Abstract
Agricultural activities have produced well-documented changes in the Florida Everglades, including establishment of a gradient in phosphorus concentrations in Water Conservation Area 2A (WCA-2A) of the northern Everglades. An effect of increased phosphorus concentrations is increased methanogenesis in the eutrophic regions compared to the oligotrophic regions of WCA-2A. The goal of this study was to identify relationships between eutrophication and composition and activity of methanogenic assemblages in WCA-2A soils. Distributions of two genes associated with methanogens were characterized in soils taken from WCA-2A: the archaeal 16S rRNA gene and the methyl coenzyme M reductase gene. The richness of methanogen phylotypes was greater in eutrophic than in oligotrophic sites, and sequences related to previously cultivated and uncultivated methanogens were found. A preferential selection for the order Methanomicrobiales was observed in mcrA clone libraries, suggesting primer bias for this group. A greater diversity within the Methanomicrobiales was observed in mcrA clone libraries than in 16S rRNA gene libraries. 16S rRNA phylogenetic analyses revealed a dominance of clones related to Methanosaeta spp., an acetoclastic methanogen dominant in environments with low acetate concentrations. A significant number of clones were related to Methanomicrobiales, an order characterized by species utilizing hydrogen and formate as methanogenic substrates. No representatives of the orders Methanobacteriales and Methanococcales were found in any 16S rRNA clone library, although some Methanobacteriales were found in mcrA libraries. Hydrogenotrophs are the dominant methanogens in WCA-2A, and acetoclastic methanogen genotypes that proliferate in low acetate concentrations outnumber those that typically dominate in higher acetate concentrations.
Methanogenesis is a terminal step and may be a primary regulator of organic matter decomposition in freshwater wetlands. Natural wetlands systems are among the most important sources of methane, emitting approximately 90 ×106 metric tons of methane per year, representing 22% of global methane emissions (6).
The Florida Everglades is one of the largest freshwater marshes in North America and historically a low-nutrient freshwater ecosystem; however, regions of this marsh are now eutrophic due to discharge of nutrient-enriched drainage water from the adjacent Everglades Agricultural Area. This nutrient discharge resulted in a gradient in phosphorus concentrations in the northern Everglades, particularly in Water Conservation Area 2A (WCA-2A) (9, 12, 30, 49). Among the most significant changes effected by the addition of phosphorus to the system was a shift from sparse stands of sawgrass (Cladium spp.) to dense stands of cattails (Typha spp.), thereby increasing the amount of plant material, and hence organic carbon, added to soils of the eutrophic regions (11). Extensive biogeochemical research has shown greater rates of microbial activities, including methanogenesis, in eutrophic zones of WCA-2A compared to more oligotrophic regions of the marsh resulting from the increased phosphorus and carbon put into the system (3, 11, 14, 66). Sulfate reduction rates are also greater in eutrophic zones, but sulfate reduction plays a minor role in anaerobic mineralization of organic matter compared with methanogenesis (3). Other terminal electron acceptors such as O2, NO3−, Fe(III), and Mn(IV) are rapidly depleted in Everglades soils and do not play a significant role in mineralization of organic matter (11, 50). Little is known of the effects of eutrophication on the composition of methanogenic assemblages, although Drake et al. (14) reported an enrichment of total anaerobes and methanogens in the eutrophic zones of WCA-2A compared to more-oligotrophic zones of the adjacent WCA-3A.
Methanogens are a specialized group of anaerobic microorganisms that utilize a narrow range of substrates, including acetate, H2-CO2, formate, and methyl compounds as electron donors (18). In freshwater ecosystems, H2-CO2, acetate, and formate are the main methanogenic precursors (54). Acetate is the primary precursor in these ecosystems, and approximately 60 to 80% of the methane is produced from acetate (7, 27, 65). Most studies on the ecology of methanogens have been conducted in soils and rhizospheres of rice paddies and anaerobic biodigesters (20, 31, 38), and little is known about methanogens in ecosystems such as freshwater marshes. Most of these studies targeted the 16S rRNA gene and mainly focused on rice paddies (5, 20, 21, 28, 31, 36, 39). An alternative approach to study methanogenic community composition is by characterization of a functional gene such as that encoding methyl coenzyme M reductase (MCR) (58), which catalyzes the last step of methanogenesis. Several studies have been conducted with this functional gene to assess diversity of methanogens in several environments, such as peat bogs (22, 37, 42), marine sediments (1), termite guts (44), landfills (40), and rice paddies (38, 39, 48), but none have focused on freshwater wetlands. Non-culture-based microbial diversity studies may provide valuable information on the composition and structure of methanogenic assemblages in freshwater ecosystems and of the effects of eutrophication on methanogenic communities. The objectives of this study were to characterize methanogen assemblages using culture-independent approaches targeting the 16S rRNA and mcr genes and to utilize this information to infer possible roles of particular constituents of these assemblages in eutrophic and oligotrophic sites of the northern Florida Everglades during two seasons, spring and summer.
MATERIALS AND METHODS
Site characteristics, sampling, and biogeochemical characterization.
Studies were conducted on samples taken from WCA-2A, as previously described (3). Samples were collected along the phosphorus gradient from the nutrient-impacted F1 cattail (Typha sp.)-dominated sites and the oligotrophic U3 sawgrass (Cladium sp.)-dominated sites. Soil cores were sampled to a depth of 30 cm and placed on ice by staff of the South Florida Water Management District and transported to the laboratory within 24 h of the collection, where they were sectioned in 10-cm increments and manually mixed. Subsamples for DNA analysis were taken and stored at −70°C until analysis. Biogeochemical characterization was conducted at the Wetland Biogeochemistry Laboratory (10, 63) and is presented in Table 1. Samples used for clone libraries were collected during April and August 2001. Methanogenesis rates were measured in samples collected during December 2001 and January and May 2002.
TABLE 1.
Characterization of 0-to-10-cm layer for eutrophic (F1) and oligotrophic (U3) Everglades soils used in the construction of methanogenic clone libraries
| Sampling site | Vegetation | Sampling time | Water table (cm) | TPb (mg/kg) | TPib (mg/kg) | TKNb (mg/kg) | Extractable TOCb (mg/kg) | MBCb (mg/kg) |
|---|---|---|---|---|---|---|---|---|
| F1 | Cattail | Spring 2001 | −9.1 | 1,536 (57) | 549 (33) | 402 (44) | 4,101 (78) | 7,638 (544) |
| Summer 2001 | 61.3 | 1,089 (205) | 388 (108) | 309 (29) | 2,798 (278) | 4,043 (1,000) | ||
| U3 | Sawgrass, | Spring 2001 | −0.9 | 277 (19) | 69 (10) | 343 (120) | 2,892 (90) | 2,066 (221) |
| slough | Summer 2001 | 62.5 | 377 (59) | 82 (15) | 305 (69) | 1,741 (376) | 2,276 (607) |
Values in parentheses represent the standard deviation of triplicate determinations.
