Abstract
Salinibacter ruber is an extremely halophilic bacterium, phylogenetically affiliated with the Flavobacterium/Cytophaga branch of the domain Bacteria. Electrospray mass analyses (negative ion) of the total lipid extract of a pure culture of S. ruber shows a characteristic peak at m/z 660 as the most prominent peak in the high-mass range of the spectrum. A novel sulfonolipid, giving rise to the molecular ion [M-H]− of m/z 660, has been identified. The sulfonolipid isolated and purified by thin-layer chromatography was shown by chemical degradation, mass spectrometry, infrared spectroscopy, and nuclear magnetic resonance analysis to have the structure 2-carboxy-2-amino-3-O-(13′-methyltetradecanoyl)-4-hydroxy-18-methylnonadec-5-ene-1-sulfonic acid. This lipid represents about 10% of total cellular lipids, and it appears to be a structural variant of the sulfonolipids found as main components of the cell envelope of gliding bacteria of the genus Cytophaga and closely related genera (W. Godchaux and E. R. Leadbetter, J. Bacteriol. 153:1238-1246, 1983) and of diatoms (R. Anderson, M. Kates, and B. E. Volcani, Biochim. Biophys. Acta 528:89-106, 1978). Since this sulfonolipid has never been observed in any other extreme halophilic microorganism, we consider the peak at m/z 660 the lipid signature of Salinibacter. This study suggests that this novel sulfonolipid may be used as a chemotaxonomic marker for the detection of Salinibacter within the halophilic microbial community in saltern crystallizer ponds and other hypersaline environments.
Salinibacter ruber is a red, extremely halophilic, aerobic bacterium, recently isolated from saltern crystallizer ponds in Alicante and Mallorca, Spain (4). The first evidence of the presence of this novel type of Bacteria came from sequencing of bacterial 16S rRNA genes from crystallizer ponds in Alicante, Spain (3). By use of fluorescent oligonucleotide probes designed to detect cells with this new phylotype, the organism was found to be rod shaped, often slightly curved, and to be quite abundant: between 5 and 25% of the total prokaryotic community in Spanish crystallizer ponds belongs to this type (3). The name “Candidatus Salinibacter” was originally given to this bacterium on the basis of these environmental studies. Thereafter, a number of extremely halophilic Bacteria have been isolated from saltern ponds in Mallorca and Santa Pola, Alicante, Spain, with 16S rRNA sequences nearly identical to the “Candidatus Salinibacter” phylotype. A formal description of the isolates as S. ruber has now been published in the taxonomic literature (4). Phylogenetically, Salinibacter belongs to the order Cytophagales. Its closest cultured relative is Rhodotermus, a genus of slightly halophilic, thermophilic Bacteria isolated from marine hot springs (1).
Salinibacter is no less halophilic than many of the archaeal halophiles of the family Halobacteriaceae. It requires at least 150 g of salt/liter for growth and grows optimally at NaCl concentrations between 200 and 300 g/liter. Examination of the physiological properties of S. ruber showed that this extremely halophilic bacterium displays a similar mode of haloadaptation to that of the Archaea of the order of Halobacteriales. In particular, Salinibacter cells contain very high concentrations of K+ ions in their cytoplasm, similar to those measured in Halobacterium salinarum cells (4, 22), and they do not accumulate organic osmotic solutes such as are used by all other known halophilic and halotolerant aerobic Bacteria. Moreover, the bulk proteins of Salinibacter were found to have a high content of acidic amino acids, a low content of basic amino acids, a low content of hydrophobic amino acids, and a high content of serine (21), as do the halophilic Archaea. Recent studies of the effects of different KCl and NaCl concentrations on four cytoplasmic enzymes have shown that Salinibacter enzymes are adapted to function in the presence of high salt concentrations, similar to most proteins of members of the family Halobacteriaceae. An important aspect of haloadaptation is the modification of membrane lipid composition that serves to preserve membrane integrity and function at high salt concentrations (25). The membrane lipids of S. ruber are typical for the bacterial domain, with glycerophospholipids containing ester-linked fatty acyl chains and not ether-linked phytanyl chains. In particular, phosphatidylcholine (PC), N,N-dimethylphosphatidyletanolamine (NN-PE), phosphatidylserine (PS), phosphatidylglycerol (PG), and bisphosphatidylglycerol (BPG) are the major membrane phospholipids in Salinibacter (23).
