Abstract
The results of empirical studies have revealed links between phytoplankton and bacterioplankton, such as the frequent correlation between chlorophyll a and bulk bacterial abundance and production. Nevertheless, little is known about possible links at the level of specific taxonomic groups. To investigate this issue, seawater microcosm experiments were performed in the northwestern Mediterranean Sea. Turbulence was used as a noninvasive means to induce phytoplankton blooms dominated by different algae. Microcosms exposed to turbulence became dominated by diatoms, while small phytoflagellates gained importance under still conditions. Denaturing gradient gel electrophoresis (DGGE) of 16S rRNA gene fragments showed that changes in phytoplankton community composition were followed by shifts in bacterioplankton community composition, both as changes in the presence or absence of distinct bacterial phylotypes and as differences in the relative abundance of ubiquitous phylotypes. Sequencing of DGGE bands showed that four Roseobacter phylotypes were present in all microcosms. The microcosms with a higher proportion of phytoflagellates were characterized by four phylotypes of the Bacteroidetes phylum: two affiliated with the family Cryomorphaceae and two with the family Flavobacteriaceae. Two other Flavobacteriaceae phylotypes were characteristic of the diatom-dominated microcosms, together with one Alphaproteobacteria phylotype (Roseobacter) and one Gammaproteobacteria phylotype (Methylophaga). Phylogenetic analyses of published Bacteroidetes 16S rRNA gene sequences confirmed that members of the Flavobacteriaceae are remarkably responsive to phytoplankton blooms, indicating these bacteria could be particularly important in the processing of organic matter during such events. Our data suggest that quantitative and qualitative differences in phytoplankton species composition may lead to pronounced differences in bacterioplankton species composition.
Studies of marine bacterioplankton diversity have provided a comprehensive inventory of the major groups of bacteria frequently observed in the water column (17, 22). Despite this, factors that determine the abundance of these bacteria remain relatively unknown. Factors strongly correlated with bulk bacterioplankton growth and abundance would be expected to be important in determining the success of different groups and species of bacteria. One such factor is phytoplankton biomass, which is frequently estimated as the concentration of chlorophyll a (Chl a) (6, 8). The concentration of Chl a is a widely used reference variable for planktonic life in aquatic environments, and the distribution of marine bacterial populations is frequently reported in relation to the spatial and temporal distribution of Chl a (18, 50, 68). However, it appears likely that bacterial community composition is influenced not only by the Chl a concentration but also by the species composition of the algal assemblage (28, 51). Phytoplankton community composition has a large impact on the survival and growth of organisms in the grazing food chain (36). However, the importance of phytoplankton community composition for the structure of the microbial food web in general, and bacterioplankton species composition in particular, is much less known.
Phytoplankton blooms dominated by diatoms typically occur in coastal seas and upwelling areas where currents bring nutrients to the photic zone. In temperate regions, spring diatom blooms typically occur when high nutrient concentrations coincide with increased solar irradiance. The species composition of algal blooms can be modified by variations in the stability of the water column. For example, extended periods of calm weather during summer may lead to blooms of dinoflagellates or filamentous cyanobacteria (3, 34). During phytoplankton bloom senescence, bacterial abundance typically increases substantially, coupled with large increases in per-cell activity in hydrolytic enzyme activity and growth rates (66). Few studies have investigated the composition of bacterioplankton assemblages in relation to naturally occurring or experimentally induced blooms of phytoplankton. However, it appears that changes in bacterial activity are associated with significant shifts in bacterioplankton species composition (48, 55). Bacteria shown to increase in abundance during the decay of algal blooms belong to such different phylogenetic groups as the Bacteroidetes phylum and the Alpha- and Gammaproteobacteria (14, 55, 61, 72). The ecology of marine representatives of these bacteria remains largely unknown. However, considering the ample phylogenetic diversity within each of these groups, it can be expected that they contain substantial physiological and metabolic variability. Thus, insights into factors that contribute to the success of defined groups of bacteria will increase understanding of marine microbial processes. Comparative analysis of the growth response of particular bacteria during algal blooms dominated by different algae or blooms occurring in different seas could provide a means to decipher the ecological roles of these bacteria.
Theoretical considerations based on hydrodynamics and diffusion rates indicate that turbulence can significantly increase the flow of nutrients to algal cells larger than 60 μm (27). The formation of chains by algae may cause an additional increase in the rate of diffusion to algal cells in turbulent water. Furthermore, turbulence may contribute to maintaining in suspension cells that would otherwise sink. Thus, it is expected that both large or chain-forming phytoplankton may be favored under turbulent conditions, while small motile phytoplankton would dominate under still conditions. Indeed, nutrient enrichment experiments with seawater incubated under different mixing regimens show that, other things being equal, turbulence results in phytoplankton assemblages dominated by diatoms larger than 50 μm (1, 13). In the same experiments, microcosms maintained under still conditions became dominated by small phytoflagellates (diameters of <20 μm). Bacteria, on the other hand, are too small to be directly affected by naturally occurring levels of turbulence (35). However, turbulence may indirectly cause changes in bacterial abundance, gross bacterial production, and bacterial cell morphology through food web interactions in the plankton community (38, 40, 46). Such interactions include turbulence-induced shifts in grazing pressure on phytoplankton and bacteria (46, 57). It has therefore been speculated that turbulence could indirectly modify the structure of the bacterial assemblage in the upper water column of the sea (40).
Given that turbulence has a significant impact on phytoplankton community composition, we set out to investigate whether such differences could be accompanied by comparable changes in the bacterioplankton assemblage. In the present study, microcosm phytoplankton blooms were induced by nutrient enrichment. Turbulent or still conditions were then used to induce phytoplankton assemblages dominated by different algae. In this way, manipulations to induce changes in phytoplankton were avoided that could otherwise also have had direct effects on the bacterioplankton species composition (e.g., addition of nutrients in different proportions or distinct algal inocula). Here we report on the growth of bacterioplankton assemblages in general, and on specific phylotypes in particular, under different phytoplankton regimens induced by turbulence. Furthermore, as a way to identify general patterns in the occurrence of bacterial phylotypes under different phytoplankton bloom conditions, we expanded our analysis and compared the identities of bacterial phylotypes in our experiments to the identities of bacteria found during phytoplankton blooms in previously published studies.
MATERIALS AND METHODS
Sampling and experimental design.
Three experiments were performed with samples of surface water. The water samples were from the northwestern Mediterranean Sea collected 1 km offshore from the Catalan coast at Masnou (41°28′N, 2°18′E, 20 km north of Barcelona, Spain). The water samples were collected from a depth of 0.5 m.
Control experiment without phytoplankton.
On 4 November 2002, seawater was collected for a dilution culture experiment to monitor the growth of bacteria under turbulent and still conditions without other components of the microbial community. For each culture, approximately 4.5 liters of sampled water to be used as growth medium was filtered through a 0.2 μm-pore-size filter (Sterivex; Millipore) using a peristaltic pump. A total of 500 ml of inoculum, prepared by gravity filtration of sampled water through 47-mm-diameter, 0.8-μm-pore-size filters (Nuclepore), was added to each culture to give a 10-fold dilution. Duplicate cultures were maintained under still (Sa and Sb) and turbulent (Ta and Tb) conditions in an environmental chamber at 17 °C ± 1°C (in situ temperature) under a 12-h light/12-h dark cycle and a light irradiance of 225 μmol of photons m−2 s−1. Cultures were maintained in 15-liter Plexiglas cylinders (24.2-cm inner diameter and 34.5-cm depth). Small-scale turbulence was generated by vertically oscillating grids, with a mesh size of 1.42 cm and bar thickness of 0.38 cm. The grid moved down to 1 cm from the bottom. The stroke length was 9 cm, and the oscillation frequency was set at 4.6 rpm. This gave an estimated energy dissipation rate of 0.050 cm2 s−3 (45). This is within the range of turbulence intensities in surface waters in coastal areas with moderate winds (29). All material in contact with the samples was rinsed with 1 M hydrochloric acid and extensively washed with MilliQ-water prior to use. The cultures received nitrogen (NaNO3), phosphorus (Na2HPO4), and silicon (NaSiO3), added to final concentrations of 16, 1, and 30 μM, respectively. Bacterial growth in the dilution cultures was monitored for 4 days.
Phytoplankton bloom experiment 1.
Seawater was collected on 30 April 2001. For each microcosm, 15 liters of seawater was filtered through a 150-μm-mesh net in order to remove large zooplankton and enclosed in eight Plexiglas cylinders (see above) within 3 h after sampling. The experiment was performed in an environmental chamber at 15°C ± 1°C (in situ temperature) under the same irradiance as in the above experiment. Inorganic nutrients consisting of N (NaNO3), P (Na2HPO4), and Si (NaSiO3) were added to final concentrations of 16, 1, and 30 μM, respectively. Microcosms received nutrients either by a single batch addition on day 0 (SB and TB) or by daily additions (SD and TD). Daily additions consisted of N, P, and Si to final concentrations of 2, 0.12, and 3.75 μM, respectively. Duplicates of each treatment were maintained under still (S) or turbulent (T) conditions until day 6, and one microcosm for each treatment was maintained until day 8. Small-scale turbulence was generated by vertically oscillating grids (see above), with a stroke length of 20 cm and an oscillation frequency of 3.75 rpm, giving an energy dissipation rate of 0.055 cm2 s−3. When the volume of water decreased due to sampling, the stroke length was corrected to maintain the initial energy dissipation rate. The final volume in the containers at the end of the experiment was approximately 5 liters.