Data are expressed per kilogram (dry weight) of soil. TP, total phosphorus; TPi, total inorganic phosphorus; TKN, total Kjeldahl nitrogen; TOC, total organic carbon; MBC, microbial biomass carbon.
Methanogenesis rate measurement.
One gram of soil from the 0-to-10-cm soil layer was mixed with 2 ml of anoxic basal carbonate yeast extract trypticase medium (60) under a N2 stream in anaerobic culture tubes that were later closed with butyl rubber stoppers and aluminum seals. The tubes were preincubated for 2 weeks before substrates were added. Acetate and formate (20 mM each) were added from anaerobic sterile concentrated stock solutions. The tubes were fitted with three-way Luer stopcocks (Cole-Parmer, Vernon Hills, Ill.) for gas sampling and incubated in the dark at 25°C with shaking at 100 rpm. Methane in the headspace was measured by gas chromatography with a Shimadzu 8A gas chromatograph equipped with a Carboxen 1000 column (Supelco, Bellefonte, Pa.) and a flame ionization detector operating at 110°C. The carrier gas was N2, and the oven temperature was 160°C. All determinations were carried out in triplicate. Headspace pressure was measured using a digital pressure indicator (DPI 705; Druck, New Fairfield, Conn.). Methane determinations were conducted in quintuplicate. Statistical analyses were performed by using JMP statistical software (version 4.04; SAS Institute, Cary, N.C.). Two-way analysis of variance was performed with substrate addition and site type as factors. Subsequently, the Tukey-Kramer honestly significant difference means test (P = 0.05) was used for comparisons of the treatments.
Nucleic acid extraction and PCR amplification.
Nucleic acids were extracted from 0.25 g of the 0-to-10-cm soil layer with UltraClean soil DNA kits (MoBio, Solana Beach, Calif.) according to the manufacturer's instructions. PCR was conducted using the primer set designed by Luton et al. (40), which amplifies a ca. 465- to 490-bp fragment of mcrA; this set consisted of primers mcrA-f (5′-GGTGGTGTMGGATTCACACARTAYGCWASCGC-3′) and mcrA-r (5′-TTCATTGCRTAGTTWGGRTAGTT-3′). The reaction mixture used for PCR amplification contained 7 μl of distilled H2O, 1 μl of each primer (10 pmol/μl), 10 μl of HotStarTaq Master Mix (QIAGEN, Valencia, Calif.), and 1 μl of diluted DNA solution. PCR amplification was carried out in a GeneAmp PCR system 2400 (Perkin-Elmer Applied Biosystems, Norwalk, Conn.). The initial enzyme activation and DNA denaturation were performed for 15 min at 95°C, followed by five cycles of 30 s at 95°C, 30 s at 55°C, and 30 s of extension at 72°C, and the temperature ramp between the annealing and extension segment was decreased to 0.1°C per s from the default 1°C per s because of the degeneracy of the primers. After this, 30 cycles were performed with the following conditions: 30 s at 95°C, 30 s at 55°C, and 30-s extension at 72°C, and a final extension of 72°C for 7 min. The PCR products were electrophoresed on 2% agarose gels in Tris-acetate-EDTA buffer to confirm amplification of expected size product.
The primer set combination 23F and 1492R was used for 16S rRNA gene amplification (2, 32). The initial enzyme activation and DNA denaturation was performed for 15 min at 95°C, followed by 35 cycles of 30 s at 94°C, 30 s at 60°C, and 60-s extension at 72°C, and a final extension of 72°C for 7 min. PCR products were electrophoresed on 0.7% agarose gels in Tris-acetate-EDTA buffer to confirm amplification of the expected size of product.
Cloning of mcrA and 16S rRNA genes and RFLP analysis.
Fresh PCR amplicons were ligated into pCRII-TOPO cloning vector and transformed into chemically competent Escherichia coli TOP10F′ cells according to the manufacturer's instructions (Invitrogen, Carlsbad, Calif.). Individual colonies of E. coli were screened by direct PCR amplification with the mcr or 16S rRNA primers using the previously described PCR programs. Random fragment length polymorphism (RFLP) analyses were conducted using the restriction enzyme TacI for mcrA and 16S rRNA gene amplification products. Digests were analyzed by agarose gel electrophoresis using 2% and 4% gels for 16S rRNA and mcrA digests, respectively. Clone libraries were analyzed by analytic rarefaction using the software aRarefactWin (version 1.3; S. Holland, Stratigraphy Lab, University of Georgia, Athens [http://www.uga.edu/∼strata/software/]).
Sequencing and phylogenetic analysis.
Selected clones were sequenced at the DNA Sequencing Core Laboratory at the University of Florida with the mcrA-f and 23F primers. A total of 76 16S rRNA gene clones were partially sequenced (approximately 500 bp). For mcrA, 52 clones were partially sequenced (480 bp, which translated into a deduced amino acid sequence of approximately 160 amino acids). Deduced amino acid sequences of the MCR α-subunit were aligned and analyzed with ClustalX version 1.81 (59). Since Luton et al. (40) did not find differences between different mcrA phylogenetic analyses, data presented here are from a neighbor-joining analysis using ClustalX 1.81. Methanopyrus kandleri sequence was used as an outgroup in the phylogenetic analysis (40). Bootstrap analysis was performed with 100 resamplings of the amino acid sequences. 16S rRNA gene sequence alignments were also evaluated with PAUP* version 4.0b8 using parsimony-based algorithms (D. L. Swofford, Sinauer Associates, Sunderland, Mass.). Several phylogenetic approaches yielded similar results with only minor changes in the placement of some sequences within the major cluster. Trees were constructed using heuristic searches with 10 random stepwise additions of taxa and by tree-bisection reconnection branch swapping. The characters were weighted to give more weight to characters with lower levels of homoplasy. Bootstrap analysis was performed with 100 resamplings. M. kandleri sequence was used as the outgroup, because this species is the suggested outgroup for the mcrA analysis (40). The nomenclature of the Archaea domain is according to the taxonomic outline of the prokaryotic genera of Bergey's Manual of Systematic Bacteriology (19).
