Abstract
We have investigated the existence and genetic organization of a functional type III secretion system (TTSS) in a mesophilic Aeromonas strain by initially using the Aeromonas hydrophila strain AH-3. We report for the first time the complete TTSS DNA sequence of an Aeromonas strain that comprises 35 genes organized in a similar disposition as that in Pseudomonas aeruginosa. Using several gene probes, we also determined the presence of a TTSS in clinical or environmental strains of different Aeromonas species: A. hydrophila, A. veronii, and A. caviae. By using one of the TTSS genes (ascV), we were able to obtain a defined insertion mutant in strain AH-3 (AH-3AscV), which showed reduced toxicity and virulence in comparison with the wild-type strain. Complementation of the mutant strain with a plasmid vector carrying ascV was fully able to restore the wild-type toxicity and virulence.
The genus Aeromonas comprises mesophilic and psychrophilic species, both motile and nonmotile. This bacterium is found in both fresh and salt water and in virtually all foods and causes a wide variety of human infections, including septicemia, wound infections, meningitis, pneumonia, and gastroenteritis (7, 12). Out of the 16 reported species within the genus, three of them, A. veronii, A. caviae, and A. hydrophila, represent more than 85% of clinical isolates (11, 13). The pathogenesis of Aeromonas has multiple factors, such as O antigens, capsule (16, 23), the S-layer (6), exotoxins such as hemolysins and enterotoxin (4, 9), and a repertoire of exoenzymes which digest cellular components such as proteases, amylases, and lipases (14, 19). These virulence determinants are involved sequentially as the bacteria colonize, gain entry, establish themselves, replicate, cause damage in host tissues, evade the host defense system, and spread, eventually killing the host. The mechanisms of action of most of these virulence factors remain unknown.
Recent studies have shown that the virulence mechanisms of various pathogens are highly similar; one such mechanism is a type III secretion system (TTSS) that plays crucial roles in host-pathogen interactions (5). The TTSS is found in some gram-negative animal and plant pathogens. This system can efficiently deliver antihost virulence determinants into the host cells, directly interfering with and altering host processes. Recently, several reports have attempted to elucidate the existence of the TTSS in Aeromonas (1, 2, 3, 21, 22). In this sense a functional TTSS located on a large thermolabile virulence plasmid has been reported for the fish-pathogenic species A. salmonicida (1, 2, 21) and in the chromosome of A. hydrophila AH-1 (22). However, in both studies the TTSS genes sequenced do not seem to correspond to a complete TTSS compared with the well-known Yersinia or Pseudomonas TTSS (5). Furthermore, dot blotting and sequencing experiments have provided evidence of the existence of other TTSS genes (ascF-ascG) in mesophilic clinical species (3), which reinforces the hypothesis that the Aeromonas TTSS sequence organization described recently is incomplete. Therefore, the purpose of the present study was to investigate and describe the complete sequence of genes that constitute the Aeromonas TTSS and its preliminary function. In addition we hoped to establish its prevalence in a set of genetically identified clinical and environmental strains of mesophilic species, i.e., A. veronii, A. caviae, and A. hydrophila, more frequently implicated in human infections.
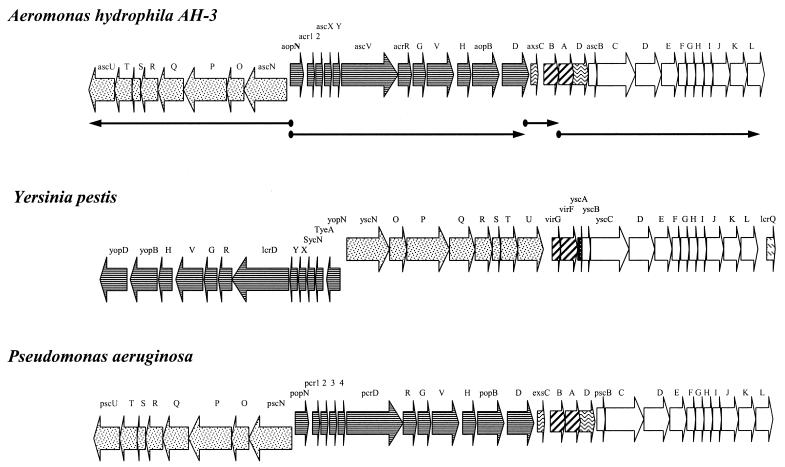
Complete A. hydrophila AH-3 TTSS gene cluster.