Phytoplankton bloom experiment 2.
Seawater samples collected on 29 April 2002 were used in an experiment with an experimental design similar to that of phytoplankton bloom experiment 1. Microcosms were enriched with different concentrations of nutrients in this experiment. Control microcosms without nutrient additions were maintained under still or turbulent conditions (S0 and T0). Enriched microcosms all received P (Na2HPO4), and Si (NaSiO3) at final concentrations of 1 and 30 μM, respectively, while the concentration of N (NaNO3) was varied between 16 μM (S16 and T16) and 24 μM (S24 and T24). Microcosms were incubated for 4 days at 15°C under a 12-h light/12-h dark cycle and irradiance as in previous experiments.
Nutrients and Chl a.
Initial concentrations of total inorganic N, PO4, and Si before nutrient addition in our experiments were ≤0.78, 0.09, and 0.20 μM, respectively. These concentrations were low compared to the nutrient concentrations added: the additions of P corresponded to maximum values after water column mixing in winter, and N was added in Redfield ratio to P. Silica was added in excess to avoid Si limitation during the experiments. The Chl a concentration was estimated fluorometrically from 20-ml samples filtered through Whatman GF/F filters. The filters were ground in 90% acetone and left in the dark at room temperature for at least 2 h. The fluorescence of the extract was measured with a Turner Designs fluorometer.
Microplankton enumeration and biomass calculation.
Samples for determination of microplankton biomass (i.e., phytoplankton and heterotrophic dinoflagellates and ciliates) in phytoplankton bloom experiment 1 were collected on day 6. In addition, heterotrophic dinoflagellates and ciliates were quantified in detail on a daily basis throughout the experiment. Abundance and biomass of heterotrophic and phototrophic dinoflagellates was determined as previously described (24). Samples were fixed with 2% glutaraldehyde (final concentration), which gave the cells a greenish color under blue fluorescence, and filtered onto black 0.8-μm-pore-size black polycarbonate filters. The filters were frozen on the sampling day and stored at −20°C to preserve Chl a autofluorescence, and the microplankton were counted within 6 months by epifluorescence microscopy at a magnification of ×600 or ×1,000. The dinoflagellates were assigned to eight groups according to their size and taxonomic relationships. Samples of microplankton other than dinoflagellates were fixed in formaldehyde buffered with hexamine (0.4% final concentration; pH 7.2). These microplankton were examined using inverted microscopy and were assigned to nine groups according to their size and taxonomic relationships by the method of Havskum and Hansen (24). At least 100 cells of each group were counted, and cell volumes were calculated from the average cell dimensions of 50 cells in each group. The dimensions of naked flagellates and ciliates were multiplied by a factor of 1.1 to compensate for shrinkage due to fixation. Cell volume was converted to cell carbon using a conversion factor of 0.13 pg of C μm−3 for thecate dinoflagellates and 0.11 pg of C μm−3 for other flagellates and ciliates. Diatom cell carbon was calculated from cell volume after subtracting the vacuole volume, assuming a plasma layer thickness of 2 μm, and the remaining volume was converted to carbon content using conversion factor 0.11 pg of C μm−3 (conversion factors used by Havskum and Hansen [24]).
Bacterial heterotrophic production and abundance.
Bacterial heterotrophic production was measured in phytoplankton bloom experiment 1, using [3H]leucine incorporation with the microcentrifuge protocol (65). For each sample, triplicate aliquots (1.2 ml) and a trichloroacetic acid-killed control were incubated with 20 nM [3H]leucine (final concentration) for 1 to 2 h at 15 or 17°C. The theoretical conversion factor of 3.1 kg of C mol−1 was used to convert leucine incorporation rates to bacterial carbon production.
In the control experiment, bacterial abundance was determined by epifluorescence microscopy. Samples were preserved with glutaraldehyde (filtered through a 0.2-μm-pore-size filter) (final concentration, 1% [wt/vol]) and were counted within 48 h. Bacteria were stained with a final concentration of 5 μg of 4′,6-diamidino-2-phenylindole (DAPI) ml−1 for 10 min and filtered onto black 0.2 μm-pore-size polycarbonate filters at a pressure of 100 to 200 mm Hg (52).
In the phytoplankton bloom experiments, bacterial abundance was determined by flow cytometry. Samples were preserved with a mixture of 1% paraformaldehyde and 0.05% glutaraldehyde (final concentrations) and stored frozen at −70°C. Cell counts were done with a Becton Dickinson FACSCalibur flow cytometer after the cells were stained with Syto13 at a final concentration of 5 μM (16).
Ectoenzymatic activity.
Hydrolytic ectoenzyme activity was determined using fluorogenic substrates by the method of Sala et al. (58). The activities of α- and β-glucosidase were assayed as the hydrolysis rate of methylumbelliferone (MUF)-α-d-glucoside and 4-MUF-β-d-glucoside, respectively. Alkaline phosphatase was assayed using 4-MUF-phosphate. Aminopeptidase was assayed using leucine 7-amido-4-methylcoumarin. Briefly, 0.9 ml of a sample was incubated with substrate (final concentration, 0.1 mM) for 60- to 90-min incubation in the dark at room temperature. The increase of fluorescence during incubation was measured with a spectrofluorometer (Shimadzu RF-540) with 365-nm-wavelength excitation and 446-nm-wavelength emission radiation. Fluorescence was transformed into activity units using a standard curve of the end product of the reaction: 4-MUF was used for α-glucosidase, β-glucosidase, and alkaline phosphatase, and 7-amino-4-methylcoumarin was used for aminopeptidase.
Collection of microbial community DNA and extraction procedure.
For extraction of community DNA, microbial biomass from approximately 700 ml of seawater was collected onto 0.2-μm-pore-size polycarbonate filters (Durapore; Millipore) by vacuum filtration at ≤150 mm Hg. Filters were stored frozen at −70°C in sucrose buffer (0.75 M sucrose, 40 mM EDTA, 50 mM Tris [pH 8.3]). In phytoplankton bloom experiment 1, samples for community DNA were collected at the beginning of the experiment (day 0) and on every second day until day 8. In the other experiments, samples were collected at the beginning and end (day 4). DNA was extracted as previously described (63). Briefly, cells were treated with lysozyme, proteinase K, and sodium dodecyl sulfate followed by phenol-chloroform-isoamyl alcohol extraction. Extracted DNA was desalted and concentrated using a Centricon-100 filter device (Millipore).
PCR and denaturing gradient gel electrophoresis (DGGE).
A 16S rRNA gene fragment (approximately 550 bp long) was amplified by PCR, using the bacterium-specific primer 358f (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG) that is complementary to positions 341 to 358 (Escherichia coli numbering) and has a GC clamp (underlined) and the universal primer 907rm [5′-CCGTCAATTC(A/C)TTTGAGTTT] that is complementary to positions 927 to 907. The reaction mixture volumes were 50 μl, containing 10 ng of template, 200 μM concentration of each deoxynucleoside triphosphate, standard 1× PCR buffer, 1.5 mM MgCl2, 0.3 μM concentration of each primer, 200 μg of bovine serum albumin, and 1 U of Taq polymerase (GIBCO BRL). After an initial denaturation step (5 min at 94°C), samples were amplified with 10 touchdown cycles, with 1 cycle consisting of denaturation (1 min at 94°C), annealing (1 min at 65 to 55 °C [temperature of 65 °C in the first cycle, with the temperature decreasing 1°C each cycle and ending at 55°C in the last cycle]), and extension (3 min at 72°C). This was followed by 20 standard cycles, with 1 cycle consisting of denaturation (1 min at 94°C), annealing (1 min at 55°C), and extension (3 min at 72°C), and a final extension step (7 min at 72°C). PCR products were quantified by agarose gel electrophoresis with a molecular size standard in the gel (Low DNA Mass ladder; GIBCO BRL).
A total of 800 ng of PCR product for each sample was loaded on a 6% (wt/vol) polyacrylamide gel (acrylamide and N,N′-methylene bisacrylamide at a ratio of 37:1) with a denaturing gradient that ranged from 40 to 80% (where 100% is defined as 7 M urea and 40% deionized formamide). Electrophoresis was performed with a DGGE-2000 system (CBS Scientific Company). The gel was run at 100 V for 16 h at 60°C in 1× TAE running buffer (40 mM Tris [pH 7.4], 20 mM sodium acetate, 1 mM EDTA). The gel was stained with the nucleic acid stain SYBR Gold (1:10,000 dilution; Molecular Probes) for 45 min, rinsed with 1× TAE running buffer, removed from the glass plate to a UV transparent gel scoop, and visualized with UV in a Fluor-S MultiImager (Bio-Rad) with the Multi-Analyst software (Bio-Rad).