Diversity index.
The cumulative number of phylotypes was calculated by fitting the rarefaction curves to a hyperbolic model with the formula y = ax/(b + x) using Datafit software (version 8.0.32; Oakdale Engineering, Oakdale, Pa.), where y represents the number of phylotypes and x is the number of individuals. Phylotypes were defined on the basis of unique restriction digestion patterns. Diversity of clone libraries was calculated using the Shannon index (45) according to the following formula: H = -Σpi(log pi) from i = 1 to n, where pi represents the proportion of a particular phylotype in the clone library and n is the total number of phylotypes.
Nucleotide sequence accession numbers.
GenBank accession numbers for partial mcrA sequences for F1 samples are AY458405 to AY458419 for spring samples and AY459307 to AY459319 for summer samples. Partial mcrA sequences for U3 samples are AY458420 to AY458427 for spring and AY460212 to AY460220 for summer samples. GenBank accession numbers for partial 16S rRNA sequences for F1 samples are AY456713 to AY456731 for spring and AY475640 to AY457657 for summer samples. Partial 16S rRNA sequences for U3 samples are AY456732 to AY456748 for spring and AY457658 to AY457673 for summer samples.
RESULTS
Methanogenic rates.
Intrinsic initial methanogenesis rates were highly affected by the addition of acetate, site eutrophication status, and the interaction effect of acetate addition and eutrophication status (two-way analysis of variance; P < 0.0001). Intrinsic methanogenic rates were five to nine times higher in F1 soils than in U3 soils (Table 2), and total methane accumulated at the end of 6 days of incubation was 4 to 10 times higher for the F1 soils than for U3 soils (P < 0.05). Addition of acetate resulted in an increase in methanogenesis rates in soils from both sites, with the exception of U3 samples taken in May 2002. The rates with acetate addition were 3 to 12 times higher for F1 than for U3 soil samples.
TABLE 2.
Potential methanogenic rates and CH4 accumulated in eutrophic (F1) and oligotrophic (U3) Everglades soils
| Date and addition | F1
|
U3
|
||
|---|---|---|---|---|
| Ratea | μmol of CH4b | Rate | μmol of CH4 | |
| Dec. 2001 | ||||
| None | 0.015 (0.002) | 2.0 (0.4) | 0.003 (0.001) | 0.2 (0.01) |
| Acetate | 0.057 (0.003) | 8.8 (0.3) | 0.007 (0.001) | 0.4 (0.04) |
| Jan. 2002 | ||||
| None | 0.026 (0.003) | 1.4 (0.2) | 0.004 (0.001) | 0.4 (0.04) |
| Acetate | 0.049 (0.005) | 7.8 (0.3) | 0.016 (0.003) | 2.9 (0.9) |
| May 2002 | ||||
| None | 0.026 (0.002) | 3.2 (0.3) | 0.003 (0.001) | 0.4 (0.1) |
| Acetate | 0.040 (0.002) | 7.1 (1.2) | 0.003 (0.001) | 0.2 (0.1) |
| Formate | 0.084 (0.005) | 7.0 (0.5) | 0.005 (0.001) | 1.2 (0.3) |
Potential methanogenic rates (in micromoles per gram per hour); for details see the section on methanogenesis in Materials and Methods. Standard errors of the means are shown in parentheses (n = 5).
Micromoles of CH4 accumulated at the end of 6 days of incubation.
Hydrogen has been shown to be an important electron donor in eutrophic zones of this marsh (61). Approximately 60% of cultured hydrogenotrophic methanogens are considered to be capable of using formate as an electron donor (18), and use of formate as a surrogate for H2-CO2 is a common practice in anaerobic digestion studies because diffusion problems from the gas headspace to the liquid media are avoided (13). We employed formate as a surrogate for H2-CO2 to test the response of these soils to H2-CO2, and we found that addition of formate resulted in a 17-fold increase in F1 samples compared with U3 soil samples.
Diversity indices for cloned mcrA and 16S rRNA sequences.
Samples from eutrophic soils were more diverse in both libraries when evaluated by either the cumulative expected phylotypes index or Shannon's index. Archaeal 16S rRNA gene libraries were more diverse than were mcrA clone libraries (Table 3).
TABLE 3.
Expected and recovered phylotypes and diversity indices for archaeal 16S rRNA and mcrA clone libraries for eutrophic and oligotrophic Everglades soils
| Clone library | No. of expected phylotypesa | No. of recovered phylotypesb | Shannon's index |
|---|---|---|---|
| Archaeal 16S rRNA | |||
| F1, summer | 36.0 ± 0.8 | 20 (38) | 1.25 |
| F1, spring | 86.7 ± 2.5 | 23 (32) | 1.29 |
| U3, summer | 28.0 ± 0.8 | 16 (38) | 1.06 |
| U3, spring | 32.8 ± 0.8 | 17 (36) | 1.10 |
| mcr | |||
| F1, summer | 22.7 ± 0.6 | 14 (35) | 1.03 |
| F1, spring | 36.1 ± 0.8 | 18 (37) | 1.12 |
| U3, summer | 13.2 ± 0.1 | 9 (29) | 0.83 |
| U3, spring | 12.9 ± 0.1 | 10 (38) | 0.83 |
Value of constant a from hyperbolic equation y = ax/(b + x) with the standard error.
Values in parentheses represent the total number of clones screened.
RFLP patterns of the 16S rRNA gene clones suggested a significant degree of sequence diversity in summer and spring samples. The number of phylotypes observed were lower than the predicted number of phylotypes according to a hyperbolic model fit to the data; however, covering all the diversity would require sequencing an extremely high number of clones, making the study economically unfeasible.
A significant degree of sequence diversity in summer and spring samples was observed with mcrA RFLP. However, diversity was lower than with the archaeal 16S rRNA gene libraries, since the mcr set of primers only targeted methanogenic prokaryotes and not the total archaeal population targeted with the archaeal primers. In the case of mcr sequences, the recovered phylotypes were more similar to the expected phylotypes than was the case for the 16S rRNA analysis (Table 3). In both cases, F1 samples exhibited a slightly higher degree of diversity as judged by the slope of the rarefaction curves (data not shown), which was in good agreement with the calculated Shannon's indices.
Phylogenetic analysis of cloned 16S rRNA sequences.
