined strain associations: inhibition phenomena and cometabolism. Appl. Microbiol. Biotechnol. 43:156-164. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, D. R., N. D. Sharma, R. Agarwal, S. M. Resnick, M. J. Schocken, D. T. Gibson, J. M. Sayer, H. Yagi, and D. M. Jerina. 1997. Bacterial dioxygenase-catalysed dihydroxylation and chemical resolution routes to enantiopure cis-dihydrodiols of chrysene. J. Chem. Soc. Perkin Trans. 1:1715-1723. [Google Scholar]
- 7.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 8.Chang, H.-K., P. Mohseni, and G. J. Zylstra. 2003. Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J. Bacteriol. 185:5871-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean-Ross, D., and C. E. Cerniglia. 1996. Degradation of pyrene by Mycobacterium flavescens. Appl. Microbiol. Biotechnol. 46:307-312. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 11.Duport, C., C. Meyer, I. Naud, and Y. Jouanneau. 1994. A new gene expression system based on a fructose-dependent promoter from Rhodobacter capsulatus. Gene 145:103-108. [DOI] [PubMed] [Google Scholar]
- 12.Ferro, M., D. Seigneurin-Berny, N. Rolland, A. Chapel, D. Salvi, J. Garin, and J. Joyard. 2000. Organic solvent extraction as a versatile procedure to identify hydrophobic chloroplast membrane proteins. Electrophoresis 21:3517-3526. [DOI] [PubMed] [Google Scholar]
- 13.Fredrickson, J. K., D. L. Balkwill, M. F. Romine, and T. Shi. 1999. Ecology, physiology, and phylogeny of deep subsurface Sphingomonas sp. J. Ind. Microbiol. Biotechnol. 23:273-283. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, D. T. 1999. Beijerinckia sp strain B1: a strain by any other name. J. Ind. Microbiol. Biotechnol. 23:284-293. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, D. T., V. Mahadevan, D. M. Jerina, H. Yogi, and H. J. Yeh. 1975. Oxidation of the carcinogens benzo[a]pyrene and benzo[a]anthracene to dihydrodiols by a bacterium. Science 189:295-297. [DOI] [PubMed] [Google Scholar]
- 16.Heitkamp, M. A., W. Franklin, and C. E. Cerniglia. 1988. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl. Environ. Microbiol. 54:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jouanneau, Y., C. Meyer, I. Naud, and W. Klipp. 1995. Characterization of an fdxN mutant of Rhodobacter capsulatus indicates that ferredoxin I serves as electron donor to nitrogenase. Biochim. Biophys. Acta 1232:33-42. [DOI] [PubMed] [Google Scholar]
- 18.Juhasz, A. L., M. L. Britz, and G. A. Stanley. 1997. Degradation of fluoranthene, pyrene, benz[a]anthracene and dibenz[a,h]anthracene by Burkholderia cepacia. J. Appl. Microbiol. 83:189-198. [Google Scholar]
- 19.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasai, Y., K. Shindo, S. Harayama, and N. Misawa. 2003. Molecular characterization and substrate preference of a polycyclic aromatic hydrocarbon dioxygenase from Cycloclasticus sp. strain A5. Appl. Environ. Microbiol. 69:6688-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan, A. A., R. F. Wang, W. W. Cao, D. R. Doerge, D. Wennerstrom, and C. E. Cerniglia. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polcyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:3577-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, E., and G. J. Zylstra. 1999. Functional analysis of genes involved in biphenyl, naphthalene, phenanthrene, and m-xylene degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 23:294-302. [DOI] [PubMed] [Google Scholar]
- 23.Krivobok, S., S. Kuony, C. Meyer, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185:3828-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leys, N. M. E. J., A. Ryngaert, L. Bastiaens, W. Verstraete, E. M. Top, and D. Springael. 2004. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:1944-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd-Jones, G., and P. C. Lau. 1997. Glutathione S-transferase-encoding gene as a potential probe for environmental bacterial isolates capable of degrading polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 63:3286-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 27.Mahaffey, W. R., D. T. Gibson, and C. E. Cerniglia. 1988. Bacterial oxidation of chemical carcinogens: formation of polycyclic aromatic acids from benz[a]anthracene. Appl. Environ. Microbiol. 54:2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller, J. G., R. Devereux, D. L. Santavy, S. E. Lantz, S. G. Willis, and P. H. Pritchard. 1997. Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Antonie Leeuwenhoek 71:329-343. [DOI] [PubMed] [Google Scholar]