Bacterial assemblage DGGE fingerprints of samples collected during phytoplankton bloom experiment 1 were used to generate a dendrogram using the Diversity Database software (Bio-Rad), applying Jaccard coefficients, Euclidean square distances, and unweighted paired group means analysis (gel image not shown). A complementary DGGE gel from this experiment was prepared to visualize details in the structure of the bacterial assemblage from days 4 to 8.
In our hands, repeated PCR and DGGE analyses consistently yielded the same banding patterns and nearly identical band intensities. The methodological limitations of the approach using PCR amplification and DGGE analysis have been discussed extensively elsewhere (33, 42, 63). The approach gives an estimate of microbial community structure in a semiquantitative manner, with variations in the banding pattern of samples indicating relative changes in the microbial community, rather than changes in the absolute abundance of particular taxa. Thus, this approach may be indicative of possible changes in bacterial community structure in response to different experimental conditions.
Sequencing of DGGE bands.
DGGE bands were excised using a sterile razor blade and eluted in 20 μl of MilliQ water overnight at 4°C, followed by a freeze-thaw cycle. A total of 5 μl of the eluate was used for reamplification with the original primer set. A portion of the PCR product was analyzed by DGGE together with the original sample to verify the correct position of the band, and in cases where more than one band was present, the target band was processed again as described above. PCR products were purified with the QIAquick PCR purification kit (QIAGEN) and quantified in an agarose gel. Ten to 20 ng of the PCR product was used for the sequencing reaction, using primer 358f without the GC clamp, with the BigDye terminator cycle-sequencing kit (Perkin-Elmer Corporation) and an ABI PRISM model 377 (v3.3) automated sequencer (performed by QIAGEN GmbH Sequencing Services).
Phylogenetic analyses.
Our 16S rRNA gene sequences were compared to existing prokaryotic sequences in GenBank (National Center for Biotechnology Information) using BLAST. Comparison of BLAST results from database searches with different regions of the sequences showed that our sequences were not chimeras. Sequences were aligned using the Pileup command in the Wisconsin Package (also called GCG package) (version 9.1; Genetics Computer Group [GCG], Madison, Wis). Phylotypes that were present in 50% or more of the samples from either the still or turbulent microcosms and that showed double or greater relative intensity in the DGGE fingerprints in one mixing regimen were considered characteristic for the treatment.
A phylogenetic tree of our 16S rRNA gene sequences was constructed using Jukes-Cantor distances with the PHYLIP package (version 3.5) (15). The tree was calculated from sequences corresponding to approximately nucleotide positions 500 to 900 (E. coli numbering) and rooted with an actinomycete as an outgroup. Sequences from representative type species of currently described genera in the Bacteroidetes phylum were included as references. Sequences deposited in GenBank from isolates and phylotypes of Bacteroidetes from previous studies of bacterioplankton species composition during natural or experimental marine algal blooms were also included in the phylogenetic tree.
Published Bacteroidetes 16S rRNA gene sequences that were derived from algal blooms and that did not overlap sufficiently with our sequences to yield a phylogenetic tree with reliable branching order were included in complementary phylogenetic trees (data not shown). These phylogenetic trees also included the Bacteroidetes reference sequences and encompassed nucleotides between approximately nucleotide positions 50 to 500 or 350 to 500 (E. coli numbering). The phylogenetic affiliations obtained from the phylogenetic trees were verified by BLAST searches with the search string limited by the terms “bacteria[ORGN] AND sp nov[WORD]” to reveal the closest matching characterized bacterial species. Pair-wise sequence comparisons were performed using the BestFit command in the GCG package. The results of our phylogenetic analyses consistently agreed with those presented in the original publications.
RESULTS
Control experiment without phytoplankton.
A dilution culture experiment, in which bacteria were grown in the absence of other components of the microbial community, was performed to determine whether turbulence per se could affect the structure of the bacterioplankton assemblage. The experiment lasted 4 days to avoid the growth of heterotrophic flagellates that might not have been eliminated by filtration of the inoculum and because bacterial abundance had increased substantially in 4 days.
Initial bacterial abundance in the seawater cultures was 0.2 × 106 cells ml−1. After 4 days of incubation, bacterial abundance had increased 10-fold to ca. 4.4 × 106 and 2.3 × 106 cells ml−1 in the turbulent (Ta and Tb) and still cultures (Sa and Sb), respectively (Fig. 1A).
FIG. 1.
Bacterial abundance (A) and DGGE profiles of bacterial assemblages (B) in the control experiment without phytoplankton. Seawater cultures with bacteria only (fraction < 0.8 μm) were maintained under turbulent (T) and still (S) conditions, with two replicates of each treatment (a and b). Values are the means ± standard deviations (error bars) of duplicate cultures. Community DNA was collected on day 0 (initial) and day 4. The sampling time is shown in days (d).
The structure of the bacterial assemblage in all seawater cultures changed substantially and in a similar way from day 0 (initial sample) until day 4 (final sample). In the final samples, the same 21 bands were detected in both the turbulent and still cultures (Fig. 1B). Only minor differences in relative band intensity between the two treatments were observed.
Phytoplankton bloom experiment 1.
Phytoplankton bloom experiment 1 was performed to study the response of the bacterioplankton species composition to different mixing regimens in the presence of the entire microbial community. The experiment lasted 8 days to allow sampling both during the induced phytoplankton blooms and their decay.
The initial Chl a concentrations were 1.5 μg liter−1. Chl a concentrations in the turbulent microcosms (TB and TD) peaked at approximately 25 μg liter−1 on day 5 (Fig. 2A) and then fell to below 10 μg liter−1 by day 8. Chl a concentrations in the still microcosms increased more slowly and reached 8 or 4 μg liter−1 in the microcosms with a single batch addition of nutrients (SB) or with daily nutrient additions (SD), respectively.
FIG. 2.
Dynamics in microbial growth variables during phytoplankton bloom experiment 1. (A) Chl a concentrations. (B) Bacterial production estimated by leucine incorporation. (C) Bacterial abundance. Data were from seawater microcosms including the microbial community (fraction < 150 μm) incubated under turbulent (T) and still (S) conditions. Microcosms received nutrients as a batch addition on day 0 (TB and SB) or as daily additions (TD and SD). Values are the means ± standard deviations (error bars) for samples from duplicate microcosms maintained until day 6. Chl a concentrations are shown in micrograms per liter, bacterial production is shown in micrograms of carbon per liter per day, and the sampling time is shown in days (d).
During the phytoplankton bloom, the chain-forming diatom Chaetoceros socialis (cell length, 8 μm; chain lengths up to a few millimeters) dominated the phytoplankton biomass in the turbulent microcosms, with a significant contribution also by Pseudonitzschia sp. and other diatoms (Table 1). These diatoms together reached a biomass of ca. 100 μg of C liter−1 in the microcosms exposed to turbulence (approximately 90% of the total algal biomass). In the still microcosms, the diatom biomass was 10-fold lower than that in the turbulent microcosms. The biomass of phytoflagellates (i.e., cells <10 μm long, mostly prasinophytes and haptophytes) was approximately 5 to 10 μg of C liter−1 in all microcosms. Thus, in the turbulent microcosms, the biomass ratio of phytoflagellates to diatoms was ca. 0.08, while in the still microcosms, the ratios were 0.37 and 0.62.
TABLE 1.
Biomass of microplankton in the still and turbulent microcosms on day 6
| Plankton group | Biomass (μg of C liter−1)a
|
|||
|---|---|---|---|---|
| TB | TD | SB | SD | |
| Diatoms | ||||
| Chaetoceros socialis | 86.23 | 126.13 | 8.74 | 11.85 |
| Pseudonitzschia sp. | 2.91 | 5.32 | 1.27 | 1.06 |
| Other diatoms | 2.98 | 5.79 | 0.12 | ND |
| Phototrophic dinoflagellates | ||||
| cf. Cachonina niei | 0.19 | 0.65 | 0.55 | 0.33 |
| Pentapharsodinium sp. | 0.62 | 0.45 | 0.26 | 0.53 |
| Scrippsiella cf. trochoidea | 1.28 | 0.40 | 1.38 | 0.81 |
| Other pigmented dinoflagellates | ND | 0.44 | 0.09 | ND |
| Other algae | ||||
| Euglenophyceae | 0.42 | ND | ND | ND |
| Dinobryon sp. | 0.20 | ND | ND | ND |
| Phytoflagellates <10 μm (prasinophytes, haptophytes, and others) | 7.56 | 9.95 | 6.28 | 4.72 |
| Heterotrophic dinoflagellates | ||||
| Gyrodinium sp. | 0.41 | 0.20 | 0.09 | 0.40 |
| Protoperidinium bipes | 3.23 | 3.23 | 0.58 | 0.28 |
| Katodinium glaucum | ND | ND | 0.59 | ND |
| Other heterotrophic dinoflagellates | 0.46 | 1.17 | 0.74 | 0.71 |
| Heterotrophic ciliates | ||||
| Tintinnids | 9.72 | 19.44 | 1.67 | 0.56 |
| Strombidium sp. | 1.54 | ND | ND | ND |
| Strombilidium sp. | 1.33 | ND | ND | ND |
Biomass values above 2 μg of C liter−1 are shown in boldface type. ND, not detected.