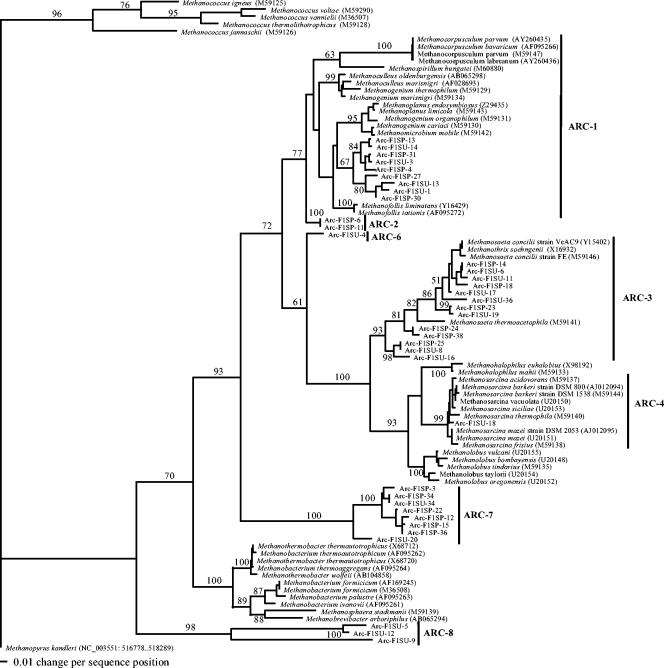
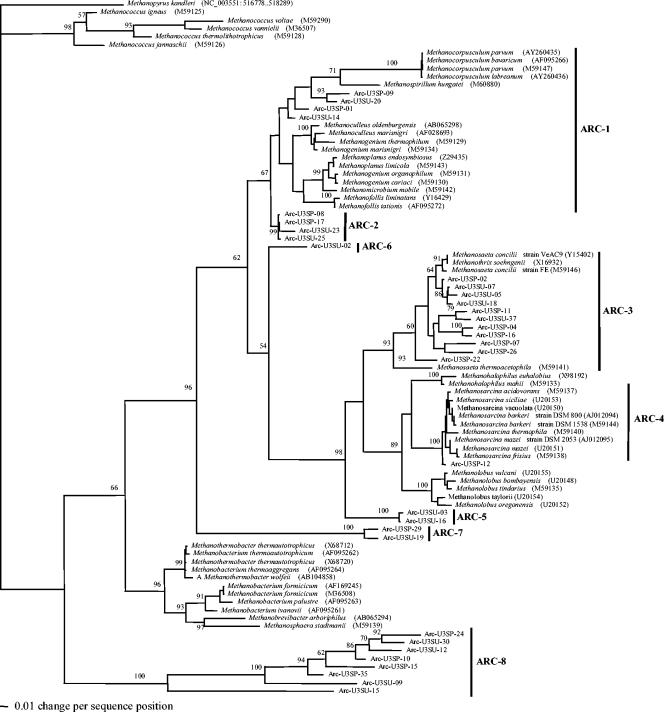
Cloned archaeal sequences were distributed in a total of eight clades: one clade in the phylum Crenarchaeota and seven clades in the phylum Euryarchaeota (Fig. 1 and 2). The overall and major clades of the archaeal phylogeny were in very good agreement with previously published archaeal phylogeny (5, 18, 20). The seven Euryarchaeota clades included methanogens of the orders Methanobacteriales, Methanomicrobiales, and Methanosarcinales and three putative clades composed of sequences from uncultured microorganisms.
FIG. 1.
Maximum parsimony tree for archaeal 16S rRNA gene sequences from eutrophic F1 sites of Everglades WCA-2A. The clones are named according to the site and time of sampling. The scale bar represents 1 nucleotide change per 100 sequence positions. Numbers at nodes represent percentages of bootstrap resamplings based on 100 replicates; only values above 50 are presented.
FIG. 2.
Maximum parsimony tree for archaeal 16S rRNA gene sequences from oligotrophic U3 sites of Everglades WCA-2A. The clones are named according to the site and time of sampling. The scale bar represents 1 nucleotide change per 100 sequence positions. Numbers at nodes represent the percentages of bootstrap resamplings based on 100 replicates; only values above 50 are presented.
The ARC-1 cluster is related to the order Methanomicrobiales and uncultivated Methanomicrobiales sequences recovered from a variety of ecosystems (16, 41, 46, 47). The ARC-2 cluster is related to sequences (26, 62) referred to in GenBank as unclassified/uncultured Archaea (environmental samples) or unclassified/uncultured Methanomicrobiales (43). The phylogenetic affiliation of these clones remains uncertain, but they may represent a deeply divergent branch of the Methanomicrobiales order. ARC-3 clusters with cultivated and uncultivated archaeal sequences related to Methanosaeta spp. recovered from a great variety of ecosystems (16, 20, 33, 39, 46-48, 55). The ARC-4 cluster was related to the genus Methanosarcina. The ARC-5 cluster was related to the Methanosarcinales order, although this cluster was deeply divergent from other Methanosarcinales. The ARC-6 cluster was composed of two clones that were deeply divergent in the Methanosarcinales order by parsimony analysis. However, these clones were placed as a very divergent deep branch grouping with the Methanomicrobiales in the neighbor-joining analysis. The GenBank BLAST report for these sequences returned representatives of the Methanomicrobiales and Methanosarcinales orders. More information is required to place these clones with a particular order. The ARC-7 cluster is deeply divergent from the Methanosarcinales and Methanomicrobiales orders, and clones related to this group have been reported in other environments (23, 29, 43, 56). The ARC-8 cluster is comprised of sequences related to the Crenarchaeota phylum. The Crenarchaeota phylum is composed of extreme hyperthermophilic prokaryotes (51, 57); however, related sequences have been found in cold and moderate-temperature environments (39, 55). No clone related to the Methanococcales or Methanobacteriales order was found in clone libraries from any season or site in this study.
Effect of eutrophication and season on the dynamics of archaeal sequences.