We selected A. hydrophila AH-3 for sequencing studies of the TTSS because it is a fish- and mouse-pathogenic strain (17) belonging to serotype O:34, which is the predominant serogroup associated with clinical isolates (15). By using A. hydrophila AH-3 genomic DNA and primers (5′-ATGGACGGCGCCATGAAGTT-3′ and 5′-TATTCGCCTTCACCCATCCC-3′) derived from the DNA sequence of ascV from A. salmonicida (a gene from the type III secretion apparatus which prevents the delivery of the AexT toxin [1, 2, 21]), we generated by PCR a 710-bp fragment which showed high identity (88%) with the A. salmonicida ascV gene. We used this DNA fragment labeled with digoxigenin to screen our gene library previously obtained from strain AH-3 (18) by colony Southern blotting. We found several clones, and with two of them we obtained the complete DNA sequence of the TTSS from strain AH-3. DNA sequencing and sequence analysis with various software programs were performed as previously described (10). The A. hydrophila AH-3 TTSS DNA sequence (GenBank accession no. AY528667) showed 35 open reading frames, organized as shown in Fig. 1, that code for 35 proteins. The nomenclature for the genes-proteins newly described in Aeromonas follows that employed for Pseudomonas aeruginosa (in parentheses), with P for Pseudomonas being substituted for A for Aeromonas; for the rest we employed the terminology used for A. salmonicida (2). The characteristics of individual proteins (amino acid residues, molecular weight, and isoelectric point) and their homologues are shown in Table 1. The first 20 proteins (from AscU to AopD) show high homology with the corresponding proteins of the A. salmonicida and A. hydrophila TTSSs previously reported (1, 2, 21, 22). However, of the last 15 proteins (from AxsC to AscL), at least eight could be considered novel TTSS proteins for Aeromonas because they have not yet been reported for the TTSS of A. salmonicida, A. veronii, or A. hydrophila (2, 22). This may be due to the fact that the genes for these 15 proteins have not been identified or may not be present in the TTSSs of the strains studied (2, 22). However, the fact that 2 and 5 of these 15 genes (ascF-ascG) have been sequenced for A. veronii (3) and A. hydrophila AH-1 (22), respectively, and recognized by dot blotting with other TTSS genes in the type strain of A. salmonicida (data not shown) indicates that this strain at least bears this additional pair of genes. Furthermore, none of the TTSSs described for Aeromonas spp. have been completely sequenced.
FIG. 1.
Genetic organization of the A. hydrophila AH-3 TTSS genes. Open reading frames and their directions of transcription are indicated by black arrows and named according to the Aeromonas gene-protein names given by us. We also show the four gene clusters determined by RT-PCRs. The complete TTSSs from A. hydrophila AH-3, Y. pestis, and P. aeruginosa are also shown. Black arrows showing the same drawing correspond to homologous genes among these bacteria.
TABLE 1.
A. hydrophila AH-3 TTSS putative proteins and their homologues in other bacteria
| Protein | Putative function | No. of amino acids | Mol wt (thousands) | pI | Protein homologues (% identity/% similarity) |
|---|---|---|---|---|---|
| AscU | Regulation of secretion | 352 | 39.36 | 8.51 | AscU, Aeromonas salmonicida (87/90) |
| AscU, Aeromonas hydrophila AH-1 (86/89) | |||||
| LscU, Photorhabdus luminescens (72/82) | |||||
| AscT | Type III secretion apparatus | 262 | 28.29 | 6.54 | AscT, Aeromonas salmonicida (72/74) |
| AscT, Aeromonas hydrophila AH-1 (69/74) | |||||
| PscT, Pseudomonas aeruginosa (56/69) | |||||
| AscS | Type III secretion apparatus | 88 | 9.66 | 5.30 | AscS, Aeromonas hydrophila AH-1 (98/100) |