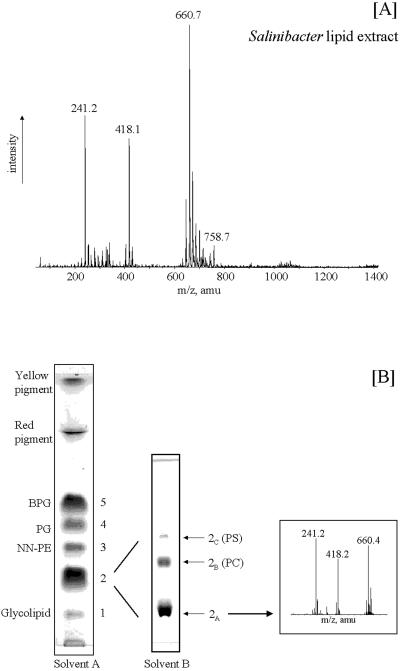
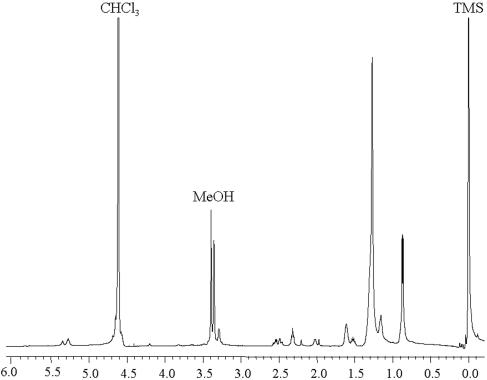
Electrospray mass spectrometry (ESI-MS) analyses (negative ion) of the total lipid extract of S. ruber (Fig. 1A) shows that the peak at m/z 660 is the most prominent peak in the high-mass range of the spectrum. In this paper we describe the identification of a novel sulfonolipid giving rise to the peak at m/z 660 and we document its structure determination by MS, infrared (IR) spectroscopy, and nuclear magnetic resonance (NMR) analysis and by identification of its degradation products. We found that the structure of the new sulfonolipid is 2-carboxy-2-amino-3-O-(13′-methyltetradecanoyl)-4-hydroxy-18-methylnonadec-5-ene-1-sulfonic acid. This lipid, which represents about 10% of the total cellular lipids, appears to be a structural variant of the sulfonolipids found as main components of the cell envelope of gliding bacteria of the genus Cytophaga and closely related genera (11) and of diatoms (2). We also show here that the novel sulfonolipid may be used as a biomarker for the presence of Salinibacter in hypersaline environments.
FIG. 1.
(A) ESI-MS spectrum (negative ion) of total lipid extract of S. ruber cells (amu, atomic mass unit); (B) TLC profile of total lipid extract in solvent A (chloroform-methanol-acetic acid-water, 85:15:10:3.5 by volume) and rechromatography in solvent B (chloroform-methanol-29.9% [wt/vol] ammonia, 65:35:5, vol/vol/vol) of lipid extract from spot 2. The chromatograms were stained by charring at 120°C after being sprayed with 5% sulfuric acid-H2O. On the right is shown the low-resolution ESI-MS (negative ion) spectrum of the isolated and purified lipid 2A.
MATERIALS AND METHODS
Materials.
All organic solvents used were commercially distilled and of the highest available purity (Sigma-Aldrich). Plates for thin layer chromatography (TLC) (Silica Gel 60A), obtained from Merck, were washed twice with chloroform-methanol (1:1, vol/vol) and activated at 120°C before use. Standard fatty acid methyl esters and tetramethylsilane (TMS) were obtained from Sigma.
Culture conditions.