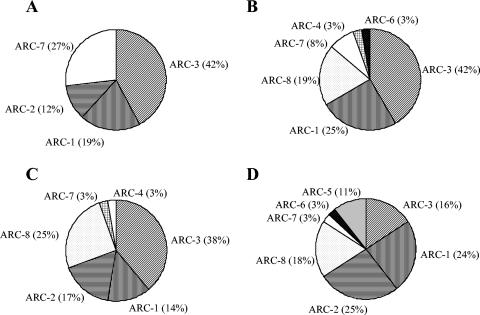
Clone libraries for F1 sites were dominated by clusters ARC-3 (Methanosaeta cluster), ARC-1 (Methanomicrobiales cluster), and ARC-7 (uncultured cluster), representing almost 80% of the clone library sequences for summer samples and 90% for spring samples (Fig. 3). The summer F1 clone library also contained ARC-8 (Crenarchaeota cluster) and minor amounts of ARC-4 sequences (Methanosarcina cluster) and ARC-6 sequences (uncultured cluster) (Fig. 3). Seasonal changes between summer and spring for eutrophic F1 sites resulted in an emergence of clones of cluster ARC-2 (uncultured cluster) and disappearance of sequences of clusters ARC-4, ARC-6, and ARC-8 from summer to spring, but clusters ARC-1, ARC-3, and ARC-7 dominated clone libraries from both sampling sites.
FIG. 3.
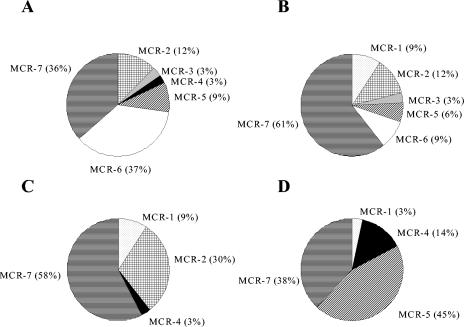
Spatial and seasonal distributions of archaeal 16S rRNA clones in eutrophic soils for spring (A) and summer (B) and oligotrophic soils for spring (C) and summer (D).
U3 clone libraries were dominated by clusters ARC-3, ARC-1, ARC-2, and ARC-8, with minor amounts of cluster ARC-7 (Fig. 3). Seasonal changes from summer to spring resulted in some changes of the relative amounts of the major cluster and in the emergence of cluster ARC-4 and disappearance of clusters ARC-5 (uncultured cluster) and ARC-6.
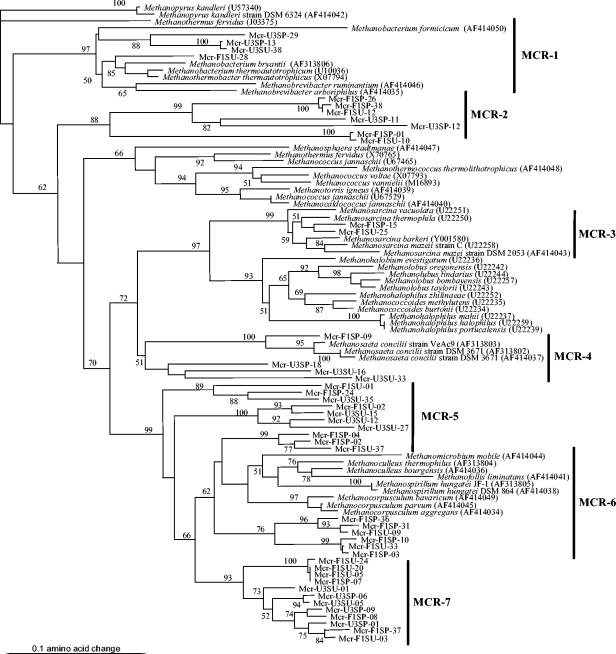
Phylogenetic analysis of cloned mcrA sequences.
Cloned MCR sequences were distributed among a total of seven clades encompassing the orders Methanobacteriales, Methanomicrobiales, and Methanosarcinales and three putative clades composed of sequences from uncultured microorganisms (Fig. 4). The overall MCR phylogeny (Fig. 4) was in very good agreement with previously published MCR phylogenies.
FIG. 4.
Neighbor-joining MCR α-subunit tree. The clones are named according to the origin and time of sampling. The scale bar represents 10% sequence divergence. Numbers at nodes represent the percentages of bootstrap resamplings based on 100 replicates; only values higher than 50 are presented.
The MCR-1 cluster was related to the order Methanobacteriales, with similar clones found in rice paddies (38) and landfill material (40). The MCR-2 cluster was composed exclusively of our clones and sequences from uncultured microorganisms (15, 25), and their phylogenetic affiliation remains uncertain. The MCR-3 cluster was related to Methanosarcina sequences, with similar sequences also recovered from landfill and rice paddy soils (38, 40). The MCR-4 cluster was related to the genus Methanosaeta of the order Methanosarcinales. Sequences Mcr-U3SP-18, U3SU-16, and U3SU-33 exhibited ca. 85% similarity to sequences of rice cluster I, outside the Methanosaeta cluster described by Lueders et al. (38); however, bootstrap analysis did not support these as a separate cluster. More sequence information would clarify the assignment of these sequences as an individual cluster. Cluster MCR-5 was composed of sequences branching deeper in the Methanomicrobiales order. Sequences clustering with this group have been also found in landfill soils (40); however, they are deeply divergent from cultured Methanomicrobiales species. Similar sequences were also obtained from a variety of environments (15, 17, 25). These sequences could be divided into two clusters but were treated as a single group here because of the lack of information on physiological characteristics that might support division of the cluster into two groups. Cluster MCR-6 was related to Methanomicrobiales, and similar sequences were obtained from landfill soils (40) and rice paddies (38). The MCR-7 cluster was related to Methanomicrobiales but in a separate cluster from cultured Methanomicrobiales.
Effect of eutrophication and season on the dynamics of MCR sequences.
Clone libraries for F1 sites were dominated by sequences in clusters MRC-7, MCR-6, and MCR-5 (related to Methanomicrobiales). The summer F1 clone library also contained MCR-1 (Methanobacteriales cluster), MCR-2 (uncultivated cluster), and minor amounts of MCR-3 sequences (Methanosarcina cluster) (Fig. 5). Seasonal changes between summer and spring for eutrophic sites resulted in an increase of MCR-6 and decrease of MCR-7, an emergence of MCR-4 clones (Methanosaeta cluster), and disappearance of MCR-1 sequences. Methanobacteriales was not recovered from archaeal libraries; however, these clones represented approximately 10% of the mcrA libraries. Screening of more clones in the archaeal libraries may yield clones within this group.
FIG. 5.
Spatial and seasonal distribution of mcrA clones in eutrophic soils for spring (A) and summer (B) and oligotrophic soils for spring (C) and summer (D).