Microzooplankton reached nearly 10-fold-higher biomass in the turbulent microcosms compared to the biomass of the still microcosms (Table 1). On day 6, a tintinnid ciliate biomass of up to 19 μg of C liter−1 was observed in the turbulent microcosms, and the biomass of the heterotrophic dinoflagellate Protoperidinium bipes was around 3 μg of C liter−1. Daily measurements of the biomass of P. bipes during the experiment showed that it increased from an initial biomass below detection to 5.54 μg of C liter−1 on day 8 in the microcosms exposed to turbulence. Observation of live samples from the microcosms showed that P. bipes was actively feeding on cells of Pseudonitzschia sp. (H. Havskum, unpublished data). In the still microcosms, the biomass of heterotrophic dinoflagellates was slightly higher than the biomass of heterotrophic ciliates.
Heterotrophic bacterial production showed an initial peak on day 2. Starting on day 4, heterotrophic bacterial production increased again in all microcosms and reached approximately 200 μg of C liter−1 day−1 toward the end of the experiment (Fig. 2B). Lower bacterial production and abundance was observed in the SB microcosms from day 6 on. Cell-specific carbon production rates ranged from 20 to 89 fg of C cell−1 day−1 in the still microcosms and 18 to 50 fg of C cell−1 day−1 in the turbulent microcosms, with the highest values from days 4 to 6.
The dynamics of bacterial abundance was similar to that of bacterial production, with an initial peak on day 2, followed by a period of decreasing abundance (Fig. 2C). Thereafter, the abundance increased again from day 5 on. On day 8, the highest abundance of bacteria was recorded in the TB microcosm (1.3 × 107 cells ml−1) and the lowest abundance was observed in the SB microcosm (2.5 × 106 cells ml−1).
The activities of alpha- and beta-glucosidase, aminopeptidase, and alkaline phosphatase remained low for the first 4 days. From day 5 on, enzyme activities increased nearly 20-fold in the turbulent microcosms and 5- to 10-fold in the still microcosms, concomitant with the peak and decay of the phytoplankton blooms (Fig. 3, left panels). The highest activities were recorded for aminopeptidase (up to 1.0 × 104 μmol liter−1 h−1) and alkaline phosphatase (up to 3.3 × 103 μmol liter−1 h−1). Bacterial cell-specific hydrolysis rates of alpha- and beta-glucosidase and aminopeptidase peaked midway through the experiment at values that were 5- to 10-fold higher than the initial rates (Fig. 3, right panels). A peak in alkaline phosphatase activity normalized to bacterial abundance was observed on day 5 after an increase of more than 20-fold. When normalized to total microbial biomass, including phytoplankton, alkaline phosphatase activity showed a ninefold increase during the last 4 days of the experiment (Fig. 3, insert).
FIG. 3.
Activities of the four hydrolytic ectoenzymes measured during phytoplankton bloom experiment 1 (left panels) and cell-specific hydrolysis rates obtained by normalizing enzyme activities to bacterial abundance (right panels). Enzyme activity is shown in micromoles per liter per hour, and the hydrolysis rate is shown in femtomoles per cell per hour. The insert graph shows alkaline phosphatase activities (APA) normalized to total bacterial and algal biomass (expressed as micromoles per microgram of C per liter). Microcosms are labeled as described in the legend to Fig. 2. Values are the means ± standard deviations (error bars) for samples from duplicate subsamples. AMA, aminopeptidase activity; d, days.
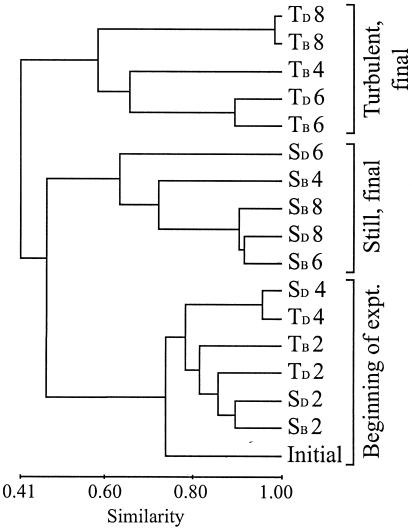
DGGE analysis of samples from days 0 to 8 revealed between 11 and 20 bands in each sample, with a total of 32 different bands (data not shown). Cluster analysis of the DGGE banding patterns from the microcosms indicated that gradual changes in the composition of the bacterial assemblages occurred during the experiment both with and without turbulence (Fig. 4). At first, bacterioplankton composition was relatively similar, as indicated by the tight clustering of the initial and day 2 samples from all microcosms. A clear division of still and turbulent samples in separate clusters was found from day 4 on for the microcosms with a batch addition of nutrients (TB and SB) and from day 6 on for the microcosms with daily nutrient additions (TD and SD). DGGE analysis showed that the structure of the bacterial assemblage in duplicate microcosms that were maintained until day 6 was highly reproducible (data not shown). The DGGE gel image of the bacterial assemblages in the microcosms from days 4 to 8 and the excised bands that were sequenced are shown in Fig. 5.
FIG. 4.
Dendrogram comparing DGGE fingerprints of the bacterial assemblages in the turbulent (T) and still (S) microcosms during phytoplankton bloom experiment 1. The subscript letters indicate batch (TB and SB) or daily (TD and SD) mode of nutrient addition. The numbers after the microcosm are the day the sample was taken. expt., experiment.
FIG. 5.
Bacterioplankton composition during phytoplankton bloom experiment 1 visualized by DGGE of PCR-amplified partial 16S rRNA genes. Bands that were cut from the gel and sequenced are indicated by their codes at the sides of the gel. Bands with S or T prefix, followed by band number, denote phylotypes mainly found in the still or turbulent microcosms, respectively. Bands with ST prefix, followed by band number, indicate ubiquitous phylotypes. All bands in Table 2 were given the prefix MED. The sampling time is shown in days (d).
The phylotypes that were abundant during the experiment were mainly affiliated with bacteria in the Bacteroidetes phylum and the Roseobacter group in the Alphaproteobacteria (Fig. 5 and Table 2). The phylogenetic affiliations of Bacteroidetes phylotypes detected in the microcosms are presented in Fig. 6.
TABLE 2.
Phylogenetic affiliation of 16S rRNA gene sequences from excised DGGE bands obtained during phytoplankton bloom experiment 1
| Phylotype and band | GenBank accession no. | Relative in GenBanka | Similarity (%) | Family | Taxonb |
|---|---|---|---|---|---|
| Still microcosm phylotypes | |||||
| MED-S1 | AY573520 | Uncultured marine bacterium AY-56; AJ298372 | 94.5 | Flavobacteriaceae | CFB |
| Cellulophaga fucicola; AJ005973 | 92.7 | ||||
| MED-S2 | AY573521 | Bacteroidetes clone CF10; AY274847 | 98.3 | Flavobacteriaceae | CFB |
| Tenacibaculum copepodarum; AB078044 | 97.6 | ||||
| MED-S3 | AY573522 | Chaetoceros socialis chloroplast; AJ319825 | 97.9 | Diatom | Algae |
| MED-S4 | AY573523 | Uncultured marine bacterium BY-71; AJ298380 | 91.1 | Cryomorphaceae | CFB |
| Cryomorphaceae bacterium UST02; AB125062 | 90.0 | ||||
| MED-S5 | AY573524 | Uncultured bacterium SB-42-DB; AJ319829 | 98.7 | Cryomorphaceae | CFB |
| Brumimicrobium glaciale; AF521195 | 93.9 | ||||
| Turbulent microcosm phylotypes | |||||
| MED-T6 | AY573525 | Uncultured marine bacterium BY-65; AJ298376 | 100.0 | Flavobacteriaceae | CFB |
| Tenacibaculum sp. strain UST99; AF465364 | 97.8 | ||||
| MED-T7 | AY573526 | Bacteroidetes clone CF2; AY274851 | 98.8 | Flavobacteriaceae | CFB |
| Bacteroidetes bacterium ANT9105; AY167316 | 92.3 | ||||
| MED-T8c | Methylophaga marina; X95459 | >90 | Piscirickettsiaceae | Gamma | |
| MED-T9 | AY573527 | Marine alpha-proteobacterium AS-21; AJ391182 | 100.0 | Roseobacter | Alpha |
| Rhodobacteraceae bacterium TL; AY177716 | 100.0 | ||||
| Ubiquitous phylotypes | |||||
| MED-ST10 | Marine eubacterium DDGE-6; AJ242823 | >96 | Roseobacter | Alpha | |
| Roseobacter sp. strain KT0202; AF305498 | >96 | ||||
| MED-ST11 | AY573528 | Roseobacter clone NAC11-3; AF245632 | 100.0 | Roseobacter | Alpha |
| Roseobacter RED68; AY136132 | 96.7 | ||||
| MED-ST12 | AY573529 | Proteobacterium clone EBAC36F02; AF268234 | 100.0 | Roseobacter | Alpha |
| Sulfitobacter pontiacus; Y13155 | 97.7 | ||||
| MED-ST13 | AY573530 | Marine bacterium ATAM407-56; AF359535 | 99.0 | Roseobacter | Alpha |
| Roseobacter gallaeciensis; Y13244 | 97.7 |
For each phylotype, the closest relative in GenBank and the closest cultured relative are shown together with their accession number (after semicolon) and sequence similarity.