S. ruber strain M31 (DSM 13855T) (4) was grown in medium containing (all concentrations in grams/liter) NaCl, 195; MgSO4 · 7H2O, 25; MgCl2 · 6H2O, 16.3; CaCl2 · 2H2O, 1.25; KCl, 5.0; NaHCO3, 0.25; NaBr, 0.625; yeast extract, 1.0 (pH 7.0). Cells were grown in a rotary shaker (180 rpm) at 35°C. Cells were harvested by centrifugation (20 min, 7,000 × g) and dried by lyophilization.
Lipid extraction.
Lipids were extracted by the method of Bligh and Dyer as modified for extreme halophiles (15). Dried cells (1 g) were rehydrated overnight at 4°C in 4 ml of water. For the lipid extraction, 15 ml of methanol-chloroform (2:1, vol/vol) was added to the cell suspension and the mixture was gently shaken for 20 min. After centrifugation, the supernatant extract was decanted into a separatory funnel, and the residue was resuspended in 19 ml of chloroform-methanol-water (1:2:0.8, vol/vol/vol). The mixture was then shaken and centrifuged; 5 ml each of chloroform and water were then added to the combined supernatants to obtain a two-phase system. After complete phase separation (requiring a few hours at room temperature), the chloroform phase, diluted with benzene, was brought to dryness under nitrogen. Dried lipids were resuspended in a small volume of chloroform and stored at −20°C.
TLC.
Cellular lipids of S. ruber were separated by preparative TLC on Silica Gel 60A plates (20 by 20 cm by 0.5 mm) in solvent A (chloroform-methanol-acetic acid-water, 85:15:10:3.5 by volume). After the silica in each band was scraped from the plate, lipids were extracted five times with chloroform-methanol (1:1, vol/vol). The combined supernatants were brought to dryness under a stream of nitrogen (15). The sulfonolipid (named component 2A) was further purified by rechromatography in solvent B (chloroform-methanol-29.9% (wt/vol) ammonia, 65:35:5, vol/vol/vol) and recovered from silica as described above. Analytical TLC was carried out on Silica Gel 60A plates (10 by 20 cm by 0.25 mm).
Lipids were detected with the following spray reagents (15): (i) molybdenum blue Sigma spray reagent for phospholipids, (ii) azure A-sulfuric acid for sulfatides, and (iii) 5% sulfuric acid, followed by charring at 120°C, for all lipids.
Acid hydrolysis.
Acid hydrolysis was performed as described elsewhere (15). The isolated and purified lipid 2A (1 to 5 mg) was deacylated in 1 M aqueous methanolic HCl (3 ml) under reflux at 70°C for 18 h in a screw-cap Teflon-lined tube. After extraction of fatty acid methyl esters with three portions (3 ml) of petroleum ether (boiling point, 40 to 60°C), the aqueous methanolic hydrolysate was diluted with chloroform and water to give a final ratio of chloroform-methanol-water of 1:1:0.9 (vol/vol/vol); after mixing, the chloroform phase, containing the residual aminosulfonate moiety, was removed and taken to dryness under nitrogen.
Alkaline hydrolysis.
Mild and strong alkaline hydrolysis of pure lipid 2A was performed as previously described (15). Dried lipid (5 mg) was dissolved in 5 ml of 0.3 N NaOH in 90% methanol and heated at 70°C for 1 h (mild hydrolysis) or 18 h (strong hydrolysis). Then the mixture was strongly acidified by addition of 6 N HCl and the free fatty acids were extracted with three portions of petroleum ether (boiling point, 40 to 60°C). The remaining aqueous methanolic phase, containing the aminosulfonate moiety, was brought to dryness under nitrogen.
Oxidation of aminosulfonate with periodate-permanganate.
Aminosulfonate (2 mg) was dissolved in a mixture of 5 ml of t-butyl alcohol and 2.5 ml of water. Then 1.25 ml of 4 mM Na2CO3 and 2.5 ml of 30 mM NaIO4 plus 2.5 mM KMnO4 were added (2, 11). The mixture was stirred vigorously for 4 h at 40°C, after which solid sodium metabisulfite was added until the solution was colorless. After removal of t-butyl alcohol under a stream of N2, the mixture was acidified with 0.2 ml of 6 N H2SO4 and extracted twice with 5-ml portions of hexane. The combined hexane phases were dried under N2, and the residue was dissolved in chloroform-methanol for ESI-MS analysis.