U3 clone libraries were more dynamic; summer samples were dominated by clones of the clusters MCR-5 and MCR-7, followed by MCR-4 and minor amounts of MCR-1 (Fig. 5). Seasonal changes from summer to spring resulted in an increase in the relative amount of MCR-7 sequences, the emergence of MCR-2 sequences, the disappearance of MCR-5 sequences (uncultured cluster), and minor changes in the frequencies of MCR-1 and MCR-4 sequences. Clearly, the dominance of clones related to Methanomicrobiales and the lower representation of Methanosaeta spp., a cluster present in significant numbers in the archaeal 16S rRNA gene libraries, may indicate a PCR bias for the mcr set of primers. Luton et al. (40) tested the set of primers with Methanosaeta and Methanosarcina spp. and obtained amplification of sequences related to these genera. However, most clones present in their libraries were related to species belonging to Methanomicrobiales and Methanobacteriales. Few clones were related to Methanosarcina spp., and none were related to Methanosaeta spp. The authors attributed these observations to PCR biases. Therefore, it seems that this set of mcr primers would be appropriate primers to study diversity of Methanobacteriales and Methanomicrobiales. The dynamics of these two methanogenic orders may be useful as indicators to assess the effects of eutrophication on microbial communities.
DISCUSSION
Although methanogenesis is one of the main processes responsible for terminal anaerobic organic matter mineralization in the Everglades, very little is known about the microbial groups involved in this process (66). Extensive biogeochemical research has shown that phosphorus loading has resulted in changes at the micro and macro levels in nutrient-impacted regions of WCA-2A (3, 11, 14, 66). However, this is the first complete report of which we are aware that has characterized assemblages of methanogens in oligotrophic and eutrophic zones of the northern Everglades. The only previous attempt to characterize methanogenic communities in the Everglades was conducted by Drake et al. (14) in similar sites. Those authors reported an enrichment of almost 6 orders of magnitude in acetoclastic methanogens in eutrophic regions. They also reported an enrichment of acetate-producing microorganisms and H2-consuming microorganisms in the eutrophic zones of the marsh. The number of H2-consuming acetogens was similar in eutrophic and oligotrophic sites, which may indicate that the increase of total H2-consuming microorganisms is due to hydrogenotrophic methanogens. These differences in microbial enumerations correspond with lower methanogenic rates observed for oligotrophic sites than for eutrophic sites. In a more recent study, our investigators found that hydrogenotrophic methanogens were 1,000 and 100 times higher than acetoclastic methanogens in eutrophic and oligotrophic soils, respectively, and relative numbers of acetoclastic methanogens were similar between eutrophic and oligotrophic sites (4). Clearly, phosphorus loading of the Everglades WCA-2A is affecting, either directly or indirectly, the population size and activity of methanogens, resulting in greater numbers and methanogenic activities in phosphorus-enriched sites. Methanogenesis rates for samples of eutrophic sites measured using formate were ca. 2.5 times higher than methanogenesis rates measured using acetate as substrate. These data suggest that hydrogenotrophic methanogens are important in the decomposition of organic matter in eutrophic Everglades soils.
Acetate utilization is restricted to two genera in the order Methanosarcinales, Methanosaeta and Methanosarcina; all other species of methanogens use H2. However, it is estimated that 70 to 80% of the methane produced in nature comes from conversion of acetate to methane by acetoclastic methanogens (27). Conrad reported that the contribution to H2 or acetate to methane production is highly variable and that the relative proportions are reversed in cases where the hydrogen contribution is higher (7). In some cases, methanogenesis is exclusively driven by hydrogen. Examples include eutrophic lakes, hot spring mats, coastal marine sediments, temperate bogs, Antarctic water bodies, and lake sediments. Recently, Horn et al. (24) reported that hydrogen is the main methanogenic precursor in acidic peat, which is in agreement with other studies conducted in acidic peats (34, 64).
There are several possible explanations for the higher contribution of hydrogen as electron donor for methanogenesis, including additional sinks or loss of acetate by nonmethanogenic microorganisms and additional pools of hydrogen, such as production of high amounts of H2 by fermentation of organic matter or geological inputs. Acetate can be consumed by nonmethanogenic microorganisms using electron acceptors such as oxygen, nitrate, ferric ion, and sulfate if these electron acceptors are available. We previously reported a slight competition for acetate between methanogens and sulfate-reducing prokaryotes in eutrophic F1 soils (3). With the exception of sulfate reduction, these alternative terminal electron-accepting processes are not considered to be significant in WCA-2A soils compared with methanogenesis.
Clones in the archaeal 16S rRNA gene libraries related to previously cultured methanogens in groups ARC-1 (Methanobacteriales), ARC-3 (Methanosaeta spp.), and ARC-4 (Methanosarcina spp.) may provide insight into the dynamics of acetate in these soils. Two methanogens related to Methanosarcina spp. and Methanosaeta spp. were detected. Methanosaeta spp. are specialists that grow only on acetate and dominate at low acetate concentrations (7 to 70 μM); Methanosarcina spp. are generalists that grow on hydrogen and methyl compounds but require acetate at much higher concentrations (0.2 to 1.2 mM) (27) than do Methanosaeta spp. The dominance of Methanosaeta-like sequences strongly suggests that acetate concentrations are low in these soils. Methanosarcina sequences were observed in small proportions, which may indicate soil niches where acetate concentrations are locally high. Cluster ARC-1 sequences (related to the Methanomicrobiales) were observed in significant numbers in all clone libraries, indicating that this order may be responsible for hydrogenotrophic methanogenesis. mcrA clone libraries were dominated by sequences related to Methanomicrobiales (clusters ARC-5, ARC-6, and ARC-7) and were highly diverse for this particular microbial group. A similar enrichment of Methanomicrobiales and Methanobacteriales, hydrogenotrophic methanogens, was reported for peat soils where hydrogen was an important methanogenic precursor (24).
The partial pressure of hydrogen can be a primary factor controlling the products of fermentation. If the hydrogen partial pressure is kept below 10−3 atm, fermentation to acetate, H2, and CO2 occurs in some systems; however, if H2 accumulates, the formation of more-reduced products such as fatty acids or alcohol is promoted (52, 67). This shift results in an increase in the pool of fatty acids (butyrate or propionate), which are syntrophically degraded to acetate and H2-CO2, increasing the pool of hydrogen for hydrogenotrophic methanogens. Hydrogen is converted to methane by species in the order Methanomicrobiales. Acetate in low concentrations may be used by sulfate-reducing prokaryotes and syntrophic acetate oxidizers (8, 35, 53, 68), which would increase the hydrogen pool. This would favor proliferation of the specialist Methanosaeta spp., rather than the generalist Methanosarcina spp.