| AscS, Aeromonas salmonicida (96/98) | |||||
| LscS, Photorhabdus luminescens (82/90) | |||||
| AscR | Type III secretion apparatus | 217 | 24.17 | 6.11 | AscR, Aeromonas salmonicida (91/90) |
| AscR, Aeromonas hydrophila AH-1 (89/90) | |||||
| PscR, Pseudomonas aeruginosa (79/88) | |||||
| AscQ | Unknown | 308 | 33.45 | 4.92 | AscQ, Aeromonas salmonicida (74/77) |
| AscQ, Aeromonas hydrophila AH-1 (63/70) | |||||
| YscQ, Yersinia spp. (43/57) | |||||
| AscP | Regulation of secretion | 408 | 44.99 | 5.07 | AscP, Aeromonas salmonicida (65/75) |
| AscP, Aeromonas hydrophila AH-1 (54/68) | |||||
| LscP, Photorhabdus luminescens (37/53) | |||||
| AscO | Regulation of secretion | 153 | 18.56 | 7.92 | AscO, Aeromonas salmonicida (49/51) |
| AscU, Aeromonas hydrophila AH-1 (43749) | |||||
| PscO, Pseudomonas aeruginosa (41/48) | |||||
| AscN | ATP synthase | 440 | 47.80 | 6.17 | AscN, Aeromonas salmonicida (95/96) |
| AscN, Aeromonas hydrophila AH-1 (95/96) | |||||
| LscN, Photorhabdus luminescens (90/94) | |||||
| AopN | Regulation of translocation | 292 | 32.03 | 5.54 | AopN, Aeromonas salmonicida (86/87) |
| AopN, Aeromonas hydrophila AH-1 (83/85) | |||||
| LopN, Photorhabdus luminescens (60/63) | |||||
| Acr1 | Translocation apparatus | 93 | 10.52 | 4.29 | Acr1, Aeromonas salmonicida (93/95) |
| Acr1, Aeromonas hydrophila AH-1 (88/96) | |||||
| LssA, Photorhabdus luminescens (70/86) | |||||
| Acr2 | Chaperone | 123 | 13.74 | 5.57 | Acr2, Aeromonas salmonicida (92/96) |
| Acr2, Aeromonas hydrophila AH-1 (83/87) | |||||
| LssN, Photorhabdus luminescens (63/77) | |||||
| AscX | Type III secretion apparatus | 121 | 13.63 | 5.84 | AscX, Aeromonas salmonicida (96/97) |
| AscX, Aeromonas hydrophila AH-1 (81/90) | |||||
| LssB, Photorhabdus luminescens (61/75) | |||||
| AscY | Type III secretion apparatus | 116 | 12.90 | 5.23 | AscY, Aeromonas salmonicida (77/79) |
| AscY, Aeromonas hydrophila AH-1 (67/69) | |||||
| LssC, Photorhabdus luminescens (57/66) | |||||
| AscV | Type III secretion apparatus | 721 | 79.26 | 6.09 | AscV, Aeromonas salmonicida (88/89) |
| AscV, Aeromonas hydrophila AH-1 (85/87) | |||||
| LssD, Photorhabdus luminescens (77/82) | |||||
| AcrR | Unknown | 151 | 16.89 | 9.22 | AcrR, Aeromonas salmonicida (89/93) |
| LcrR, Yersinia spp. (56/68) | |||||
| AcrR, Aeromonas hydrophila AH-1 (53/66) | |||||
| AcrG | Regulation of low-calcium response | 94 | 10.52 | 5.87 | AcrG, Aeromonas salmonicida (90/94) |
| PcrG, Pseudomonas aeruginosa (47/63) | |||||
| AcrG, Aeromonas hydrophila AH-1 (44/59) | |||||
| AcrV | Protective antigen, anti-host factor | 361 | 40.14 | 5.22 | AcrV, Aeromonas salmonicida (74/81) |
| LssV, Photorhabdus luminescens (41/53) | |||||
| AcrV, Aeromonas hydrophila AH-1 (38/60) | |||||
| AcrH | Chaperone | 167 | 18.53 | 4.32 | AcrH, Aeromonas salmonicida (86/89) |
| LssH, Photorhabdus luminescens (63/76) | |||||
| AcrH, Aeromonas hydrophila AH-1 (57/72) | |||||
| AopB | Translocation apparatus | 390 | 40.24 | 9.17 | AopB, Aeromonas salmonicida (63/73) |
| LopB, Photorhabdus luminescens (38/56) | |||||
| AopB, Aeromonas hydrophila AH-1 (32/45) | |||||
| AopD | Translocation apparatus | 299 | 32.17 | 6.21 | AopD, Aeromonas salmonicida (57/69) |
| AopD, Aeromonas hydrophila AH-1 (52/71) | |||||
| YopD, Yersinia spp. (40/59) | |||||
| AxsC (ExsC) | Unknown | 147 | 16.72 | 4.59 | HscY, Aeromonas hydrophila AH-1 (79/87) |
| LscY, Photorhabdus luminescens (70/80) | |||||
| ExsC, Pseudomonas aeruginosa (60/84) | |||||
| AxsB (ExsB) | Regulation of secretion | 133 | 14.86 | 9.22 | AscX, Aeromonas hydrophila AH-1 (55/67) |
| LscW, Photorhabdus luminescens (36/56) | |||||