CFB, Cytophaga-Flavobacterium-Bacteroides or Bacteroidetes phylum; Alpha, Alphaproteobacteria; Gamma, Gammaproteobacteria.
Sequence of low quality and was not submitted to GenBank.
FIG. 6.
Phylogenetic tree depicting relationships among partial 16S rRNA gene sequences of Bacteroidetes phylotypes detected during phytoplankton bloom experiment 1 (shown in boldface type) compared to type species of representative genera in the phylum. The approximate position of phylotype MED-S4 is indicated by the short boldface broken line; the sequence was not included in the alignment due to its shorter length. Phylotypes found in natural and experimental algal blooms in previous articles are also included. Phylotype codes are as follows: S and T (this paper), AY and BY (61), AWS (72), BB (5), and DI (J. Pinhassi, unpublished data). Phylotype and isolate names are followed by their GenBank accession number. The scale bar depicts 0.1 substitution per nucleotide position.
All four phylotypes unique to or mainly found in the still microcosms belonged to the Bacteroidetes (Fig. 5 and Table 2). The phylotypes MED-S1, -S2, and -S4 were absent in the initial samples and appeared from day 4 on. MED-S1 clustered with members of the family Flavobacteriaceae but was only distantly related to previously reported sequences. The very intense band in the still microcosms corresponding to phylotype MED-S2 was affiliated with bacteria in the genus Tenacibaculum in the Flavobacteriaceae (Fig. 6). This phylotype showed high sequence similarity to clone CF10 obtained from a 16S rRNA gene clone library from water collected from the Delaware estuary (31). Phylotype MED-S4 showed low sequence similarities (<90%) to previously reported bacteria in the family Cryomorphaceae. Phylotype MED-S5, which was present in the initial sample, was maintained in the still microcosms but disappeared with turbulent conditions. This phylotype was related to bacteria in the newly described genus Brumimicrobium in the Cryomorphaceae (7). Furthermore, MED-S5 was closely related to a phylotype associated with the diatom Dytilum brightwellii from the Gulf of Mexico (60). The DGGE band MED-S3 showed high similarity to a chloroplast sequence of the diatom Chaetoceros socialis (the dominant algal species during the experiment).
The bacterial assemblages in the turbulent microcosms were characterized by two phylotypes affiliated with bacteria in the Bacteroidetes phylum (MED-T6 and -T7), one Gammaproteobacteria phylotype (MED-T8), and a Roseobacter phylotype (MED-T9) (Fig. 5 and Table 2). The MED-T6 phylotype was present in the initial samples and was maintained in turbulent microcosms, while it disappeared in the still microcosms. MED-T6 showed a sequence similarity of 96.6% to the MED-S2 phylotype that was dominant in the still microcosms, and both probably represent separate species in the genus Tenacibaculum (Fig. 6). MED-T7 appeared as a novel phylotype in the microcosms exposed to turbulence. It clustered with members of the genera Cellulophaga and Zobellia in the Flavobacteriaceae, but the relatively low sequence similarity indicated that this phylotype may represent a novel genus. MED-T8 was affiliated with methylotrophic Gammaproteobacteria able to process methylated sulfur compounds. MED-T9 was 96.5% similar to Roseobacter litoralis and showed >96% sequence similarity to many bacterial isolates and sequences from 16S rRNA gene clone libraries from various seas.
At least four different Roseobacter group phylotypes (MED-ST10, -ST11, -ST12, and -ST13) were members of the microcosm bacterial assemblages independent of mixing regimen (Fig. 5). The sequence similarity among these phylotypes was >94.5%. The sequence similarity among all members affiliated with the Roseobacter group is ≥90% (21), indicating that a limited subset of this group was present in our experiment. MED-ST10 and -ST13 were new phylotypes that appeared in both the still and turbulent microcosms. MED-ST11 and -ST12 were present in the initial samples and were maintained in all microcosms throughout the experiment. MED-ST11 was identical to clone NAC11-3 found in the North Atlantic (21), and it was 96.7% similar to an isolate that was abundant in unenriched seawater cultures from the Red Sea (49). MED-ST13 was closely related to the taxonomically defined species Roseobacter gallaeciensis.
Phytoplankton bloom experiment 2.
Phytoplankton bloom experiment 2 was performed to verify that the different bacterioplankton species compositions during phytoplankton blooms generated under turbulent and still conditions were reproducible. The experiment lasted 4 days, since the phytoplankton blooms evolved faster than in phytoplankton bloom experiment 1.
Initial Chl a concentrations were 3 μg liter−1 in all microcosms (Fig. 7A). Peaks in Chl a concentrations were reached on day 2 in the enriched microcosms. The highest Chl a concentrations, 24 μg liter−1, were observed in the turbulent microcosms (T16 and T24). Chl a concentrations in the still microcosms (S16 and S24) on day 2 peaked at 16 μg liter−1. Chl a in the unenriched microcosms (T0 and S0) remained around 3 μg liter−1 during the experiment.
FIG. 7.
Time course of Chl a concentration (A) and bacterial abundance (B) in phytoplankton bloom experiment 2. DGGE profiles of the bacterial assemblages (C). Seawater microcosms including the microbial community (fraction < 150 μm) were incubated under turbulent (T) or still (S) conditions. Microcosms without nutrient additions (T0 and S0) and enriched with N, P, and Si (T16, 24, and S16, 24). Values are the means ± standard deviations (error bars) of pooled data from the enriched microcosms. Community DNA was collected on day 0 (initial) and on day 4. Chl a concentration is shown in micrograms per liter. The sampling time is shown in days (d).
Initial bacterial abundance in the microcosms was 1.8 × 106 cells ml−1 (Fig. 7B). After 24 h, bacteria declined from 1.5 × 106 to 2.0 × 106 cells ml−1 to 0.5 to 0.7 × 106 cells ml−1. Bacteria remained low until day 4, when the abundance in all microcosms increased to approximately 1.1 × 106 cells ml−1.
DGGE analysis indicated the presence of 15 to 17 phylotypes in each of the microcosms, with a total of 25 different bands (Fig. 7C). In the microcosms exposed to turbulence (T16 and T24), a very strong band was located in the upper part of the gel, while nine bands with similar and low intensities were found in the lower part of the gel. In the still microcosms (S16 and S24), four bands were visible in the upper part of the gel (Fig. 7C). In these samples, seven bands identical to those in the turbulent microcosms were visible in the lower part of the gel, two of which were relatively intense. The DGGE gel patterns from the microcosms without nutrient addition (S0 and T0) showed some similarities to those from the enriched still microcosms, containing several bands in the upper region of the gel, and a higher intensity of some of the multiple bands in the lower part of the gel (Fig. 7C).
DISCUSSION
Both theoretical and empirical evidence indicate that the growth of marine bacterioplankton is mostly unaffected by turbulence (27, 40, 46). This is mainly due to the small size of bacteria, which precludes significant gains in bacterial nutrient uptake even with relatively high turbulence. Our control experiment without phytoplankton confirmed that the structures of the bacterial assemblages growing under turbulent or still conditions were generally similar. Nevertheless, some differences in bacterial abundance were observed, similar to those found in a recent freshwater study (2). Many phytoplankton groups, however, are sufficiently large to be directly affected by turbulence, and the mixing regimen plays an important role in structuring phytoplankton communities (1, 3, 13, 23). In accordance with previous studies (1, 13), we found that diatoms and phytoflagellates increased their share of the total phytoplankton biomass under turbulent and still conditions, respectively. Although the direct effect of turbulence on bacteria is limited, significant effects of turbulence on gross bacterial production and abundance are observed in the presence of phytoplankton (38, 40, 46). Here we show that changes in the phytoplankton composition between microcosms maintained under turbulent or still conditions were paralleled by shifts in the composition of the bacterial community. The differences in bacterioplankton were apparent from both the appearance and disappearance of unique phylotypes and changes in the relative abundance (i.e., band intensity) of ubiquitous phylotypes. This implies that changes in phytoplankton community composition and other subsequent associated changes in the microbial food web caused the differences in the bacterioplankton species composition. Our results support previous speculations that the composition of algal assemblages is an important factor in determining the development of different bacterial populations in waters with contrasting phytoplankton (28, 51).