- 29.Nadalig, T., N. Raymond, N. M. Gilewicz, H. Budzinski, and J. C. Bertrand. 2002. Degradation of phenanthrene, methylphenanthrenes and dibenzothiophene by a Sphingomonas strain 2mpII. Appl. Microbiol. Biotechnol. 59:79-85. [DOI] [PubMed] [Google Scholar]
- 30.Nam, J. W., H. Nojiri, T. Yoshida, H. Habe, H. Yamane, and T. Omori. 2001. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci. Biotechnol. Biochem. 65:254-263. [DOI] [PubMed] [Google Scholar]
- 31.Pinyakong, O., H. Habe, and T. Omori. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J. Gen. Appl. Microbiol. 49:1-19. [DOI] [PubMed] [Google Scholar]
- 32.Pinyakong, O., H. Habe, N. Supaka, P. Pinpanichkarn, K. Juntongjin, T. Yoshida, K. Furihata, H. Nojiri, H. Yamane, and T. Omori. 2000. Identification of novel metabolites in the degradation of phenanthrene by Sphingomonas sp. strain P2. FEMS Microbiol. Lett. 191:115-121. [DOI] [PubMed] [Google Scholar]
- 33.Pinyakong, O., H. Habe, T. Yoshida, H. Nojiri, and T. Omori. 2003. Identification of three novel salicylate 1-hydroxylases involved in the phenanthrene degradation of Sphingobium sp. strain P2. Biochem. Biophys. Res. Commun. 301:350-357. [DOI] [PubMed] [Google Scholar]
- 34.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schneider, J., R. Grosser, K. Jayasimhulu, W. Xue, and D. Warshawsky. 1996. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl. Environ. Microbiol. 62:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, T., J. K. Fredrickson, and D. L. Balkwill. 2001. Biodegradation of polycyclic aromatic hydrocarbons by Sphingomonas strains isolated from the terrestrial subsurface. J. Ind. Microbiol. Biotechnol. 26:283-289. [DOI] [PubMed] [Google Scholar]
- 38.Sokolovska, I., P. Wattiau, P. Gerin, and S. N. Agathos. 2002. Biodegradation of fluorene at low temperature by a psychrotrophic Sphingomonas sp. L-138. Chem. Pap.-Chem. Zvesti. 56:36-40. [Google Scholar]
- 39.Stolz, A., C. Schmidt-Maag, E. B. M. Denner, H. J. Busse, T. Egli, and P. Kampfer. 2000. Description of Sphingomonas xenophaga sp. nov. for strains BN6(T) and N,N which degrade xenobiotic aromatic compounds. Int. J. Syst. Evol. Microbiol. 50:35-41. [DOI] [PubMed] [Google Scholar]
- 40.Story, S. P., S. H. Parker, S. S. Hayasaka, M. B. Riley, and E. L. Kline. 2001. Convergent and divergent points in catabolic pathways involved in utilization of fluoranthene, naphthalene, anthracene, and phenanthrene by Sphingomonas paucimobilis var. EPA505. J. Ind. Microbiol. Biotechnol. 26:369-382. [DOI] [PubMed] [Google Scholar]
- 41.Vila, J., Z. Lopez, J. Sabate, C. Minguillon, A. M. Solanas, and M. Grifoll. 2001. Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp strain AP1: actions of the isolate on two- and three-ring polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 67:5497-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuilleumier, S. 1997. Bacterial glutathione S-transferases: what are they good for? J. Bacteriol. 179:1431-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter, U., M. Beyer, J. Klein, and H.-J. Rehm. 1991. Degradation of pyrene by Rhodococcus sp. UW1. Appl. Microbiol. Biotechnol. 34:671-676. [Google Scholar]
- 44.Wattiau, P., L. Bastiaens, R. van Herwijnen, L. Daal, J. R. Parsons, M. E. Renard, D. Springael, and G. R. Cornelis. 2001. Fluorene degradation by Sphingomonas sp. LB126 proceeds through protocatechuic acid: a genetic analysis. Res. Microbiol. 152:861-872. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, S. C., and K. C. Jones. 1993. Bioremediation of soil contaminated with polynuclear aromatic hydrocarbons (PAHs): a review. Environ. Pollut. 81:229-249. [DOI] [PubMed] [Google Scholar]
- 46.Yen, K. M., and C. M. Serdar. 1988. Genetics of naphthalene catabolism in pseudomonads. Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]
- 47.You, I. S., D. Ghosal, and I. C. Gunsalus. 1991. Nucleotide sequence analysis of the Pseudomonas putida PpG7 salicylate hydroxylase gene (nahG) and its 3′-flanking region. Biochemistry 30:1635-1641. [DOI] [PubMed] [Google Scholar]
- 48.Zylstra, G. J., and E. Kim. 1997. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 19:408-414. [DOI] [PubMed] [Google Scholar]