Abstract
The distribution of eight putative adhesins that are not encoded in the locus for enterocyte effacement (LEE) in 139 Shiga toxin-producing Escherichia coli (STEC) of different serotypes was investigated by PCR. Five of the adhesins (Iha, Efa1, LPFO157/OI-141, LPFO157/OI-154, and LPFO113) are encoded in regions corresponding to genomic O islands of E. coli EDL933, while the other three adhesins have been reported to be encoded in the STEC megaplasmid of various serotypes (ToxB [O157:H7], Saa [O113:H21], and Sfp [O157:NM]). STEC strains were isolated from humans (n = 54), animals (n = 52), and food (n = 33). They were classified into five seropathotypes (A through E) based on the reported occurrence of STEC serotypes in human disease, in outbreaks, and in the hemolytic-uremic syndrome (M. A. Karmali, M. Mascarenhas, S. Shen, K. Ziebell, S. Johnson, R. Reid-Smith, J. Isaac-Renton, C. Clark, K. Rahn, and J. B. Kaper, J. Clin. Microbiol. 41:4930-4940, 2003). The most prevalent adhesin was that encoded by the iha gene (91%; 127 of 139 strains), which was distributed in all seropathotypes. toxB and efa1 were present mainly in strains of seropathotypes A and B, which were LEE positive. saa was present only in strains of seropathotypes C, D, and E, which were LEE negative. Two fimbrial genes, lpfAO157/OI-141 and lpfAO157/OI-154, were strongly associated with seropathotype A. The fimbrial gene lpfAO113 was present in all seropathotypes except for seropathotype A, while sfpA was not present in any of the strains studied. The distribution of STEC adhesins depends mainly on serotypes and not on the source of isolation. Seropathotype A, which is associated with severe disease and frequently is involved in outbreaks, possesses a unique adhesin profile which is not present in the other seropathotypes. The wide distribution of iha in STEC strains suggested that it could be a candidate for vaccine development.
Shiga toxin (Stx)-producing Escherichia coli (STEC) strains can cause a broad spectrum of human disease, including diarrhea, hemorrhagic colitis, and the life-threatening hemolytic-uremic syndrome (HUS) (20). E. coli O157:H7 is by far the most prevalent serotype associated with large outbreaks and sporadic cases of hemorrhagic colitis and HUS in many countries (10). However, more than 100 serotypes have a similar pathogenic potential in humans (2).
Although the main virulence property of STEC is the production of one or more types of Stx (Stx1, Stx2, or Stx variants), adherence to the intestinal epithelium and colonization of the gut are also important components of pathogenesis. Some STEC serotypes, considered to be highly virulent in humans, harbor a large pathogenicity island, termed the locus for enterocyte effacement (LEE). This locus is associated with intimate adherence to epithelial cells, the initiation of host signal transduction pathways, and the formation of attaching and effacing intestinal lesions (19). LEE appears to confer enhanced virulence, since LEE-positive STEC serotypes (such as O157:H7, O26:H11, O111:NM, and O145:NM) are much more commonly associated with outbreaks and HUS than are LEE-negative serotypes (14, 28). However, the presence of the LEE is not essential for pathogenesis, as a number of cases of severe STEC disease, including HUS, as well as occasional outbreaks were caused by LEE-negative strains (17, 25). Additional virulence factors, including adhesins encoded outside of the LEE (26) and a plasmid-encoded enterohemolysin (34), may play a role in pathogenesis.
A major task of STEC research is the development of vaccines against STEC. Immunization with antigens that promote colonization would prevent infection, whereas immunization with Stx would prevent the pathological effects of toxin-mediated manifestations of disease. Vaccines targeting adhesins to block the colonization of either humans or animal reservoirs would be effective in controlling STEC infection, because adhesion is the initial step in pathogenesis. With more LEE-negative STEC strains being reported, investigations of adhesins encoded outside of the LEE have been carried out by several groups (22, 26, 40). The genome sequence of E. coli O157:H7 strain EDL933 has revealed multiple regions in the chromosome that may have a putative role in adherence (29, 36). A thorough understanding of the mechanisms used by STEC to adhere to epithelial cells and to colonize animals has yet to emerge.
Several proteins were proposed to be novel adhesion factors; these include ToxB (a protein identified from large, 93-kb plasmid pO157 and required for full expression of adherence of O157:H7 strain Sakai) (42), Saa (an autoagglutinating adhesin identified in LEE-negative strains) (26), Sfp (sorbitol-fermenting enterohemorrhagic E. coli [EHEC] O157 fimbriae) (4), Iha (adherence-conferring protein similar to Vibrio cholerae IrgA) (35, 40), Efa1 (EHEC factor for adherence) (22), and LPF (long polar fimbriae; closely related to LPF of Salmonella enterica serovar Typhimurium) (7, 45). These putative adhesins are encoded either in the large plasmid harbored by STEC strains or in unique DNA segments of E. coli EDL933 called O islands (OIs). ToxB, Sfp fimbriae, and Saa are plasmid encoded, and an association between the presence of saa and enterohemolysin gene ehxA was reported (26). Iha is encoded in OI-43 and OI-48, which are identical and contain 106 open reading frames (ORFs). Efa1 is encoded in OI-122, which was recently reported by Karmali et al. to be associated with STEC serotypes that are linked to epidemic and/or serious disease (16). In O157:H7 strains, the LPF OI-141 and OI-154 operons are present. LPFO157/OI-141 was reported by Torres et al. (45) to play a role in adherence. On the other hand, Doughty et al. (7) suggested that LPF of O113:H21, encoded in OI-154 (LPFO113), functions as an adhesin in LEE-negative isolates of STEC. The aim of this study was to establish the prevalence of putative adhesins that are not encoded in the LEE region in STEC strains of different serotypes and of human, animal, and food origins.
MATERIALS AND METHODS
Bacterial strains.