ESI-MS.
All MS analyses were carried out in loop injection mode with dried lipid extracts that were dissolved in chloroform-methanol (1:1, vol/vol). Samples (2 μl), injected via a 10-μl loop, were transferred into the MS electrospray interface (ESI) with a flow rate of 0.1 ml/min of chloroform-methanol (1:1, vol/vol) delivered by a Varian 9012 chromatographic system.
Low-resolution mass spectra were obtained with an API 165 mass spectrometer (Applied Biosystem/MSD Sciex) equipped with a turbo ion spray interface. Interface conditions (negative ions) were as follows: nebulizer gas (air), 1.2 liters/min; curtain gas (nitrogen), 1 liter/min; needle voltage, −5,000 V; declustering potential, −150 V; and focusing potential, −200 V.
High-resolution mass spectra were obtained with a QSTAR hybrid Qq-TOF mass spectrometer (Applied Biosystem/MSD Sciex) equipped with a turbo ion spray interface. Interface conditions (negative ions) were as follows: nebulizer gas (air), 1.2 liters/min; curtain gas (nitrogen), 1 liter/min; needle voltage, −4,500 V; declustering potential, −50 V; focusing potential, −300 V. Accurate mass measurements (four decimal figures) were carried out by obtaining averaged spectra of loop injection peaks and then by calibrating them with two ions of known molecular structure present in the same spectra. Errors associated with such determinations were usually within 4 ppm. Accurate MS-MS measurements (four decimal figures) were carried out by first fragmenting the target ions at proper collision energy (usually 35 eV) and then applying the above-described procedure using the [M-H]− ion (whose accurate mass was previously measured) and the HSO3−ion (m/z 80.9652) as calibrating ions.
NMR spectroscopy.
1H-NMR spectra of isolated lipids were taken in CDCl3-CD3OD (4:3, vol/vol; final lipid concentration, about 2 mM). All NMR analyses were performed on an AM 500 Bruker instrument. 1H chemical shifts are given relative to TMS.
IR spectroscopy.
IR spectra were taken in KBr disks (about 1 mg of lipid per 150 mg of KBr), using a Perkin-Elmer FT-IT 1710 IR spectrophotometer.
Gas chromatography-MS.
Analysis of fatty acid methyl esters, prepared by methanolysis of the pure sulfonolipid, was performed with a Shimadzu GCMS-QP5000 gas chromatograph equipped with a Supelco SpB-1 (methylsilicone) column (30 m by 0.25 mm by 0.25 μm).
RESULTS
Isolation and purification of the m/z 660 lipid.
ESI-MS analysis (negative ion) of the total lipid extract of a pure culture of S. ruber strain M31 (Fig. 1A) shows that the peak at m/z 660 is the most prominent peak in the high-mass range of the spectrum.
In order to identify the lipid component giving rise to the peak at m/z 660, we isolated individual lipid components of the extract by TLC (Fig. 1B). After development in an acidic solvent system (solvent A), each component was recovered from the silica as described in Materials and Methods. ESI-MS (−) analyses of individual lipid components revealed that BPG (spot 5), PG (spot 4), NN-PE (spot 3), and an unknown glycolipid (spot 1) were present in the lipid extract (Fig. 1B). Only the ESI-MS spectrum of lipid 2 showed a major peak at m/z 660. The purity of this component was checked by rechromatography in a neutral solvent system (solvent B). Figure 1B shows three components to be present in the original spot. Each component was isolated and purified for further investigation. ESI-MS (−) analyses showed that the peak at m/z 660 was present only in the spectrum of component 2A (Fig. 1B) and that the other two components were PC and PS. The yield of lipid 2A was 10.5 mg/100 mg of total cellular lipids.
Chromatographic behavior.