| VirG, Yersinia spp. (32/51) | |||||
| ExsB, Pseudomonas aeruginosa (24/41) | |||||
| AxsA (ExsA) | Transcriptional activator | 271 | 30.82 | 6.18 | AscA, Aeromonas hydrophila AH-1 (87/93) |
| LscA, Photorhabdus luminescens (74/83) | |||||
| ExsA, Pseudomonas aeruginosa (65/76) | |||||
| AxsD (ExsD) | Putative regulator | 271 | 31.53 | 5.57 | AscZ, Aeromonas hydrophila AH-1 (68/81) |
| LscZ, Photorhabdus luminescens (44/60) | |||||
| ExsD, Pseudomonas aeruginosa (36/51) | |||||
| AscB (PscB) | Putative chaperone | 141 | 15.72 | 5.43 | AscB, Aeromonas hydrophila AH-1 (76/80) |
| LscB, Photorhabdus luminescens (56/68) | |||||
| YscB, Yersinia spp. (45/61) | |||||
| PscB, Pseudomonas aeruginosa (41/56) | |||||
| AscC (PscC) | Secretin | 618 | 67.83 | 5.07 | YscC, Yersinia spp. (73/85) |
| LscC, Photorhabdus luminescens (67/79) | |||||
| PscC, Pseudomonas aeruginosa (66/79) | |||||
| AscD (PscD) | Type III secretion apparatus | 433 | 48 | 6.15 | LscD, Photorhabdus luminescens (50/65) |
| PscD, Pseudomonas aeruginosa (46/64) | |||||
| YscD, Yersinia spp. (45/63) | |||||
| AscE (PscE) | Translocation apparatus | 67 | 7.48 | 4.93 | SctE, Photorhabdus luminescens (47/70) |
| PscE, Pseudomonas aeruginosa (48/64) | |||||
| YscE, Yersinia spp. (35/60) | |||||
| AscF (PscF) | Translocation apparatus (needle) | 81 | 9.02 | 6.55 | AscF, Aeromonas veronii (82790) |
| PscF, Pseudomonas aeruginosa (79/80) | |||||
| LscF, Photorhabdus luminescens (70/79) | |||||
| AscG (PscG) | Chaperone | 117 | 12.94 | 5.04 | AscG, Aeromonas veronii (75/77) |
| VP1693, Vibrio parahaemolyticus (49/60) | |||||
| YscG, Yersinia enterocolitica (49/58) | |||||
| AscH (PscH) | Unknown | 183 | 20.69 | 5.71 | PscH, Pseudomonas aeruginosa (50/68) |
| YscH, (yopR), Yersinia spp. (38/57) | |||||
| LscH, Photorhabdus luminescens (37/54) | |||||
| AscI (PscI) | Chaperone | 112 | 12.04 | 4.48 | LscI, Photorhabdus luminescens (56/72) |
| PscI, Pseudomonas aeruginosa (61/72) | |||||
| YscI, Yersinia spp. (46/60) | |||||
| AscJ (PscJ) | Type III secretion apparatus | 246 | 27 | 7.01 | LscJ, Photorhabdus luminescens (73/84) |
| YscJ, Yersinia spp. (73/80) | |||||
| PscJ, Pseudomonas aeruginosa (70/82) | |||||
| AscK (PscK) | Unknown | 207 | 22.61 | 6.59 | LscK, Photorhabdus luminescens (47/57) |
| YscK, Yersinia spp. (46/59) | |||||
| PscK, Pseudomonas aeruginosa (42/54) | |||||
| AscL (PscL) | Unknown | 221 | 24.71 | 5.20 | LscL, Photorhabdus luminescens (73/86) |
| YscL, Yersinia spp. (68/84) | |||||
| PscL, Pseudomonas aeruginosa (61/77) |
Four putative promoter regions have been identified upstream of the genes (ascN, aopN, axsC [exsC], and axsA [exsA]), and four rho-independent terminator sequences have been identified downstream (ascU, aopD, axsB [exsB], and ascL [pscL]), showing four putative clusters as in Fig. 1. The four clusters have been confirmed using reverse transcription-PCR (RT-PCR) with specific primers derived from the DNA sequence. The total RNA was extracted with Trizol reagent (Invitrogen), and to ensure that the RNA was devoid of contaminating DNA, the preparation was treated with RNase-free RQ1 DNase (Invitrogen) for 1 h. The isolated RNA was used as a template in RT-PCRs, with the use of the SuperScript One-Step RT-PCR system (Invitrogen) according to the manufacturer's instructions. Using DNA sequencing-derived oligonucleotides and RT-PCRs, we found amplification between ascU to ascN, aopN to aopD, axsC (exsC) to axsB (exsB), and axsA (exsA) to ascL. However, no amplifications were obtained with oligonucleotide pairs from ascN to aopN, aopD to axsC (exsC), and axsB (exsB) to axsA (exsA), thus confirming the four putative clusters of this entire TTSS region (Fig. 1).