A total of 139 STEC strains isolated from humans (n = 54), animals (n = 52), and food (n = 33) were studied. The 130 Argentinean strains isolated during the surveillance of HUS and diarrheal disease and food and animal surveys were submitted to the National Reference Laboratory for phenotypic and genotypic characterizations (5, 6, 8, 9, 11). Six strains were kindly supplied by B. E. C. Guth, Departamento de Microbiologia, Imunologia e Parasitologia, Universidade Federal de São Paulo, São Paulo, Brazil (11). Three strains were isolated from patients with diarrhea in Japan as part of an ongoing study of diarrheagenic E. coli. The selection of strains was generally random, but all strains belonging to the same serotype were selected to ensure that they were isolates from different patients or animals that were not linked temporally. Strains belonging to the same serotype were tested for XbaI macrorestriction enzyme digestion patterns by pulsed-field gel electrophoresis (11) to ensure that they were distinct.
Standard E. coli strains for preparing antisera to all of the serogroups between O174 and O181 were obtained from the Statens Serum Institut (Copenhagen, Denmark). Positive control strains for Shiga toxin typing were E. coli strains EDL933 (stx1 and stx2), 92-3580 (stx2vh-a), and 93-016 (stx2vh-b) (provided by D. Woodward, National Microbiology Laboratory, Canadian Science Centre for Human and Animal Health, Winnipeg, Manitoba, Canada) and E. coli strain EH250 (stx2vh-d) (provided by D. Piérard, Department of Microbiology, Academisch Ziekenhuis, Vrije Universiteit Brussels, Brussels, Belgium). Additional positive control strains for putative adhesins were strains 493/89 (13) (provided by T. Whittam, National Food Safety and Toxicology Center, Michigan State University), 93-016 (provided by D. Woodward), and 434-1 from our strain collection (30). The negative control strain was E. coli K-12 strain JM109 (Promega, Madison, Wis.).
Serotyping.
The serotypes of the STEC strains were determined by using either commercially available O and H antisera (Denka Seiken Co., Tokyo, Japan) or antisera prepared at the National Institute of Infectious Diseases, Tokyo, Japan, at the Centers for Disease Control and Prevention, Atlanta, Ga., or at the National Microbiology Laboratory, Canadian Science Centre for Human and Animal Health (23).
Seropathotype classification.
The strains were classified into five seropathotypes (A through E) as described by Karmali et al. (16). Briefly, seropathotypes A and B included the serotypes proposed by Karmali et al., with the exception that we also included serotype O26:NM in seropathotype B. Seropathotypes C, D, and E included diverse serotypes that were assigned accoding to the criteria of low incidence but an association with severe disease (seropathotype C), low incidence and no association with severe disease (seropathotype D), and nonhuman only (seropathotype E).
Determination of genes encoding Shiga toxin, intimin, enterohemolysin, and putative adhesins by PCR.
All primers used in this study are listed in Table 1. Template DNA was prepared as described previously (44). The presence of the stx1 and stx2 genes was identified by a multiplex PCR with the primers described by Pollard et al. (32). For the differentiation of Stx2 variants, the genotyping method of Tyler et al. (46) was extended to include the primers and restriction enzymes described by Piérard et al. (31).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Target(s) | Amplicon size (bp) | Reference or source |
|---|---|---|---|---|
| Stx1R | AGC GAT GCA GCT ATT AAT AA | stx1 | 130 | 32 |
| Stx1F | GAA GAG TCC GTG GGA TTA CG | |||
| Stx2FC | TTA ACC ACA CCC CAC CGG GCA GT | stx2 | 346 | 32 |
| Stx2R | GCT CTG GAT GCA TCT CTG GT | |||
| VT2-c | AAG AAG ATG TTT ATG GCG GT | stx2, stx2vh-a, stx2vh-b | 285 | 46 |
| VT2-d | CAC GAA TCA GGT TAT GCC TC | |||
| VT2v-1 | CAT TCA GAG TAA AAG TGG CC | stx2vh-a, stx2vh-b, stx2d-Ount, stx2d-OX3a | 385 | 46 |
| VT2v-2 | GGG TGC CTC CCG GTG AGT TC | |||
| VT2-e | AAT ACA TTA TGG GAA AGT AAT A | stx2, stx2vh-a, stx2vh-b, stx2d-Ount, stx2d-OX3a | 348 | 31 |
| VT2-f | TAA ACT GCA CTT CAG CAA AT | |||
| SK1 | CCC GAA TTC GGC ACA AGC ATA AGC | eae | 864 | 15 |
| SK2 | CCC GGA TCC GTC TCG CCA GTA TTC G | |||
| hlyA1 | GGT GCA GCA GAA AAA GTT GTA G | ehxA | 1,551 | 34 |
| hlyA4 | TCT CGC CTG ATA GTG TTT GGT A | |||
| SAADF | CGT GAT GAA CAG GCT ATT GC | saa | 119 | 27 |
| SAADR | ATG GAC ATG CCT GTG GCA AC | |||
| iha-I | CAG TTC AGT TTC GCA TTC ACC | iha | 1,305 | 35 |
| iha-II | GTA TGG CTC TGA TGC GAT G | |||
| toxB.911F | ATA CCT ACC TGC TCT GGA TTG A | toxB | 602 | 39 |
| toxB.1468R | TTC TTA CCT GAT CTG ATG CAG C | |||
| 88T14 | GAG ACT GCC AGA GAA AG | efa1 | 479 | 22 |
| 88T9 | GGT ATT GTT GCA TGT TCA G | |||
| lpfA-F | ATG AAG CGT AAT ATT ATA G | lpfAO113 | 573 | 7 |
| lpfA-R | TTA TTT CTT ATA TTC GAC | |||
| sfpA-U | AGC CAA GGC CAA GGG ATT ATT A | sfpA | 440 | 4 |
| sfpA-L | TTA GCA ACA GCA GTG AAG TCT C | |||
| lpfO141-F | CTG CGC ATT GCC GTA AC | lpfAO157/OI-141 | 412 | 38 |
| lpfO141-R | ATT TAC AGG CGA GAT CGT G | |||
| O154-FCT | GCA GGT CAC CTA CAG GCG GC | lpfAO157/OI-154 | 525 | This study |
| O154-RCT | CTG CGA GTC GGC GTT AGC TG | |||
| O141-F | AAA AGT GTG GGG AAA GAG TG | OI-141 | 6,000a | 7 |
| O141-R | AGC AGA AAG TAT TGC GTG AG | |||
| O154-F | CTG GCA AAA TCG GTA ACG GT | OI-154 | 6,900a | 7 |
| O154-R | CCA CCG GAA GAA CCG AT |
Approximate sizes from amplification of the regions corresponding to OI-141 and OI-154 in the genomes of O157:H7 strains.