Lipid 2A had a low mobility in both solvent A (Rf, 0.26) and solvent B (Rf, 0.24). It gave a negative test on TLC plates for sugar and phosphate but stained positively with azure A, indicating the presence of a sulfate or sulfonate group in the molecule.
IR spectrum.
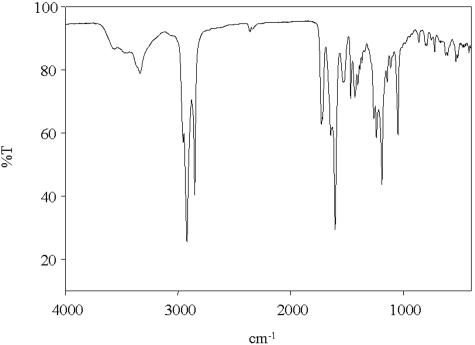
The IR spectrum of the pure sulfonolipid (Fig. 2) shows a complex of intense bands in the sulfonate region at 1,260 cm−1 (asymmetric SO2 stretch) and 1,045 cm−1 (symmetric SO2 stretch), along with characteristic peaks for NH2 at 3,335 cm−1 (N-H stretch) and at 1,650 cm−1 (N-H deformation). Two prominent absorption bands at 1,727 and 1,605 cm−1 (C=O stretch) imply the presence of an ester C=O and an ionized carboxyl group, respectively. IR bands for isopropyl groups were present at 1,365 cm−1 and 1,385 cm−1. This spectrum was similar to that obtained for N-acylaminosulfonates isolated from Flexibacter sp. (11).
FIG. 2.
IR spectrum of isolated and purified lipid 2A (1 mg on KBr disk).
ESI-MS analyses.
Accurate ESI-MS measurements, obtained by calibrating the averaged spectrum with two ions of known molecular structure present in the same spectrum, revealed the [M-H]− to have an m/z 660.4494, corresponding to the C35H66NO8S molecular formula (calculated m/z, 660.4514; error, 3.1 ppm).
The low-resolution ESI-MS spectrum of isolated and purified lipid 2A (Fig. 1B) appears quite similar to that of the total lipid extract of S. ruber cells, showing three main peaks: the molecular ion [M-H]− at m/z 660 and two fragment ions at m/z 241 and 418.
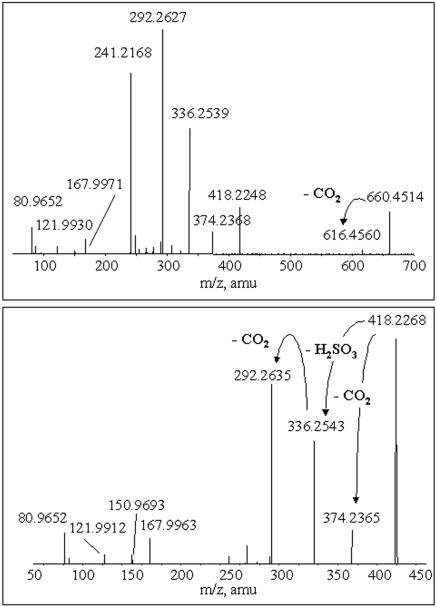
In order to obtain more information, we also performed high-resolution ESI-MS-MS analyses of the [M-H]− ion (nominal m/z 660) and of its fragment ion at nominal m/z 418. Figure 3 (upper panel) and Table 1 show the high-resolution product ion spectrum of the m/z 660 ion and the accurate mass measurement results for their principal fragments, respectively. The presence of a fragment at m/z 81 (HSO3−) and the absence of the ion at m/z 97 indicate that the molecule has a sulfonate group and not sulfate. Also, the absence of peaks at m/z 79 and 97 (diagnostic for the presence of phosphate groups) is in agreement with the negative test for phosphate. The fragment at m/z 616.4560, resulting from the neutral loss of CO2, indicates the presence of a carboxylic group in the lipid molecule. The fragments at m/z 418.2248 and 241.2168 result from the neutral loss of a C15:0 fatty acid and from the C15:0 fatty acid itself. Their presence suggests the presence of a bound ester, as confirmed by hydrolysis experiments (see the next section).
FIG. 3.