Phosphorus enrichment plays a major role in the carbon cycle of the Everglades by increasing the input of organic matter, which consequently increases the activity and number of methanogens in these soils. The diversity of hydrogenotrophic methanogenic assemblages was also affected by eutrophication, which may indicate a selection of different hydrogenotrophic methanogens driven by different levels of hydrogen between eutrophic and oligotrophic sites of the Everglades. We are currently studying the response of other microbial groups, including syntrophic bacteria (4) and fermentative bacteria (61), to eutrophication, which will provide greater insight into the role of hydrogen as a major substrate of methanogenic communities in this type of ecosystem.
Acknowledgments
This work was supported by grant DEB-0078368 from the National Science Foundation.
We are thankful to Joe Prenger and Yu Wang, Wetland Biogeochemistry Laboratory, Soil and Water Science Department, and Sue Newman, South Florida Water Management District, for their assistance with field sampling, sample coordination, and preparation.
Footnotes
Florida Agricultural Experimental Station journal series no. R-10329.
REFERENCES
- 1.Bidle, K. A., M. Kastner, and D. H. Bartlett. 1999. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP site 892B). FEMS Microbiol. Lett. 177:101-108. [DOI] [PubMed] [Google Scholar]
- 2.Burggraf, S., K. O. Stetter, P. Rouviere, and C. R. Woese. 1991. Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst. Appl. Microbiol. 14:346-351. [DOI] [PubMed] [Google Scholar]
- 3.Castro, H., K. R. Reddy, and A. Ogram. 2002. Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68:6129-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chauhan, A., A. Ogram, and K. R. Reddy. 2004. Syntrophic-methanogenic associations along a nutrient gradient in the Florida Everglades. Appl. Environ. Microbiol. 70:3475-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cicerone, R. J., and R. S. Oremland. 1988. Biogeochemical aspects of atmospheric methane. Global Biogeochem. Cycles 2:299-327. [Google Scholar]
- 7.Conrad, R. 1999. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 28:193-202. [Google Scholar]
- 8.Cord-Ruwisch, R., D. R. Lovley, and B. Schink. 1998. Growth of Geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl. Environ. Microbiol. 64:2232-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craft, C. B., and C. J. Richardson. 1993. Peat accretion and phosphorus accumulation along a eutrophication gradient in the northern Everglades. Biogeochemistry 22:133-156. [Google Scholar]
- 10.D'Angelo, E. M., and K. R. Reddy. 1999. Regulators of heterotrophic microbial potentials in wetland soils. Soil Biol. Biochem. 31:815-830. [Google Scholar]
- 11.DeBusk, W. F., and K. R. Reddy. 1998. Turnover of detrital organic carbon in a nutrient-impacted Everglades marsh. Soil Sci. Soc. Am. J. 62:1460-1468. [Google Scholar]
- 12.DeBusk, W. F., K. R. Reddy, M. S. Koch, and Y. Wang. 1994. Spatial distribution of soil nutrients in a northern Everglades marsh: Water Conservation Area 2A. Soil Sci. Soc. Am. J. 58:543-552. [Google Scholar]
- 13.Dolfing, J., and W. G. B. M. Bloemen. 1985. Activity measurements as a tool to characterize the microbial composition of methanogenic environments. J. Microbiol. Methods 4:1-12. [Google Scholar]
- 14.Drake, H. L., N. G. Aumen, C. Kuhner, C. Wagner, A. Grieshammer, and M. Schmittroth. 1996. Anaerobic microflora of Everglades sediments effects of nutrients on population profiles and activities. Appl. Environ. Microbiol. 62:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Earl, J., G. Hall, R. W. Pickup, D. A. Ritchie, and C. Edwards. 2003. Analysis of methanogen diversity in a hypereutrophic lake using PCR-RFLP analysis of mcr sequences. Microbial Ecol. 46:270-278. [DOI] [PubMed] [Google Scholar]
- 16.Ficker, M., K. Krastel, S. Orlicky, and E. Edwards. 1999. Molecular characterization of a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 65:5576-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galand, P. E. 2002. Depth related diversity of methanogen Archaea in Finnish oligotrophic fen. GenBank direct submission. http://www.ncbi.nlm.nih.gov/. [DOI] [PubMed]
- 18.Garcia, J. L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic phylogenetic and ecological diversity of methanogenic Archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 19.Garrity, G. M., M. Winters, and D. B. Searles. 2001. Taxonomic outline of the prokaryotic genera. Bergey's manual of systematic bacteriology. http://www.cme.msu.edu/bergeys/april2001-genus.