The LEE-encoded intimin gene, eae, was detected by PCR with primers SK1 and SK2 as described by Karch et al. (15). The plasmid-carried enterohemolysin gene (ehxA) was identified by PCR with primers hlyA1 and hlyA4 as described by Schmidt et al. (34).
To study the presence of eight putative adhesin genes (iha, toxB, sfpA, saa, efa1, lpfAO113, lpfAO157/OI-141, and lpfAO157/OI-154), PCR amplifications were carried out with 30-μl reaction mixtures containing PCR buffer, 200 μM each deoxynucleoside triphosphate, 6.25 pmol of each primer, and 1 U of HotStart Taq DNA polymerase (Qiagen, Hilden, Germany). Five sets of primer mixtures were prepared: set 1 contained primers for the detection of iha, toxB, and saa (triplex PCR); set 2 contained primers for the detection of efa1; set 3 contained primers for the detection of sfpA; set 4 contained primers for the detection of lpfAO113; and set 5 contained primers for the detection of lpfAO157/OI-141 and lpfAO157/OI-154 (duplex PCR). Cycling conditions for the triplex PCR consisted of an initial activation step at 95°C for 15 min, followed by 25 cycles, each consisting of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1.5 min. There was a final extension step at 72°C for 10 min. Cycling conditions for the duplex PCR were similar to those for the triplex PCR, but the annealing temperature was increased to 55°C. Cycling conditions for sfpA, lpfAO113, and efa1 were as previously described (4, 7, 22) but were preceded by an initial activation step at 95°C for 15 min.
Detection of OI-141 and OI-154 and cloning of OI-141.
The presence of OI-141 and OI-154 was analyzed with the primers reported by Doughty et al. (7) (Table 1 and Fig. 1) and SP-Taq DNA polymerase for long and efficient PCR (COSMO, Seoul, Korea). PCR conditions were 95°C for 2 min; 30 cycles of 95°C for 20 s, 50°C for 40 s, and 68°C for 6 min; and extension for 10 min at 68°C. The 6-kbp PCR product obtained from strain 843/02 (O26:H11) with primers O141-F and O141-R was cloned into vector pCR2.1 for determination of the DNA sequence.
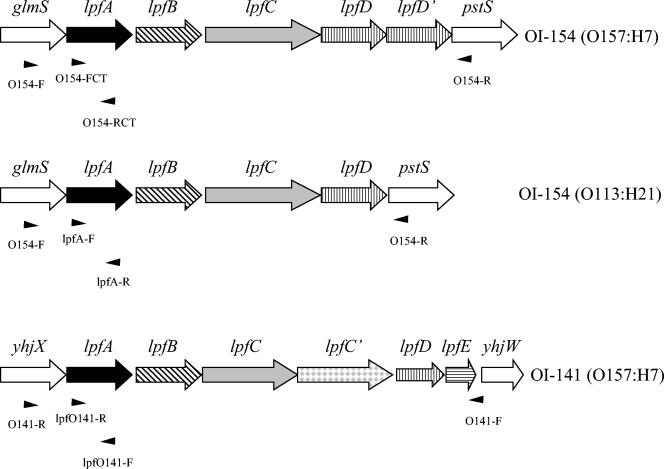
FIG. 1.
Genetic organization of LPF gene clusters encoded in OI-141 and OI-154 of serotypes O157:H7 and O113:H21 (7, 45). Predicted functions for the proteins encoded by the LPF operons are major fimbrial subunit for LpfA, chaperone for LpfB, outer membrane usher chaperone for LpfC, outer membrane usher protein for LpfC′, minor fimbrial subunit for LpfD, and fimbrial subunit for LpfE. OI-154 of O157:H7 contains two copies of lpfD (the second one is presented as lpfD′). Annealing sites for the primers used in the PCR analysis are indicated.
Nucleotide sequence and analysis.
Plasmid DNA was prepared for sequence analysis by using a Qiagen-tip 100 (Qiagen) according to the manufacturer's instructions. Sequencing of the major fimbrial subunit (lpfAO26) was performed by the dideoxynucleotide triphosphate chain termination method with an ABI PRISM 310 genetic analyzer and a BigDye Terminator Cycle Sequencing FS Ready Reaction kit (Applied Biosystems). The program BLAST 1.4.9 was used to search for related sequences in databases.
Nucleotide sequence accession number.
The nucleotide sequence of lpfAO26 will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession number AB161111.
RESULTS
Strain characterization.
Seropathotypes, serotypes, sources of isolation, and Shiga toxin genotypes of the strains are shown in Table 2. All strains belonging to the same serotype had distinct macrorestriction enzyme digestion patterns on pulsed-field gel electrophoresis (data not shown). Twenty-two O157:H7 strains and 2 O157:NM non-sorbitol-fermenting strains were included in seropathotype A. Seropathotype B comprised 26 strains belonging to serotypes O26:H11, O26:NM, O103:H2, O111:NM, O121:H19, and O145:NM. Seropathotype C comprised 48 strains of 15 serotypes. Serotypes O174:H28 and O178:H19, which were not previously associated with HUS, were also included in seropathotype C. Twenty-two strains of 13 serotypes were included in seropathotype D. Nineteen strains of 14 serotypes were included in seropathotype E.
TABLE 2.