High-resolution product ion spectrum of the molecular ion at m/z 660 (upper panel) and of the fragment ion at m/z 418 (lower panel). amu, atomic mass unit.
TABLE 1.
Molecular formulas and accurate mass measurements of the principal fragments of the m/z 660 ion and its fragment of m/z 418 obtained by high-resolution MS-MS experimentsa
| Fragment no. reported in fragmentation pathway (Fig. 6) | Molecular formula | Calculated mass (Da) | Measured mass, Da (mass relative error, ppm) for m/z 660 ion | Measured mass, Da (mass relative error, ppm) for fragment of m/z 660 ion (m/z 418) |
|---|---|---|---|---|
| 1 | C34H66NO6S | 616.4616 | 616.4560 (9) | Not present |
| 2 | C20H36NO6S | 418.2268 | 418.2248 (5) | Used for calibration |
| 3 | C19H36NO4S | 374.2370 | 374.2368 (0.7) | 374.2365 (1.5) |
| 4 | C20H34NO3 | 336.2544 | 336.2539 (1.5) | 336.2543 (0.4) |
| 5 | C19H34NO | 292.2645 | 292.2627 (6.5) | 292.2635 (3.7) |
| 6 | C15H29O2 | 241.2173 | 241.2168 (2.1) | Not present |
| 7 | C3H6NO5S | 167.9972 | 167.9971 (0.7) | 167.9963 (5.5) |
| 8 | C3H3O5S | 150.9706 | 150.9715 (5.5) | 150.9693 (9) |
| 9 | C2H4NO3S | 121.9917 | 121.9930 (10) | 121.9912 (4.4) |
| 10 | C3H4NO2 | 86.0247 | 86.0248 (0.6) | 86.0238 (11) |
| 11 | C4H6NO | 84.0455 | 84.0463 (9.7) | Not present |
Ions used for mass calibration were as follows: HSO3− ion of m/z 80.9652 and [M-H]− ion m/z 660.4514 (m/z 660 ion); HSO3− ion of m/z 80.9652 and ion at m/z 418.2268 (fragment of m/z 660 ion).
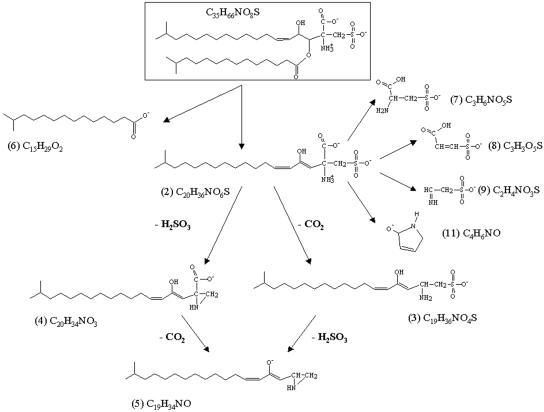
The high-resolution product ion spectrum of the m/z 418 ion and the corresponding accurate mass measurements for their principal fragments (Fig. 3, lower panel, and Table 2, respectively) revealed fragments at m/z 374.2368 (neutral loss of CO2), 336.2539 (neutral loss of H2SO3), 292.2627 (neutral loss of CO2 and H2SO3), and 80.9652 (HSO3−). It is important to point out that the peak at m/z 241, observed in the product ion spectrum of the m/z 660 ion, does not originate from the fragment at m/z 418. The ion at m/z 121.9930, indicating the presence of the taurine backbone (2-aminoethane sulfonic acid), has previously been observed in mass spectra of other kinds of capnine (8). The proposed fragmentation pathway for lipid 2A that accounts for the molecular formulas obtained by accurate mass measurements is reported in Fig. 6.
TABLE 2.