Algal classes differ substantially in their biochemical composition, in terms of C/N/P ratios and in terms of relative proportions of cellular protein, fatty acids, and nucleic acids (53, 70). Differences in the biochemical composition of dominant phytoplankton could cause differences in the stoichiometry of particulate and dissolved organic matter and in the availability of mineral nutrients in seawater, all of which could profoundly affect the growth of bacteria (37). Studies of algal cultures, including diatoms and dinoflagellates, have suggested the existence of specific associations between phytoplankton and bacterioplankton species, possibly due to the differential release of organic matter from the algae (25, 60). To this end, van Hannen et al. showed that enrichment of lake water continuous cultures with detritus from either a green alga or cyanobacterium resulted in the growth of different bacterial assemblages (71). These findings are in agreement with general conclusions concerning the importance of the quality of the organic matter for bacterial community composition and activity (10, 11, 32, 48, 49). This suggests the possibility of finding common patterns in the distribution of particular bacterial groups or species during blooms dominated by different phytoplankton.
Quantitative and qualitative differences in planktonic grazer populations have important consequences both for the pattern of nutrient regeneration and the release of dissolved organic matter. Furthermore, higher nutrient recycling rates resulting from increased grazing rates on phytoplankton under turbulent conditions have been suggested to affect bacterial growth, possibly in combination with decreased grazing rates on bacteria (46, 47). We observed greater microzooplankton biomass in the microcosms dominated by diatoms (turbulent microcosms) than in the microcosms with a higher proportion of phytoflagellates (still microcosms). Moreover, ciliates accounted for a majority of the microzooplankton in the diatom blooms, while heterotrophic dinoflagellates were the dominant grazers in blooms with higher proportion of phytoflagellates. Thus, variation in substrate supply to bacteria due to differences in phytoplankton community composition could have been further amplified by different grazer assemblages.
A majority of the bacterial phylotypes identified in our study belonged to the Roseobacter group (Alphaproteobacteria) or the Bacteroidetes phylum. Phylotypes belonging to these groups had different patterns in our microcosms. Whereas most of the Roseobacter phylotypes were ubiquitous, pronounced differences were observed in the occurrence of Bacteroidetes phylotypes between the microcosms dominated by diatoms and those with a higher proportion of phytoflagellates. This could indicate that these bacterial groups responded to different stimuli in our experiments. Possibly, Roseobacter phylotypes represented a response to the nutrient additions, while the Bacteroidetes phylotypes were responding more to the turbulence-induced differences in the phytoplankton.
Bacteria in the Roseobacter group are widespread and abundant in the oceans and coastal areas of the world (20, 64). Members of the Roseobacter group have 16S rRNA gene sequence similarities of ≥90% (21), and novel genera in this group are continuously being described. Many characteristics have been ascribed to the marine members of the Alphaproteobacteria. For example, they are important in mediating transformations of dimethylated sulfur compounds (41), and members of the Roseobacter group have been suggested to be active colonizers of particles under algal bloom conditions (55). Pinhassi and Berman (49) found that several Roseobacter species became dominant in cultures of unamended seawater collected from the oligotrophic eastern Mediterranean Sea and the Red Sea, suggesting that these bacteria are good competitors under low-nutrient conditions. Most of the Roseobacter phylotypes detected in the present study were found throughout our microcosms and were affiliated with different representatives of the group, such as Roseobacter gallaeciensis, Sulfitobacter pontiacus, and the Atlantic Ocean clone library sequence NAC11-3. The persistence and growth of a substantial subset of Roseobacter phylotypes in our microcosms may at first suggest that these are generalist bacteria. However, in a detailed study of a Roseobacter clade-affiliated cluster, which includes the clone NAC11-3, Selje et al. (64) described significant differences in the geographic distribution of phylotypes that differed by only a few base pairs in the 16S rRNA gene. Future comprehensive phylogenetic analyses and genome comparisons of members of the Roseobacter group will clarify whether the large variety of characteristics ascribed to these bacteria can be explained by a high degree of phenotypic differentiation by phylogenetically relatively closely related bacteria.
The Bacteroidetes phylum is highly diverse; at this time, it consists of 12 families and 79 described genera. Several of these taxa contain representatives from marine environments, documented by culture-independent techniques or by cultured isolates. Bacteria belonging to the Bacteroidetes are abundant in seawater (12, 19, 30), and previous studies have emphasized their importance under algal bloom conditions (48, 55). However, it remains unclear whether particular taxa within the Bacteroidetes are more commonly found than others in the marine environment (31, 44) and whether certain growth conditions selectively favor certain taxa in this phylum. Two phylotypes (MED-S4 and -S5) belonging to the family Cryomorphaceae were unique to the microcosms with a higher proportion of phytoflagellates, although they formed relatively faint bands on the DGGEs. The most intense bands in the upper region of the DGGE were identified as phylotypes belonging to the family Flavobacteriaceae. Two Flavobacteriaceae phylotypes (MED-S1 and -S2) were dominant in the microcosms with a higher proportion of phytoflagellates. Two other members of this family were dominant in the microcosms dominated by diatoms. These phylotypes, MED-T6 and -T7, were identical to the BY-65 and BY-66 phylotypes, respectively (Fig. 6), which were originally detected in 1996 during a diatom bloom experiment performed 200 km northeast of the site of the present experiment (61). This surprisingly consistent response to diatom bloom conditions of a few Flavobacteriaceae phylotypes in the Mediterranean Sea could indicate that some bacteria in this family harbor physiological or ecological traits that make them tightly coupled to diatom species.
Bacteria responding to experimental treatments in a single location may be particular to that location rather than representing general trends in the growth potential of different bacteria. Therefore, we extended our phylogenetic analysis to include 16S rRNA gene sequences of Bacteroidetes phylotypes and isolates from all published studies of marine bacterioplankton diversity associated with natural or experimental algal blooms (Table 3 and Fig. 6). Our analyses showed that even though the Bacteroidetes phylum can be divided into at least 12 families, as much as 80% of the Bacteroidetes phylotypes in these studies (among a total of 63 sequences) belonged to one single family, the Flavobacteriaceae. In particular, phylotypes affiliated with the Tenacibaculum and Cellulophaga or Zobellia genera were frequently found. Other recurrently detected genera in this family included Chryseobacterium, Polaribacter, Gelidibacter, and Psychroserpens. Therefore, we suggest that members of the family Flavobacteriaceae represent bacterial populations that play particularly important roles during and after algal bloom events.
TABLE 3.
Number of 16S rRNA gene sequences affiliated with the family Flavobacteriaceae compared to other families in the phylum Bacteroidetesa
| System | Location | No. of phylotypes
|
Reference | |
|---|---|---|---|---|
| Flavobacteriaceae | Other Bacteroidetes | |||
| Natural algal blooms | ||||
| Dinoflagellates | Southern California Bight | 10 | 2 | 14 |
| Diatoms | Chukchi Sea (Arctic) | 3 | 0 | 72 |
| Experimental blooms | ||||
| Diatoms | Southern California Bight | 3 | 1 | 55 |
| Diatoms | NW Mediterranean Seab | 6 | 5 | 61 |
| Diatoms or flagellates | NW Mediterranean Sea | 4 | 2 | This paper |
| Flagellates | Mexican Gulf | 3 | 2 | J. Pinhassi (unpublished data) |
| Decomposition expts | ||||
| Protein (bovine serum albumin) | Southern California Bight | 7 | 0 | 48 |
| Phaeocystis mucopolysaccharides | North Sea and Balsfjord | 4 | 1 | 26 |
| Diatom debris | Southern California Bight | 10 | 0 | 5 |
| Total no. | 50 | 13 | ||
The 16S rRNA gene sequences were derived from studies of marine bacterioplankton diversity associated with natural and experimental algal blooms and decomposition experiments with natural bacterial assemblages. Sequences from both phylotypes detected by culture-independent techniques and from cultured isolates are included.
NW, northwestern.
At this time, efforts are being made to revise the taxonomy of the Bacteroidetes phylum using phylogenetic analysis of the 16S rRNA gene. This has contributed to the recent redefinition of the family Flavobacteriaceae (4). As a consequence of this revision, bacteria previously named Cytophaga and found throughout the phylum have been renamed, and the type species of the genus Cytophaga is actually well separated from the Flavobacteriaceae. Bacteria in the Flavobacteriaceae have menaquinone 6 (MK-6) as a major respiratory quinone, and this family has therefore been called the marine MK-6 group (67). Suzuki et al. (67) observed that the members of Flavobacteriaceae in their study were unable to use nitrate or ammonium as nitrogen sources, indicating that they required an organic nitrogen source for growth. If confirmed, this observation would substantiate previous findings that bacteria in this group show a high growth capacity when dissolved proteins are abundant (9, 48), which may contribute to explaining why bacteria in the family Flavobacteriaceae are recurrently found during the decay of algal blooms.