Seropathotypes, serotypes, sources, and stx genotypes of the strains included in this study
| Seropathotype | Serotype | No. of strains
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tested | Isolated from the following source:
|
With the following stx genotype:
|
|||||||||||
| Humana | Animal | Food | 1 | 2 | 2cb | 2d | 1 + 2 | 1 + 2c | 2 + 2c | 1 + 2 + 2c | |||
| A | O157:H7 | 22 | 12 (10) | 3 | 7 | 0 | 4 | 3 | 0 | 0 | 4 | 11 | 0 |
| O157:NM | 2 | 2 (1) | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| B | O26:H11 | 7 | 4 (1) | 1 | 2 | 5 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| O26:NM | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O103:H2 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O111:NM | 4 | 4 (2) | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | |
| O121:H19 | 2 | 2 (1) | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O145:NM | 11 | 10 (5) | 1 | 0 | 1 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | |
| C | O8:H2 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| O8:H19 | 8 | 2 (1) | 4 | 2 | 0 | 4 | 1 | 0 | 1 | 1 | 1 | 0 | |
| O8:H21 | 3 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | |
| O20:H19 | 3 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | |
| O25:H2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O48:H21 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| O91:H21 | 8 | 3 (1) | 5 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 2 | 1 | |
| O112:H2 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O113:H21 | 6 | 2 (1) | 3 | 1 | 1 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O145:H25 | 1 | 1 (1) | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O161:NM | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| O174:H21 | 6 | 2 | 1 | 3 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | |
| O174:H28 | 2 | 1 (1) | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |
| O174:NM | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O178:H19 | 4 | 1 (1) | 3 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | |
| D | O15:H27 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| O22:H16 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O58:H40 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O82:H8 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| O103:H7 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O104:H7 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O113:H7 | 5 | 0 | 0 | 5 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 0 | |
| O116:NM | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O116:H21 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| O128:H12 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O146:H28 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O171:H2 | 4 | 1 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | |
| O174:HNT | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | |
| E | O2:H25 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| O2:H28 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O2:HNT | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| O8:H16 | 4 | 0 | 3 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O8:H49 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O39:H14 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O39:H49 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O44:H8 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O46:H38 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| O65:H48 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| O113:H19 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |
| O117:H21 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| O136:H19 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O161:H2 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
Numbers in parentheses indicate numbers of strains isolated from HUS cases.
2c includes stx variants 2vh-a and 2vh-b.
A close relationship was found between serotype and the eae gene, because strains of the same serotype were either all eae positive or all eae negative (Table 3). In contrast, differences were observed within the same serotype with respect to the Shiga toxin genotype and the presence of the ehxA gene (Tables 2 and 3).
TABLE 3.
Serotype distribution of putative adhesin genes, eae, and ehxA
| Seropathotype | Serotype | No. of strains
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tested | Found positive by PCR for:
|
|||||||||||
| iha | toxB | efa1 | sfpA | saa | lpfAO113 | lpfAO157/OI-141 | lpfAO157/OI-154 | eae | ehxA | |||
| Control (EDL933) | O157:H7 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Control (493/89) | O157:NM | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| Control (93-016) | O113:H21 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Control (434-1) | O2:H8 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 |
| Control (JM109) | E. coli K-12 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A | O157:H7 | 22 | 22 | 22 | 22 | 0 | 0 | 0 | 22 | 22 | 22 | 22 |
| O157:NM | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 2 | 2 | 2 | 2 | |
| B | O26:H11 | 7 | 7 | 6 | 7 | 0 | 0 | 7 | 0 | 0 | 7 | 7 |
| O26:NM | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| O103:H2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| O111:NM | 4 | 4 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 4 | 4 | |
| O121:H19 | 2 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 2 | 2 | |
| O145:NM | 11 | 11 | 11 | 11 | 0 | 0 | 0 | 11 | 0 | 11 | 11 | |
| C | O8:H2 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| O8:H19 | 8 | 4 | 0 | 0 | 0 | 3 | 8 | 0 | 0 | 0 | 6 | |
| O8:H21 | 3 | 3 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 1 | |
| O20:H19 | 3 | 3 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 1 | |
| O25:H2 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O48:H21 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O91:H21 | 8 | 8 | 0 | 0 | 0 | 6 | 8 | 0 | 0 | 0 | 6 | |
| O112:H2 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O113:H21 | 6 | 5 | 0 | 0 | 0 | 5 | 6 | 0 | 0 | 0 | 5 | |
| O145:H25 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | |
| O161:NM | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| O174:H21 | 6 | 6 | 0 | 0 | 0 | 1 | 6 | 0 | 0 | 0 | 1 | |
| O174:H28 | 2 | 2 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 2 | |
| O174:NM | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O178:H19 | 4 | 4 | 0 | 0 | 0 | 3 | 4 | 0 | 0 | 0 | 3 | |
| D | O15:H27 | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 |
| O22:H16 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O58:H40 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O82:H8 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O103:H7 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O104:H7 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O113:H7 | 5 | 5 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 0 | 1 | |
| O116:NM | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O116:H21 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O128:H12 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O146:H28 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O171:H2 | 4 | 4 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 1 | |
| O174:HNT | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| E | O2:H25 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| O2:H28 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O2:HNT | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O8:H16 | 4 | 3 | 0 | 0 | 0 | 3 | 4 | 0 | 0 | 0 | 1 | |
| O8:H49 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O39:H14 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O39:H49 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O44:H8 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| O46:H38 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | |
| O65:H48 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| O113:H19 | 2 | 2 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | |
| O117:H21 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | |
| O136:H19 | 2 | 2 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 2 | |
| O161:H2 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
Prevalence of putative adhesins in different seropathotypes.
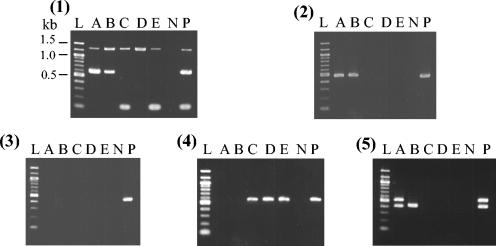
Figure 2 shows the PCR amplification products of the eight adhesin genes studied for control and representative strains of different seropathotypes. The distribution of putative adhesin genes, eae, and ehxA in different serotypes and seropathotypes is shown in Table 3. The most prevalent adhesins among all seropathotypes were those encoded by the iha (127 of 139 strains; 91%) and lpfAO113 (101 of 139 strains; 73%) genes. The 12 iha-negative strains belonged to serotypes O8:H19 (4 strains), O121:H19 (2 strains), and O8:H16, O58:H40, O103:H2, O113:H21, O116:NM, and O145:H25 (1 strain each). The 38 lpfAO113-negative strains belonged to serotypes O157:H7 (22 strains), O157:NM (2 strains), O145:NM (11 strains), and O65:H48, O103:H2, and O174:HNT (1 strain each).