As can be observed in Table 1 and Fig. 1, the A. hydrophila AH-3 TTSS is similar in gene pair organization to the P. aeruginosa TTSS despite individual gene-protein homology sometimes being higher with Yersinia pestis TTSS genes-proteins than with those of P. aeruginosa (in the last 15 genes not previously described for A. salmonicida), thus showing that in this study we have obtained for the first time the complete arrangement of TTSS genes of Aeromonas. A comparison of the overall genetic organization between different TTSSs defined three subgroups that match similarities in the order of the large genetic blocks. Subgroup 2 includes Yersinia spp. and P. aeruginosa, and according to our findings Aeromonas TTSS should also be included in this subgroup. The overall G+C content of the A. hydrophila AH-3 TTSS region (58.3%) is in the range of the genomic G+C content (57 to 62%) of mesophilic Aeromonas strains.
Distribution of the TTSS genes among mesophilic Aeromonas strains.
Taking advantage of the complete DNA sequence obtained, by PCR we prepared two more digoxigenin-labeled DNA probes from both edges of the TTSS by using AH-3 chromosomal DNA: the first one was 4,815 bp (ascN to ascT) with oligonucleotides 5′-TATCGAAGCTGATCTGGGGG-3′ and 5′-ATGGCAATAAGCAGCGGG-3′, and the second one was 4,141 bp (ascC to ascJ, names in Aeromonas given by us) with oligonucleotides 5′-GCGCTCTCCATCATCGAC-3′ and 5′-CCACGTCGGATTCTTCAAC-3′. The three labeled DNA probes (ascV, ascN to ascT, and ascC to ascJ) were used in a dot blot assay to screen genomic DNAs from clinical and environmental Aeromonas strains (n = 182) to establish their prevalence. The environmental strains (n = 65) were isolated mainly from water samples (n = 54) and shellfish (n = 11) by using ampicillin dextrin agar or Tergitol agar, while the clinical strains (n = 117) were isolated from blood agar supplemented or not with ampicillin depending upon their intestinal (n = 74) or extraintestinal (n = 43) origin. All the strains were identified to the species level by 16S ribosomal DNA restriction fragment length polymorphism analysis (8). The results are summarized in Table 2. As can be observed, all the mesophilic Aeromonas strains testing positive or negative for one probe were also positive or negative for the rest of the DNA probes. In addition the TTSS was less prevalent in environmental strains (26%) than in clinical strains (56%). This is to our knowledge the first study that comparatively evaluates the prevalence of TTSS genes in a representative number of genetically identified clinical and environmental Aeromonas strains.
TABLE 2.
Distributions by dot blotting of TTSS genes in clinical and environmental mesophilic Aeromonas strains
| Species | Strain type | No. of positive strains/no. of strains tested
|
||
|---|---|---|---|---|
| ascN to ascT | ascV | ascC to ascJ | ||
| A. hydrophila | Clinical | 28/35 | 28/35 | 28/35 |
| Environmental | 4/25 | 4/25 | 4/25 | |
| A. veronii | Clinical | 32/40 | 32/40 | 32/40 |
| Environmental | 9/20 | 9/20 | 9/20 | |
| A. caviae | Clinical | 5/40 | 5/40 | 5/40 |
| Environmental | 4/20 | 4/20 | 4/20 | |
| A. jandaei | Clinical | 1/2 | 1/2 | 1/2 |
Mutant isolation in ascV and characterization.