We have interpreted the changes in bacterioplankton species composition primarily as a consequence of the differences in dominant phytoplankton groups, which in turn were dependent on the mixing regimen. It is also possible that the total phytoplankton biomass played a role, since nearly three- to sevenfold-higher peaks in Chl a were recorded under turbulent conditions compared to still conditions. Even though the higher bacterial abundance in the final samples in the microcosms with pronounced diatom blooms could have resulted from the higher phytoplankton biomass achieved under turbulence, cell-specific bacterial growth rates and enzymatic activities reached relatively similar levels in all microcosms. Differences in the structure of the bacterial assemblage were also evident in our second bloom experiment, where peak Chl a concentrations were only 50% higher under turbulent conditions compared to still conditions. We note that studies of horizontal variability in bacterial community structure repeatedly find relatively similar bacterial assemblages over substantial distances despite apparent differences in Chl a concentrations (54, 56). At the same time, considerable variability in bacterial diversity is commonly found with depth, which may depend largely on the stratification of phytoplankton populations (see reference 51 and references therein).
For most of the year, the growth of bacteria in northwestern Mediterranean waters is primarily limited by the availability of P (59, 69). The first peak in bacterial production and abundance observed in phytoplankton bloom experiment 1 may therefore be interpreted as a direct effect of the addition of nutrients. The subsequent increase in bacterial production and abundance from day 5 on coincided with the peak and subsequent senescence of the algal blooms. Concomitantly, alpha- and beta-glucosidase and aminopeptidase activities indicated an increase of nearly an order of magnitude or more in the rate of bacterial utilization of carbohydrates and protein. The increase in phosphatase activity could result from the depletion of inorganic phosphate (data not shown) coupled with increased availability of organically bound P. Monitoring bacterioplankton species composition during the experiment showed that the initial bacterial assemblage evolved into different final assemblages in the microcosms dominated by diatoms (turbulent microcosms) and in the microcosms with a greater proportion of phytoflagellates (still microcosms). These findings corroborate previous results obtained primarily from mesocosm experiments with water collected off Scripps Pier, California (39, 48, 55), which showed that changes in bacterial growth and activity during algal blooms were due mainly to shifts in bacterioplankton species composition.
Experimental phytoplankton bloom cycles usually take place within a week or two. The rapid shifts in bacterial activity and community composition during such experiments result from a rapid release of large quantities of organic matter and nutrients due to algal lysis and/or intense grazing during the decay phase of the blooms. Although sudden mass lysis of phytoplankton blooms in the sea has been reported (43), phytoplankton succession is mostly a gradual process, with changes taking place on a time scale of several weeks (23). As a consequence, the opportunity for rapid shifts in bacterioplankton species composition could be limited by a relatively low supply rate of organic matter and/or inorganic nutrients to bacteria under natural algal bloom conditions. This could explain in part why the rate of seasonal succession in bacterioplankton is relatively slow (i.e., several weeks), despite the high growth potential of marine bacteria (50, 62). Still, we find it likely that a sustained release of organic matter and nutrients by live algal cells or by grazers in the long term could profoundly influence the growth of particular bacterial populations, whether under algal bloom or nonbloom conditions. Thus, the greater length of time that natural phytoplankton blooms persist compared to that of experimental phytoplankton blooms could result in larger differences in the bacterioplankton species associated with natural than with experimental blooms.
We have shown that changes in phytoplankton community composition were accompanied by shifts in the composition of the bacterial assemblages. The most pronounced changes were observed for bacterial populations belonging to the phylum Bacteroidetes, principally members of the family Flavobacteriaceae, indicating that flavobacteria could be particularly important in the processing of organic matter during algal blooms.
Acknowledgments
We thank José M. González for knowledgeable advice on phylogenetic analysis. We greatly appreciate the valuable critical and helpful comments on the manuscript by Tom Berman, Josep M. Gasol, Connie Lovejoy, Carlos Pedrós-Alió, and two anonymous reviewers.
This work was supported in part by the European Union (EVK3-CT-2000-00022 NTAP and EVK3-CT-2002-00078 BASICS) and by a fellowship grant cofinanced by the Spanish Secretaría de Estado de Educación y Universidades and the European Social Fund to J.P.
REFERENCES
- 1.Arin, L., C. Marrasé, M. Maar, F. Peters, M.-M. Sala, and M. Alcaraz. 2002. Combined effects of nutrients and small-scale turbulence in a microcosm experiment. I. Dynamics and size of osmotrophic plankton. Aquat. Microb. Ecol. 29:51-61. [Google Scholar]
- 2.Bergstedt, M., M. M. Hondzo, and J. B. Cotner. 2004. Effects of small scale fluid motion on bacterial growth and respiration. Freshwater Biol. 49:28-40. [Google Scholar]
- 3.Berman, T., and B. Shteinman. 1998. Phytoplankton development and turbulent mixing in Lake Kinneret (1992-1996). J. Plankton Res. 20:709-726. [Google Scholar]
- 4.Bernardet, J.-F., Y. Nakagawa, and B. Holmes. 2002. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int. J. Syst. Evol. Microbiol. 52:1049-1070. [DOI] [PubMed] [Google Scholar]
- 5.Bidle, K. D., and F. Azam. 2001. Bacterial control of silicon regeneration from diatom detritus: significance of bacterial ectohydrolases and species identity. Limnol. Oceanogr. 46:1606-1623. [Google Scholar]
- 6.Bird, D. F., and J. Kalff. 1984. Empirical relationships between bacterial abundance and chlorophyll concentration in fresh and marine waters. Can. J. Fish. Aquat. Sci. 41:1015-1023. [Google Scholar]
- 7.Bowman, J. B., C. M. Nichols, and J. A. E. Gibson. 2003. Algoriphagus ratkowskyi gen. nov., sp. nov., Brumimicrobium glaciale gen. nov., sp. nov., Cryomorpha ignava gen. nov., sp. nov., and Crocinitomix catalasitica gen. nov., sp. nov., novel flavobacteria isolated from various polar habitats. Int. J. Syst. Evol. Microbiol. 53:1343-1355. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. J., S. Findlay, and M. L. Pace. 1988. Bacterioplankton production in fresh and saltwater ecosystems: a cross-system overview. Mar. Ecol. Prog. Ser. 43:1-10. [Google Scholar]
- 9.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covert, J. S., and M. A. Moran. 2001. Molecular characterization of estuarine bacterial communities that use high- and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127-139. [Google Scholar]
- 11.Eilers, H., J. Pernthaler, and R. Amann. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glöckner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German Bight and their seasonal contributions to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estrada, M., M. Alcaraz, and C. Marrasé. 1987. Effects of turbulence on the composition of phytoplankton assemblages in marine microcosms. Mar. Ecol. Prog. Ser. 38:267-281. [Google Scholar]
- 14.Fandino, L. B., L. Riemann, G. F. Steward, R. A. Long, and F. Azam. 2001. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 23:119-130. [Google Scholar]
- 15.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package. Cladistics 5:164-166. [Google Scholar]
- 16.Gasol, J. M., and P. A. del Giorgio. 2000. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64:197-224. [Google Scholar]
- 17.Giovannoni, S., and M. Rappé. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 18.Giovannoni, S. J., M. S. Rappé, K. L. Vergin, and N. L. Adair. 1996. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc. Natl. Acad. Sci. USA 93:7979-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton composition of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagström, Å., T. Pommier, F. Rohwer, K. Simu, W. Stolte, D. Svensson, and U. L. Zweifel. 2002. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 68:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris, G. P. 1986. Phytoplankton ecology, structure, function and fluctuation. Cambridge University Press, Cambridge, United Kingdom.