FIG. 2.
PCR analysis of adhesin genes in representative strains of different seropathotypes (A to E) and controls. (Panel 1) Triplex PCR for detection of iha (1,305 bp), toxB (602 bp), and saa (119 bp). (Panel 2) PCR for efa1 (479 bp). (Panel 3) PCR for sfpA (440 bp). (Panel 4) PCR for lpfAO113 (573 bp). (Panel 5) Duplex PCR for lpfAO157/OI-141 (412 bp) and lpfAO157/OI-154 (525 bp). Lanes: L, 100-bp ladder (New England Biolabs); A, O157:H7; B, O145:NM; C, O91:H21; D, O171:H2; E, O8:H16; N, negative control (strain JM109); P, positive control (strain EDL933 for iha, toxB, efa1, lpfAO157/OI-141, and lpfAO157/OI-154; strain 93-016 for lpfAO113; and strain 434-1 for saa).
Seropathotype A was positive for iha, toxB, efa1, lpfAO157/OI-154, and lpfAO157/OI-141. One characteristic of seropathotype A was the presence of a specific LPF gene cluster in OI-154, as reflected by the PCR results for lpfAO157/OI-154. In seropathotype B, most of the strains were positive for iha, toxB, and efa1, with the exception that strains of serotypes O111:NM and O103:H2 and one strain of serotype O26:H11 were toxB negative and strains of serotypes O121:H19 and O103:H2 were iha negative. In seropathotype B, strains of serotypes O26:H11, O26:NM, O111:NM, and O121:H19 contained lpfAO113, and strains of serotype O145:NM did not contain lpfAO113 but contained lpfAO157/OI-141. Serotype O103:H2 did not contain any of the lpfA genes investigated. On the other hand, strains of seropathotypes C, D, and E, which were eae negative (with the exception of O145:H25 in seropathotype C), did not harbor lpfAO157/OI-154, lpfAO157/OI-141, toxB, or efa1. One strain of serotype O145:H25 (eae positive) was the only one in seropathotype C that was efa1 positive. The adhesin gene present in all seropathotypes was iha.
There was a correlation between the presence of the ehxA and saa genes for seropathotypes C, D, and E. Among 43 strains positive for ehxA, 38 were positive for saa; among 42 strains positive for saa, 38 were positive for ehxA. The five ehxA-positive saa-negative strains belonged to serotypes O8:H19 (three strains) and O15:H27 and O145:H25 (one strain each). The four saa-positive ehxA-negative strains belonged to serotypes O8:H16 (two strains) and O8:H2 and O117:H21 (one strain each).
Prevalence of adhesins according to source of isolation.
Overall, there was no relationship between the source of isolation and the prevalence of adhesins (Tables 2 and 3). To better understand this relationship, we further analyzed serotypes O157:H7, O26:H11, O8:H19, O113:H21, and O174:H21 represented by strains of human, animal, and food origins (Table 4). Serotypes O157:H7, O26:H11, O113:H21, and O174:H21 showed similar profiles of adhesins regardless of the source of isolation. Human isolates of serotype O8:H19 were iha and saa negative, in spite of the presence of ehxA. We found that this serotype showed an stx genotype dependency in the distribution of adhesins, since four O8:H19 stx2 strains (two from human, one from animal, and one from food sources) were iha negative. Three of these O8:H19 stx2 strains harbored the ehxA gene but were saa negative.
TABLE 4.
Distribution of putative adhesins according to source of isolation
| Serotype | Source | No. of strains
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tested | Found positive by PCR for:
|
|||||||||
| iha | toxB | efa1 | sfpA | saa (ehxA) | lpfAO113 | lpfAO157/OI-141 | lpfAO157/OI-154 | |||
| O157:H7 | Human | 12 | 12 | 12 | 12 | 0 | 0 (12) | 0 | 12 | 12 |
| Animal | 3 | 3 | 3 | 3 | 0 | 0 (3) | 0 | 3 | 3 | |
| Food | 7 | 7 | 7 | 7 | 0 | 0 (7) | 0 | 7 | 7 | |
| O26:H11 | Human | 4 | 4 | 3 | 4 | 0 | 0 (4) | 4 | 0 | 0 |
| Animal | 1 | 1 | 1 | 1 | 0 | 0 (1) | 1 | 0 | 0 | |
| Food | 2 | 2 | 2 | 2 | 0 | 0 (2) | 2 | 0 | 0 | |
| O8:H19 | Human | 2 | 0 | 0 | 0 | 0 | 0 (2) | 2 | 0 | 0 |
| Animal | 4 | 3 | 0 | 0 | 0 | 2 (2) | 4 | 0 | 0 | |
| Food | 2 | 1 | 0 | 0 | 0 | 1 (2) | 2 | 0 | 0 | |
| O113:H21 | Human | 2 | 2 | 0 | 0 | 0 | 2 (2) | 2 | 0 | 0 |
| Animal | 3 | 2 | 0 | 0 | 0 | 2 (2) | 3 | 0 | 0 | |
| Food | 1 | 1 | 0 | 0 | 0 | 1 (1) | 1 | 0 | 0 | |
| O174:H21 | Human | 2 | 2 | 0 | 0 | 0 | 0 (0) | 2 | 0 | 0 |
| Animal | 1 | 1 | 0 | 0 | 0 | 0 (0) | 1 | 0 | 0 | |
| Food | 3 | 3 | 0 | 0 | 0 | 1 (1) | 3 | 0 | 0 | |
Number of strains positive for ehxA by PCR.
Analysis of OI-141 and cloning of lpfAO26.