We also constructed an A. hydrophila AH-3-defined insertion mutation in gene ascV. Briefly, we used the previously described oligonucleotides (5′-ATGGACGGCGCATGAAGTTS-3′ and 5′-TATTCGCCTTCACCCATCCC-3′) to amplify an internal ascV DNA fragment (710 bp). This DNA fragment was ligated to the vector pGEM-T (Promega) and transformed in Escherichia coli DH5α. The DNA fragment was recovered by SalI-NcoI double digestion; blunt ended with Klenow fragment; ligated to EcoRI-digested, blunt-ended, dephosphorylated pSF100 (20); and transformed into E. coli MC1061 (λpir) to generate plasmid pFS-AcsV. Plasmid pFS-AcsV was isolated, transformed into E. coli SM10 (λpir), and transferred by conjugation from E. coli SM10 to an A. hydrophila AH-3 rifampin-resistant mutant (from our laboratory collection) as previously described (17, 18). Kanamycin- and rifampin-resistant transconjugants arising from pFS-AcsV integration were obtained and analyzed by Southern blot hybridization with an ascV DNA probe to obtain a defined insertion ascV mutant (AH-3AscV) as previously described (17, 18). The 50% lethal doses of the mutant and wild-type strains were evaluated using rainbow trout (12 to 20 g) maintained in 20-liter static tanks and albino Swiss female mice (5 to 7 weeks old) in all cases injected intraperitoneally with 0.05 and 0.25 ml of the test samples (approximately 109 viable cells), respectively, as previously described (17, 18). Mutant AH-3AscV had decreased virulence as shown by significantly higher 50% lethal doses in rainbow trout (106.7) and mice (108.5) than those of the wild-type strain (105.3 and 107.4, respectively). Mutant AH-3AscV showed a clearly reduced cytotoxic effect on different eukaryotic cell lines (epithelioma papillosum of carp [EPC] or HEp-2 cells) in comparison with the wild-type strain. At 2 h postinfection with the wild-type strain (AH-3 or the rifampin-resistant mutant), approximately 50% of the eukaryotic cells from the monolayer (EPC or HEp-2 cells) became rounded and were detached from the well. At the same time after infection no morphological changes in the eukaryotic cells (EPC or HEp-2 cells) were observed when inoculated with mutant AH-3AscV; however, some morphological changes and detachment (<50%) were observed after 4 h postinfection. No complementation with the plasmid vector alone (pACYC184) was achieved, while a full complementation (restoring the same lethal doses and cytotoxic effects as those of the wild-type strain) was obtained with the plasmid vector with the complete ascV gene. Data were analyzed by a one-way analysis of variance; P values of <0.05 were considered significant.
This is the first report of the complete TTSS DNA sequence of an Aeromonas strain (A. hydrophila AH-3) that comprises 35 genes. From the results obtained using the different DNA probes, which cover the entire TTSS region, it seems logical to conclude that the mesophilic Aeromonas strains that possess TTSS genes have a complete TTSS. Due to the fact that some of the mesophilic Aeromonas strains tested do not show any plasmid DNA and that the G+C content of the TTSS is in the range of the genomic G+C content of the genus, we suggest that the TTSS on mesophilic Aeromonas strains is located in the chromosome and not on a plasmid as in A. salmonicida (1, 2, 21), as with the A. hydrophila AH-1 TTSS (22). As in the clinical strains with other pathogenic features, this characteristic virulence trait (the presence of TTSS) is more frequent in A. veronii and A. hydrophila (80% of strains) than in A. caviae (13% of strains) (Table 2). Finally, to judge by the results obtained with mutant AH-3AscV, the A. hydrophila AH-3 TTSS is required for the virulence of this strain. Since the bacterium's virulence is known to be multifactorial in many cases, the presence of TTSS in Aeromonas strains seems to be an important factor related to their pathogenicity.
Acknowledgments
This work was supported by grants from the Plan Nacional de I + D (Ministerio de Ciencia y Tecnología, Spain), the Generalitat de Catalunya, and the Spanish Ministry of Health (FIS 03/1183). S.V. received a predoctoral fellowship from the Generalitat de Catalunya.
We also thank Maite Polo for her technical assistance.