- 24.Havskum, H., and A. S. Hansen. 1997. Importance of pigmented and colourless nano-sized protists as grazers on nanoplankton in a phosphate-depleted Norwegian fjord and in enclosures. Aquat. Microb. Ecol. 12:139-151. [Google Scholar]
- 25.Hold, G. L., E. A. Smith, M. S. Rappé, E. W. Maas, E. R. B. Moore, C. Stroempl, J. R. Stephen, J. I. Prosser, T. H. Birkbeck, and S. Gallacher. 2001. Characterization of bacterial communities associated with toxic and non-toxic dinoflagellates. FEMS Microbiol. Ecol. 37:161-173. [Google Scholar]
- 26.Janse, I., G. Zwart, M. J. E. C. van der Maarel, and J. C. Gottschal. 2000. Composition of the bacterial community degrading Phaeocystis mucopolysaccharides in enrichment cultures. Aquat. Microb. Ecol. 22:119-133. [Google Scholar]
- 27.Karp-Boss, L., E. Boss, and J. E. Jumars. 1996. Nutrient fluxes to planktonic osmotrophs in the presence of fluid motion. Oceanogr. Mar. Biol. Annu. Rev. 37:71-107. [Google Scholar]
- 28.Kerkhof, L. J., M. A. Voytek, R. M. Sherrell, D. Millie, and O. Schofield. 1999. Variability in bacterial community structure during upwelling in the coastal ocean. Hydrobiologia 401:139-148. [Google Scholar]
- 29.Kiøorboe, T., and E. Saiz. 1995. Planktivorous feeding in calm and turbulent environments, with emphasis on copepods. Mar. Ecol. Prog. Ser. 122:135-145. [Google Scholar]
- 30.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 31.Kirchman, D. L., L. Yu, and M. T. Cottrell. 2003. Diversity and abundance of uncultured Cytophaga-like bacteria in the Delaware Estuary. Appl. Environ. Microbiol. 69:6587-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kisand, V., R. Cuadros, and J. Wikner. 2002. Phylogeny of culturable estuarine bacteria catabolizing riverine organic matter in the northern Baltic Sea. Appl. Environ. Microbiol. 68:379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kisand, V., and J. Wikner. 2003. Limited resolution of 16S rDNA DGGE caused by melting properties and closely related DNA sequences. J. Microbiol. Methods 54:183-191. [DOI] [PubMed] [Google Scholar]
- 34.Kononen, K., J. Kuparinen, K. Mäkelä, J. Laanemets, J. Pavelson, and S. Nommann. 1996. Initiation of cyanobacterial blooms in a front region at the entrance to the Gulf of Finland, Baltic Sea. Limnol. Oceanogr. 41:98-112. [Google Scholar]
- 35.Lazier, J. R. N., and K. H. Mann. 1989. Turbulence and the diffusive layers around small organisms. Deep-Sea Res. 36:1721-1733. [Google Scholar]
- 36.Legendre, L., and F. Rassoulzadegan. 1995. Plankton and nutrient dynamics in marine waters. Ophelia 41:153-172. [Google Scholar]
- 37.Maar, M., L. Arin, R. Simó, M.-M. Sala, F. Peters, and C. Marrasé. 2002. Combined effects of nutrients and small-scale turbulence in a microcosm experiment. II. Dynamics of organic matter and phosphorous. Aquat. Microb. Ecol. 29:63-72. [Google Scholar]
- 38.Malits, A., F. Peters, M. Bayer, C. Marrasé, A. Zoppini, O. Guadayol, and M. Alcaraz. Effects of small-scale turbulence on bacteria: a matter of size. Microb. Ecol., in press. [DOI] [PubMed]
- 39.Martinez, J., D. C. Smith, G. F. Steward, and F. Azam. 1996. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat. Microb. Ecol. 10:223-230. [Google Scholar]
- 40.Moeseneder, M. M., and G. J. Herndl. 1995. Influence of turbulence on bacterial production in the sea. Limnol. Oceanogr. 40:1466-1473. [Google Scholar]
- 41.Moran, M. A., J. M. Gonz ález, and R. P. Kiene. 2003. Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol. J. 20:1-14. [Google Scholar]
- 42.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 43.Napier, I. R. 1995. Growth and collapse of a spring phytoplankton bloom in the Firth of Clyde, Scotland. Mar. Biol. 123:189-195. [Google Scholar]
- 44.O'Sullivan, L. A., K. E. Fuller, E. M. Thomas, C. M. Turley, J. C. Fry, and A. J. Weightman. 2004. Distribution and culturability of the uncultivated “AGG58 cluster” of the Bacteroidetes phylum in aquatic environments. FEMS Microbiol. Ecol. 47:359-370. [DOI] [PubMed] [Google Scholar]
- 45.Peters, F., and T. Gross. 1994. Increased grazing rates of microplankton in response to small-scale turbulence. Mar. Ecol. Prog. Ser. 115:299-307. [Google Scholar]
- 46.Peters, F., C. Marrasé, J. M. Gasol, M. M. Sala, and L. Arin. 1998. Effects of turbulence on bacterial growth mediated through food web interactions. Mar. Ecol. Prog. Ser. 172:293-303. [Google Scholar]
- 47.Peters, F., C. Marrasé, H. Havskum, F. Rassoulzadegan, J. Dolan, M. Alcaraz, and J. M. Gasol. 2002. Turbulence and the microbial food web: effects on bacterial losses to predation and on community structure. J. Plankton Res. 24:321-331. [Google Scholar]
- 48.Pinhassi, J., F. Azam, J. Hemphälä, R. A. Long, J. Martinez, U. L. Zweifel, and Å. Hagström. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 49.Pinhassi, J., and T. Berman. 2003. Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69:199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinhassi, J., and Å.. Hagström. 2000. Seasonal succession in marine bacterioplankton. Aquat. Microb. Ecol. 21:245-256. [Google Scholar]
- 51.Pinhassi, J., A. Winding, S. Binnerup, U. L. Zweifel, B. Riemann, and Å. Hagström. 2003. Spatial variability in bacterioplankton community composition at the Skagerrak-Kattegat front. Mar. Ecol. Prog. Ser. 255:1-13. [Google Scholar]
- 52.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 53.Quigg, A., Z. F. Finkel, A. J. Irwin, Y. Rosenthal, T.-Y. Ho, J. R. Reinfelder, O. Schofield, F. M. M. Morel, and P. G. Falkowski. 2003. The evolutionary inheritance of elemental stoichiometry in marine phytoplankton. Nature 425:291-294. [DOI] [PubMed] [Google Scholar]
- 54.Riemann, L., and M. Middelboe. 2002. Stability of bacterial and viral community compositions in Danish coastal waters as depicted by DNA fingerprinting techniques. Aquat. Microb. Ecol. 27:219-232. [Google Scholar]
- 55.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riemann, L., G. F. Steward, L. B. Fandino, L. Campbell, M. R. Landry, and F. Azam. 1999. Bacterial community composition during two consecutive NE monsoon periods in the Arabian Sea studied by denaturing gradient gel electrophoresis (DGGE). Deep Sea Res. Part II 46:1791-1811. [Google Scholar]
- 57.Rothchild, B. J., and T. R. Osborn. 1988. Small-scale turbulence and plankton contact rates. J. Plankton Res. 10:465-474. [Google Scholar]
- 58.Sala, M. M., M. Karner, L. Arin, and C. Marrasé. 2001. Measurement of ectoenzyme activities as an indication of inorganic nutrient imbalance in microbial communities. Aquat. Microb. Ecol. 23:301-311. [Google Scholar]
- 59.Sala, M. M., F. Peters, J. M. Gasol, C. Pedrós-Alió, C. Marrasé, and D. Vaqué. 2002. Seasonal and spatial variations in the nutrient limitation of bacterioplankton growth in the northwestern Mediterranean. Aquat. Microb. Ecol. 27:47-56. [Google Scholar]
- 60.Schäfer, H., B. Abbas, H. Witte, and G. Muyzer. 2002. Genetic diversity of “satellite” bacteria present in cultures of marine diatoms. FEMS Microbiol. Ecol. 42:25-35. [DOI] [PubMed] [Google Scholar]
- 61.Schäfer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 62.Schauer, M., V. Balagué, C. Pedrós-Alió, and R. Massana. 2003. Seasonal changes in the taxonomic composition of bacterioplankton in a coastal oligotrophic system. Aquat. Microb. Ecol. 31:163-174. [Google Scholar]
- 63.Schauer, M., R. Massana, and C. Pedrós-Alió. 2000. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol. 33:51-59. [DOI] [PubMed] [Google Scholar]
- 64.Selje, N., M. Simon, and T. Brinkhoff. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445-448. [DOI] [PubMed] [Google Scholar]
- 65.Smith, D. C., M. Simon, A. L. Alldredge, and F. Azam. 1992. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139-142. [Google Scholar]
- 66.Smith, D. C., G. F. Steward, R. A. Long, and F. Azam. 1995. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep-Sea Res. 42:75-97. [Google Scholar]
- 67.Suzuki, M., Y. Nakagawa, S. Harayama, and S. Yamamoto. 2001. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int. J. Syst. Evol. Microbiol. 51:1639-1652. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki, M. T., C. M. Preston, F. P. Chavez, and E. F. DeLong. 2001. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquat. Microb. Ecol. 24:117-127. [Google Scholar]
- 69.Thingstad, T. F., U. L. Zweifel, and F. Rassoulzadegan. 1998. P limitation of heterotrophic bacteria and phytoplankton in the northwest Mediterranean. Limnol. Oceanogr. 43:88-94. [Google Scholar]
- 70.van den Hoek, C., D. G. Mann, and H. M. Jahns. 1995. Algae: an introduction to phycology. Cambridge University Press, Cambridge, United Kingdom.
- 71.van Hannen, E. J., W. Mooij, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yager, P. L., T. L. Connelly, B. Mortazavi, K. E. Wommack, N. Bano, J. E. Bauer, S. Opsahl, and J. T. Hollibaugh. 2001. Dynamic bacterial and viral response to an algal bloom at subzero temperatures. Limnol. Oceanogr. 46:790-801. [Google Scholar]