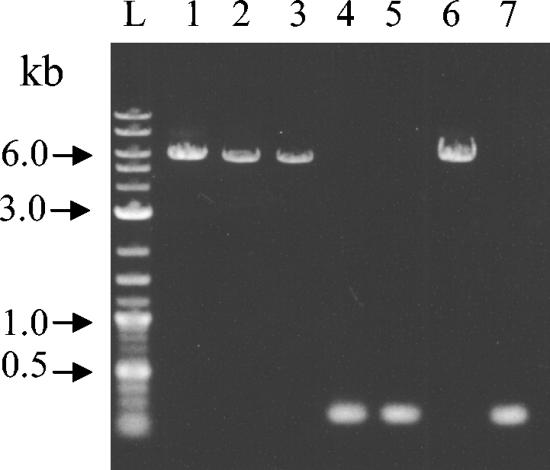
Since lpfAO157/OI-141 was present only in serotypes O157:H7, O157:NM, and O145:NM, we investigated the presence of OI-141 in strains of other serotypes. Strains harboring OI-141 showed an amplicon of ∼6 kbp, while strains without OI-141 showed an amplicon of 200 bp (Fig. 3). OI-141 was present in all strains of seropathotype B, with the exception of two strains belonging to serotype O121:H19. In seropathotypes C, D, and E, OI-141 was present in 17 of 48 strains (35%), 9 of 22 strains (41%), and 8 of 19 strains (42%), respectively. DNA sequence analysis of the PCR product from strain 843/02 (O26:H11) revealed the presence of an lpfA gene, which was designated lpfAO26. The predicted amino acid sequence from lpfAO26 showed 89% similarity to the sequence of LpfAR141 of rabbit enteropathogenic E. coli (EPEC) (21). LpfAO26 also showed 75% similarity to LpfAO157/OI-141 (accession number AE005581), 57% similarity to LpfAO157/OI-154 (accession number AE005604), and 52% similarity to LpfAO113 (accession number AY057066).
FIG. 3.
PCR analysis of OI-141 in representative strains of different seropathotypes (A to E) and controls. Lanes: L, 2-log ladder (New England Biolabs); 1, O157:H7 (seropathotype A); 2, O103:H2 (seropathotype B); 3, O178:H19 (seropathotype C); 4, O174:HNT (seropathotype D); 5, O65:H48 (seropathotype E); 6, strain EDL933 (positive control); 7, strain JM109, negative control.
Analysis of OI-154.
We investigated the presence of OI-154 in two strains (one strain each of serotypes O65:H48 and O174:HNT) that were negative for lpfAO113, lpfAO157/OI-141, lpfAO157/OI-154, and OI-141. These two strains were also found negative for OI-154 in a PCR analysis with primers O154-F and O154-R.
DISCUSSION
Studies on putative STEC adhesins so far have been confined to a few serotypes. In view of the increasing number of reports of non-O157 STEC infections, there is now a need for comprehensive data on the prevalence of adhesins in STEC strains of diverse serotypes. In this study, we investigated by PCR the prevalence of eight putative adhesins among a wide range of serotypes, which were divided into five seropathotypes according to incidence and associations with outbreaks and with complicated diseases. A variety of in vitro and in vivo data support the role of intimin in adherence to epithelial cells. However, because intimin-negative strains belonging to serotypes such as O113:H21, O91:H21, and O8:H19 were isolated from HUS patients, we wanted to determine whether there is a common adhesin among seropathotypes A, B, and C and whether there is a difference between seropathotypes A, B, C, and D (isolated from humans) and seropathotype E (comprised of nonhuman STEC serotypes). Our study investigated only the presence of the adhesin genes; therefore, studies on expression, function, and antigenic diversity need to be confirmed before vaccine work can be undertaken.
Our results for efa1, toxB, sfpA, and lpfA O157/OI-141 extended the observations of previous reports that investigated the distribution of some of these adhesins in a limited number of strains (1, 4, 16, 22, 38, 39). To our knowledge, this is the first report on the prevalence of lpfA in OI-154 of serotype O157:H7. Interestingly, the nucleotide sequence of lpfAO157/OI-154 seems to be unique to seropathotype A. Although we did not analyze the biological significance of this unique segment, it might confer some advantage in colonization over the lpfA sequences found in other serotypes.
Efa1 was first reported to be an adhesin in an O111:NM clinical isolate (22), and recently Stevens et al. (37) reported that Efa1 also influences the colonization of the bovine intestine by STEC serotypes O5 and O111. Klapproth et al. (18) reported the identification in EPEC of a lymphotoxin gene (lifA) which shows 99.9% similarity to efa1. LifA also contributes to the adherence of EPEC to epithelial cells (1). We investigated the presence of efa1, which is present as an ORF of 9,669 bp in O111:NM, with primers directed at the 5′ end. PCR for efa1 was positive for all eae-positive strains, in agreement with the results of previous reports (16, 22). Other studies with PCRs and DNA probes directed at the center and at the 3′ end of efa1 showed that serotypes O157:H7 and O145:NM are efa1 negative (13, 18, 38). The recent genome sequencing of EHEC O157:H7 has revealed a truncated chromosomal form of efa1 that comprises the first 2 kb of efa1 divided into two ORFs, efa-O157a and efa-O157b (12, 29). A random mutagenesis study revealed that a transposon insertion into efa-O157a resulted in decreased adherence of O157:H7 strain Sakai to epithelial cells (41). Our results and the results of previous reports suggested that efa1 in O145:NM could be truncated as in O157:H7.
Interestingly, O145:NM is the serotype that most resembled serotype O157:H7 in terms of the prevalence of putative adhesins, since it is the only serotype that possessed lpfAO157/OI-141. We wondered whether other serotypes harbor a variant LPF gene cluster in OI-141. PCR with primers external to OI-141 showed that other serotypes indeed had OI-141. The presence of OI-141 was strongly correlated with seropathotypes A and B, similar to the association between OI-122 and STEC serotypes linked to epidemics and/or serious diseases (16). The nucleotide sequence of lpfAO26 was different from those of previously reported STEC lpfA genes. LpfAO26 had only two amino acids that were different from those in recently reported LpfAR141 (21), which is involved in the early stages of rabbit EPEC-mediated diarrhea. PCR with primers specific for lpfAO26 showed that the O103:H2 strain that was negative for the three lpfA genes studied possessed lpfAO26 (data not shown). Our results suggested that at least four LPF variants exist in STEC strains.