- 20.Groβkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groβkopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog feat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 24.Horn, M. A., C. Matthies, K. Kusel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hougaard, E., and P. Westermann. 2000. The archaeal populations in anaerobic digesters investigated by comparison of 16S rDNA, rRNA and the gene mcrI encoding the mRNA of methyl coenzyme M reductase. GenBank direct submission. http://www.ncbi.nlm.nih.gov/
- 26.Huang, L. N., H. Zhou, Y. Q. Chen, S. Luo, C. Y. Lan, and L. H. Qu. 2002. Diversity and structure of the archaeal community in the leachate of a full-scale recirculating landfill as examined by direct 16S rRNA gene sequence retrieval. FEMS Microbiol. Lett. 214:235-240. [DOI] [PubMed] [Google Scholar]
- 27.Jetten, M. S. M., A. J. M. Stams, and A. J. B. Zehnder. 1992. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanotrix soehngenii and Methanosarcina spp. FEMS Microbiol. Ecol. 88:181-197. [Google Scholar]
- 28.Joulian, C., B. Ollivier, B. K. C. Patel, and P. A. Roger. 1998. Phenotypic and phylogenetic characterization of dominant culturable methanogens isolated from ricefield soils. FEMS Microbiol. Ecol. 25:135-145. [Google Scholar]
- 29.Jurgens, G., F. O. Glockner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Munster. 2000. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 30.Koch, M. S., and K. R. Reddy. 1992. Distribution of soil and plant nutrients along a trophic gradient in the Florida Everglades. Soil Sci. Soc. Am. J. 56:1492-1499. [Google Scholar]
- 31.Kudo, Y., T. Nakajima, T. Miyaki, and H. Oyaizu. 1997. Methanogen flora of paddy soils in Japan. FEMS Microbiol. Ecol. 22:39-48. [Google Scholar]
- 32.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 33.Lanoil, B. D., R. Sassen, M. T. La Duc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lansdown, J. M., P. D. Quay, and S. L. King. 1992. CH4 production via CO2 reduction in a temperate bog: a source of C-13-depleted CH4. Geochim. Cosmochim. Acta 56:3493-3503. [Google Scholar]
- 35.Lee, M. J., and S. H. Zinder. 1988. Isolation and characterization of a thermophilic bacterium which oxidizes acetate in syntrophic association with a methanogen and which grows acetogenically on H2-CO2. Appl. Environ. Microbiol. 54:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann-Richter, S., R. Groβkopf, W. Liesack, P. Frenzel, and R. Conrad. 1999. Methanogenic archaea and CO2-dependent methanogenesis on washed rice roots. Environ. Microbiol. 1:159-166. [DOI] [PubMed] [Google Scholar]
- 37.Lloyd, D., K. L. Thomas, A. Hayes, B. Hill, B. A. Hales, C. Edwards, J. R. Saunders, D. A. Ritchie, and M. Upton. 1998. Micro-ecology of peat: minimally invasive analysis using confocal laser scanning microscopy, membrane inlet mass spectrometry and PCR amplification of methanogen-specific gene sequences. FEMS Microbiol. Ecol. 25:179-188. [Google Scholar]
- 38.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 39.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 41.McHugh, S., M. Carton, T. Mahony, and V. O'Flaherty. 2003. Methanogenic population structure in a variety of anaerobic bioreactors. FEMS Microbiol. Lett. 219:297-304. [DOI] [PubMed] [Google Scholar]
- 42.Nercessian, D., M. Upton, D. Lloyd, and C. Edwards. 1999. Phylogenetic analysis of peat bog methanogen populations. FEMS Microbiol. Lett. 173:425-429. [DOI] [PubMed] [Google Scholar]
- 43.Nusslein, B., K. J. Chin, W. Eckert, and R. Conrad. 2001. Evidence for anaerobic syntrophic acetate oxidation during methane production in the profundal sediment of subtropical Lake Kinneret (Israel). Environ. Microbiol. 3:460-470. [DOI] [PubMed] [Google Scholar]
- 44.Ohkuma, M., S. Noda, K. Horikoshi, and T. Kudo. 1995. Phylogeny of symbiotic methanogens in the gut of the termite Reticulitermes speratus. FEMS Microbiol. Lett. 134:45-50. [DOI] [PubMed] [Google Scholar]
- 45.Peet, R. K. 1974. The measurement of species diversity. Annu. Rev. Ecol. Syst. 5:285-307. [Google Scholar]
- 46.Purdy, K. J., M. A. Munson, D. B. Nedwell, and T. M. Embley. 2002. Comparison of the molecular diversity of the methanogenic community at the brackish and marine ends of a UK estuary. FEMS Microbiol. Ecol. 39:17-21. [DOI] [PubMed] [Google Scholar]
- 47.Purdy, K. J., D. B. Nedwell, and T. M. Embley. 2003. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 69:3181-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramakrishnan, B., T. Lueders, P. F. Dunfield, R. Conrad, and M. W. Friedrich. 2001. Archaeal community structures in rice soils from different geographical regions before and after initiation of methane production. FEMS Microbiol. Ecol. 37:175-186. [Google Scholar]
- 49.Reddy, K. R., R. D. DeLaune, W. F. DeBusk, and M. S. Koch. 1993. Long-term nutrient accumulation rates in the Everglades. Soil Sci. Soc. Am. J. 57:1147-1155. [Google Scholar]
- 50.Reddy, K. R., J. R. White, A. Wright, and T. Chua. 1999. Influence of phosphorus loading on microbial processes in soil and water column of wetlands, p. 249-273. In K. R. Reddy, G. A. O'Connor, and C. L. Schelske (ed.), Phosphorus biogeochemistry in subtropical ecosystems. Lewis Publishers, New York, N.Y.
- 51.Reysenbach, A. L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schink, B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schnurer, A., B. Schink, and B. H. Svensson. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int. J. Syst. Bacteriol. 46:1145-1152. [DOI] [PubMed] [Google Scholar]
- 54.Schutz, H., W. Seiler, and R. Conrad. 1989. Processes involved in formation and emission of methane in rice paddies. Biogeochemistry 7:33-53. [Google Scholar]
- 55.Stein, L. Y., G. Jones, B. Alexander, K. Elmund, C. Wright-Jones, and K. H. Nealson. 2002. Intriguing microbial diversity associated with metal-rich particles from a freshwater reservoir. FEMS Microbiol. Ecol. 42:431-440. [DOI] [PubMed] [Google Scholar]
- 56.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teske, A., K. U. Hinrichs, V. Edgcomb, A. D. Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 59.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Touzel, J. P., and G. Albagnac. 1983. Isolation and characterization of Methanococcus mazei strain MC3. FEMS Microbiol. Lett. 16:241-245. [Google Scholar]
- 61.Uz, I., A. Ogram, and K. R. Reddy. 2003. Composition and activities of cellulolytic and fermentative bacteria in impacted and nonimpacted wetland soils, abstr. N-080, p. 412. Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., Washington, D.C.
- 62.Uz, I., M. E. Rasche, T. G. Townsend, A. V. Ogram, and A. S. Lindner. 2003. Characterization of methanogenic and methanotrophic assemblages in landfill samples. Proc. R. Soc. Lond. B 270:S202-S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, J. R., and K. R. Reddy. 1999. Influence of nitrate and phosphorus loading on denitrifying enzyme activity in Everglades wetland soils. Soil Sci. Soc. Am. J. 63:1945-1954. [Google Scholar]
- 64.Williams, R. T., and R. L. Crawford. 1984. Methane production in Minnesota peatlands. Appl. Environ. Microbiol. 47:1266-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolfe, R. S. 1996. 1776-1996: Alessandro Volta's combustible air. ASM News 62:529-534. [Google Scholar]
- 66.Wright, A. L., and K. R. Reddy. 2001. Heterotrophic microbial activity in northern Everglades wetland soils. Soil Sci. Soc. Am. J. 65:1856-1864. [Google Scholar]
- 67.Zinder, S. H. 1984. Microbiology of anaerobic conversion of organic wastes to methane: recent developments. ASM News 50:294-298. [Google Scholar]
- 68.Zinder, S. H., and M. Koch. 1984. Non-acetoclastic methanogenesis from acetate: acetate oxidation by thermophilic syntrophic coculture. Arch. Microbiol. 138:263-272. [Google Scholar]