REFERENCES
- 1.Burr, S. E., K. Stuber, T. Wahli, and J. Frey. 2002. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184:5966-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burr, S. E., K. Stuber, and J. Frey. 2003. The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion system. J. Bacteriol. 185:6583-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chacón, M. R., L. Soler, E. A. Groisman, J. Guarro, and M. J. Figueras. 2004. Type III secretion system in clinical Aeromonas isolates. J. Clin. Microbiol. 42:1285-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty, T., M. A. Montenegro, S. C. Sanyal, R. Helmuth, E. Bulling, and K. N. Timmis. 1984. Cloning of enterotoxin gene from Aeromonas hydrophila provides conclusive evidence of production of a cytotoxic enterotoxin. Infect. Immun. 46:435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis, G. R., and F. Van Gijesem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 6.Dooley, J. S. G., and T. J. Trust. 1988. Surface protein composition of Aeromonas hydrophila strains virulent for fish: identification of a surface array protein. J. Bacteriol. 170:499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueras, M. J., J. Guarro, and A. Martinez-Murcia. 2000. Clinically relevant Aeromonas species. Clin. Infect. Dis. 30:988-989. [DOI] [PubMed] [Google Scholar]
- 8.Figueras, M. J., L. Soler, M. R. Chacón, J. Guarro, and A. J. Martínez-Murcia. 2000. Extended method for discrimination of Aeromonas spp. by 16S rDNA-RFLP. Int. J. Syst. Evol. Microbiol. 50:2069-2073. [DOI] [PubMed] [Google Scholar]
- 9.Howard, S. P., S. Macintyre, and J. T. Buckley. 1996. The genus Aeromonas, p. 267-286. In B. Austin, M. Altwegg, P. J. Gosling, and S. Joseph (ed.), Toxin. John Wiley and Sons, Singapore, Singapore.
- 10.Izquierdo, L., N. Coderch, N. Piqué, E. Bedeni, M. M. Corsaro, S. Merino, S. Fresno, J. M. Tomás, and M. Regué. 2003. The Klebsiella pneumoniae wabG gene: role in biosynthesis of the core lipopolysaccharide and virulence. J. Bacteriol. 185:7213-7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 12.Janda, J. M. 2001. Aeromonas and Plesiomonas, p. 1237-1270. In M. Sussman (ed.), Molecular medical microbiology. Academic Press, San Diego, Calif.
- 13.Joseph, S. W., and A. M. Carnahan. 2000. Update on the genus Aeromonas. ASM News 66:218-233. [Google Scholar]
- 14.Leung, K. Y., and R. M. W. Stevenson. 1988. Tn5-induced protease-deficient strains of Aeromonas hydrophila with reduced virulence for fish. Infect. Immun. 56:2639-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merino, S., S. Camprubí, and J. M. Tomás. 1993. Incidence of Aeromonas sp. serotypes O:34 and O:11 among clinical isolates. Med. Microbiol. Lett. 2:48-55. [Google Scholar]
- 16.Merino, S., X. Rubires, A. Aguilar, J. F. Guillot, and J. M. Tomás. 1996. The role of the O-antigen lipopolysaccharide on the colonization in vivo of the germfree chicken gut by Aeromonas hydrophila serogroup O:34. Microb. Pathog. 20:325-333. [DOI] [PubMed] [Google Scholar]
- 17.Merino, S., A. Aguilar, M. M. Nogueras, M. Regué, S. Swift, and J. M. Tomás. 1999. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect. Immun. 67:4008-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nogueras, M. M., S. Merino, A. Aguilar, V. J. Benedí, and J. M. Tomás. 2000. Cloning, sequencing and role in serum susceptibility of porin II from mesophilic Aeromonas sp. Infect. Immun. 68:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pemberton, J. M., S. P. Kidd, and R. Schmidt. 1977. Secreted enzymes of Aeromonas. FEMS Microbiol. Lett. 152:1-10. [DOI] [PubMed] [Google Scholar]
- 20.Rubirés, X., F. Saigi, N. Piqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179:7581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuber, K., S. E. Burr, M. Braun, T. Wahli, and J. Frey. 2003. Type III secretion genes in Aeromonas salmonicida subsp. salmonicida are located on a large thermolabile virulence plasmid. J. Clin. Microbiol. 41:3854-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu, H. B., P. S. S. Rao, H. C. Lee, S. Vilches, S. Merino, J. M. Tomás, and K. Y. Leung. 2004. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 72:1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, Y. L., E. Arakawa, and K. Y. Leung. 2002. Novel Aeromonas hydrophila PPD134/91 genes involved in O-antigen and capsule biosynthesis. Infect. Immun. 70:2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]