ToxB showed 47% similarity to Efa1 and is considered to be a plasmid-encoded efa1 homologue. ToxB was reported to be involved in the full adherence phenotype of O157 strain Sakai by promoting the production and/or secretion of type III secretion proteins encoded in the LEE (EspA, EspB, and Tir) (42). Therefore, its association with the presence of eae, which is also located in the LEE, is not surprising. However, a homology search showed that ToxB possesses several distinctive, if not separable, domains each associated with the full production of type III secretion proteins, adherence to epithelial cells, or inhibition of lymphocyte activation (42). We wondered whether ToxB could function as an adhesin without the presence of LEE-encoded factors. The PCR results obtained with primers directed at the 5′ end of toxB showed that the gene was never present in eae-negative strains. Six eae-positive strains were toxB negative but possessed efa1. Because of the similarity between Efa1 and ToxB, Efa1 may be able to compensate functionally for the absence of ToxB.
Saa is encoded by a gene located on the large plasmid of some eae-negative STEC strains. The saa-specific PCR primers used allowed the amplification of a 119-bp portion of the gene which is absolutely conserved among diverse STEC strains (27). The presence of saa showed a strong association with ehxA in eae-negative strains of different serotypes, as proposed by Paton et al. (26). Although some strains were only either saa or ehxA positive, these results could be explained by the high variability of the large STEC plasmids (3). On the contrary, Sfp fimbriae seemed to be restricted only to sorbitol-fermenting O157:NM strains (4), since all strains were found to be negative when primers directed at the major fimbrial subunit gene sfpA were used.
Recently, Osek et al. (24) studied the prevalence of lpfAO113 (encoded in OI-154) in a limited number of strains. Osek et al. concluded that lpfAO113 is closely associated with eae-negative STEC. Our results showed that lpfAO113 is present in all STEC serotypes, with the exception of O157:H7, O157:NM, O103:H2, O145:NM, O174:HNT, and O65:H48. Interestingly, serotype O65:H48 has never been associated with human disease and did not contain any LPF gene cluster in OI-141 or OI-154 (data not shown), suggesting a role for LPF in human infection. Furthermore, the LPF gene cluster of OI-154 was identified when genomic subtraction was used with O91:H21 (HUS isolate) and O6:H10 (bovine isolate) to identify DNA sequences that might encode factors involved in virulence (33).
The most widely distributed STEC adhesin gene was iha. Iha is encoded by an ORF of 2,088 bp which is located in OI-43 and OI-48. These two OIs are identical and are 0.5 Mb apart on the EDL933 genome, with OI-43 being inserted close to the serX tRNA gene and OI-48 being inserted close to the serW tRNA gene (43). In eae-negative STEC strains, iha is located in the locus of proteolytic activity island inserted in the selC locus (35). PCR analysis with primers amplifying the central 1,305-bp region of iha showed that 91% of the strains were iha positive. Our results suggested that iha is conserved although located at different chromosomal loci in O157:H7 and eae-negative STEC strains.
In conclusion, this study showed that (i) seropathotype A possesses a unique profile of adhesins which was not present in other seropathotypes; (ii) lpfAO113 was present in all seropathotypes except for seropathotype A; (iii) OI-141 and OI-154 harbored LPF gene clusters with variable sequences; (iv) the distribution of STEC adhesins depends mainly on serotypes and not on the source of isolation; and (v) iha is a common adhesin gene in all seropathotypes, suggesting that Iha is necessary but not sufficient for human infection. Thus, Iha could be a candidate for vaccine development, although its role as an adhesin in serotypes other than O157:H7 should be proved experimentally.
Acknowledgments
We thank Beatriz E. C. Guth, David Woodward, Denis Piérard, and Thomas Whittam for generously providing the STEC strains used in this study; Ariela Baschkier, Eduardo Manfredi, Ana Garbini, and Natalia Martinez for technical assistance; and Kazumichi Tamura and Haruo Watanabe for helpful discussions.
This work was partially supported by grants from Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET) (PIP no. 0020/98) and Fundación Alberto J. Roemmers, Buenos Aires, Argentina.
REFERENCES
- 1.Badea, L., S. Doughty, L. Nicholls, J. Sloan, R. M. Robins-Browne, and E. L. Hartland. 2003. Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34:205-215. [DOI] [PubMed] [Google Scholar]
- 2.Bettelheim, K. A. 2003. Non-O157 Verotoxin-producing Escherichia coli: a problem, paradox, and paradigm. Exp. Biol. Med. 228:333-344. [DOI] [PubMed] [Google Scholar]
- 3.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 4.Brunder, W., A. S. Khan, J. Hacker, and H. Karch. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H−. Infect. Immun. 69:4447-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinen, I., J. D. Tanaro, E. S. Miliwebsky, H. L. Lound, G. M. Chillemi, S. Ledri, A. Baschkier, M. Scarpin, E. Manfredi, and M. Rivas. 2001. Isolation and characterization of Escherichia coli O157:H7 from retail meats in Argentina. J. Food Prot. 64:1346-1351. [DOI] [PubMed] [Google Scholar]
- 6.Chinen, I., J. L. Otero, E. S. Miliwebsky, M. L. Roldán, A. Baschkier, G. M. Chillemi, C. Nóboli, L. Frizzo, and M. Rivas. 2003. Isolation and characterisation of Shiga toxin-producing Escherichia coli O157:H7 from calves in Argentina. Res. Vet. 74:283-286. [DOI] [PubMed] [Google Scholar]
- 7.Doughty, S., J. Sloan, V. Bennet-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gioffré, A., L. Meichtri, E. Miliwebsky, A. Baschkier, G. Chillemi, M. I. Romano, S. Sosa Estani, A. Cataldi, R. Rodriguez, and M. Rivas. 2002. Detection of Shiga toxin-producing Escherichia coli by PCR in cattle in Argentina. Evaluation of two procedures. Vet. Microbiol. 87:301-313. [DOI] [PubMed] [Google Scholar]
- 9.Gomez, D., E. Miliwebsky, C. Fernandez Pascua, A. Baschkier, E. Manfredi, M. Zotta, F. Nario, A. Piquín, M. Sanz, A. Etcheverría, N. Padola, A. Parma, and M. Rivas. 2002. Aislamiento y caracterización de Escherichia coli productor de toxina Shiga en hamburguesas supercongeladas y quesos de pasta blanda. Rev. Argent. Microbiol. 34:66-71. [PubMed] [Google Scholar]
- 10.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 11.Guth, B. E. C., I. Chinen, E. Miliwebsky, A. M. F. Cerqueira, G. Chillemi, J. R. C. Andrade, A. Baschkier, and M. Rivas. 2003. Serotypes and Shiga toxin genotypes among Escherichia coli isolated from animals and food in Argentina and Brazil. Vet. Microbiol. 92:335-349. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 13.Janka, A., M. Bielaszewska, U. Dobrindt, and H. Karch. 2002. Identification and distribution of the enterohemorrhagic Escherichia coli factor for adherence (efa1) gene in sorbitol-fermenting Escherichia coli O157:H−. Int. J. Med. Microbiol. 292:207-214. [DOI] [PubMed] [Google Scholar]
- 14.Jenkis, C., G. A. Willshaw, J. Evans, T. Cheasty, H. Chart, D. J. Shaw, G. Dougan, G. Frankel, and H. R. Smith. 2003. Subtyping of virulence genes in verocytotoxin-producing Escherichia coli (VTEC) other than serogroup O157 associated with disease in the United Kingdom. J. Med. Microbiol. 52:941-947. [DOI] [PubMed] [Google Scholar]
- 15.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Ölschläger, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karmali, M. A., M. Mascarenhas, S. Shen, K. Ziebell, S. Johnson, R. Reid-Smith, J. Isaac-Renton, C. Clark, K. Rahn, and J. B. Kaper. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keskimäki, M., R., Ikäheimo, P. Kärkkäinen, F. Scheutz, R. L. Puohiniemi, and A. Siitonen. 1997. Shiga toxin producing Escherichia coli serotype OX3:H21 causing hemolytic uremic syndrome. Clin. Infect. Dis. 24:1278-1279. [DOI] [PubMed] [Google Scholar]
- 18.Klapproth, J. M., I. C. Scaletsky, B. P. McNamara, L. C. Lai, C. Malstrom, S. P. James, and M. S. Donnenberg. 2000. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 68:2148-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogencity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K12. Mol. Microbiol. 2:399-407. [DOI] [PubMed] [Google Scholar]
- 20.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton, H. J., J. Sloan, V. Bennett-Wood, L. M. Adams, R. M. Robins-Browne, and E. L. Hartland. 2004. Contribution of long polar fimbriae to the virulence of rabbit-specific enteropathogenic Escherichia coli. Infect. Immun. 72:1230-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 23.Orskov, F., and I. Orskov. 1984. Serotyping of Escherichia coli. Methods Microbiol. 14:43-112. [Google Scholar]
- 24.Osek, J., M. Weiner, and E. L. Hartland. 2003. Prevalence of the lpfO113 gene cluster among Escherichia coli O157 isolates from different sources. Vet. Microbiol. 96:259-266. [DOI] [PubMed] [Google Scholar]
- 25.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paton, A. W., and J. C. Paton. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. Kirkpatrick, G. Posfal, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 30.Phantouamath, B., N. Sithivong, S. Insisiengmay, N. Higa, C. Toma, N. Nakasone, and M. Iwanaga. 2003. The incidence of Escherichia coli having pathogenic genes for diarrhea: a study in the People's Democratic Republic of Lao. Jpn. J. Infect. Dis. 56:103-106. [PubMed] [Google Scholar]
- 31.Piérard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard, D. R., W. M. Johnson, H. Lior, S. D. Tyler, and K. R. Rozee. 1990. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J. Clin. Microbiol. 28:540-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pradel, N., S. Leroy-Setrin, B. Joly, and V. Livrelli. 2002. Genomic subtraction to identify and characterize sequences of Shiga toxin-producing Escherichia coli O91:H21. Appl. Environ. Microbiol. 68:2316-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, H., W.-L. Zhang, U. Hemmrich, S. Jelacic, W. Brunder, P. I. Tarr, U. Dobrindt, J. Hacker, and H. Karch. 2001. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69:6863-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens, M. P., P. M. van Diemen, G. Frankel, A. D. Phillips, and T. S. Wallis. 2002. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70:5158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szalo, I. M., F. Goffaux, V. Pirson, D. Pièrard, H. Ball, and J. Mainil. 2002. Presence of bovine enteropathogenic (EPEC) and enterohaemorrhagic (EHEC) Escherichia coli of genes encoding for putative adhesins of human EHEC strains. Res. Microbiol. 153:653-658. [DOI] [PubMed] [Google Scholar]
- 39.Tarr, C. L., T. M. Large, C. L. Moeller, D. W. Lacher, P. I. Tarr, D. W. Acheson, and T. S. Whittam. 2002. Molecular characterization of a serotype O121:H19 clone, a distinct Shiga toxin-producing clone of pathogenic Escherichia coli. Infect. Immun. 70:6853-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2001. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tatsuno, I., H. Kimura, A. Okutani, K. Kanamaru, H. Abe, S. Nagai, K. Makino, H. Shinagawa, M. Yoshida, K. Sato, J. Nakamoto, T. Tobe, and C. Sasakawa. 2000. Isolation and characterization of mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect. Immun. 68:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taniguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, D. E., M. Rooker, M. Keelan, L. Ng, I. Martin, N. T. Perna, N. T. V. Burland, and F. R. Blattner. 2002. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184:4690-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toma, C., Y. Lu, N. Higa, N. Nakasone, I. Chinen, A. Baschkier, M. Rivas, and M. Iwanaga. 2003. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J. Clin. Microbiol. 41:2669-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyler, S. D., W. M. Johnson, H. Lior, G. Wang, and K. R. Rozee. 1991. Identification of verotoxin type 2 variant B subunit genes in Escherichia coli by the polymerase chain reaction and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 29:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]