Molecular formulas and accurate mass measurements of the principal fragments of hydrolyzed lipid of m/z 436 obtained by high-resolution MS-MS experimentsa
| Molecular formula | Calculated mass (Da) | Measured mass (Da) | Mass relative error (ppm) |
|---|---|---|---|
| C20H36NO6S | 418.2268 | 418.2297 | 6.7 |
| C19H38NO5S | 392.2476 | 392.2418 | 15 |
| C20H36NO4 | 354.2649 | 354.2649 | 0.2 |
| C20H34NO3 | 336.2544 | 336.2566 | 6.5 |
| C19H34NO | 292.2645 | 292.2631 | 5.1 |
| C17H33O3 | 285.2435 | 285.2426 | 3.2 |
| C5H8NO6S | 210.0077 | 210.0062 | 7.5 |
| C3H6NO5S | 167.9972 | 167.9961 | 6.6 |
| C3H3O5S | 150.9706 | 150.9699 | 5 |
| C5H6NO3 | 128.0353 | 128.0347 | 4.8 |
| C2H4NO3S | 121.9917 | 121.9915 | 2 |
| C4H6NO | 84.0455 | 84.0459 | 4.9 |
Mass calibration performed with HSO3− ion of m/z 80.9652 and [M-H]− ion of m/z 436.2374.
FIG. 6.
Proposed structure and fragmentation pathway for lipid 2A. All molecular formulas reported in the figure were obtained by accurate mass measurements (see Table 1).
1H-NMR spectrum of the isolated sulfonolipid.
The 1H-NMR spectrum of lipid 2A (Fig. 4) showed aliphatic proton signals at 0.88 and 1.27 ppm, indicating that the compound has two long hydrocarbon chains with an isopropyl group at the end. The spectrum also shows signals due to two amine protons (-NH2 at 1.62 ppm), a methylene sulfonate (CH2SO3 at 3.47 and 3.51 ppm), and two olefinic protons (CH=CH at 5.26 and 5.34 ppm). It is worth noting that the absence in the IR spectrum (Fig. 2) of an intense absorption band at 980 cm−1, characteristic for trans-double bonds, suggests a cis configuration for the double bond in question. Integration of aliphatic proton signals is consistent with that expected for the structure proposed in Fig. 6.
FIG. 4.
1H-NMR spectrum of isolated and purified lipid 2A. Two milligrams of pure sulfonolipid was dissolved in 700 μl of CDCl3-CD3OD (4:3, vol/vol). Chemical shifts are given relative to TMS as an internal standard.
Analysis of the products of acid hydrolysis.
Strong acid methanolysis with 1 N HCl-10 M H2O-CH3OH of lipid 2A gave two products. The first was the methyl ester of a fatty acid. It was extracted with petroleum ether and identified by gas chromatography-MS as 13-methyl tetradecanoic acid methyl ester. The second product (extracted by the method of Bligh and Dyer) was analyzed by IR spectroscopy and ESI-MS. The IR spectrum (not shown) showed the presence of sulfonate (bands at 1,045 and 1,201 cm−1) and, interestingly, one carboxyl group (C=O stretch at 1,739 cm−1). The amine absorption band at 1,650 cm−1 (N-H deformation) was present, while the band of N-H stretch (3,300 cm−1) was not visible because of the presence of an intense band at 3,401 cm−1, probably due to free hydroxyl groups. The low-resolution ESI-MS spectrum (negative ions) of the aminosulfonate produced by strong acidic hydrolysis showed two intense ions at m/z 436 and 450 that could be attributed to the molecular ion of aminosulfonate (436 = 660-225: loss of fatty acyl moiety) and its methylated form.
Analysis of the products of alkaline hydrolysis.
To avoid the formation of methylated products and to establish if the fatty acid was linked by an amide or an ester bond, we performed alkaline hydrolysis under mild and strong conditions. Both mild and strong alkaline hydrolysis of pure sulfonolipid, performed in 0.3 M methanolic NaOH, for 1 and 18 h, respectively, released a single fatty acid in the ether phase, indicating the presence of an ester bond. This fatty acid was confirmed to be methylmyristic acid (measured m/z, 241.2163; calculated m/z, 241.2173; error, 4.2 ppm) by high-resolution ESI-MS analysis. The aminosulfonate remaining in the methanol phase was analyzed by ESI-MS and NMR. Peaks in the aliphatic proton region (0.88 and 1.18 ppm) in the 1H-NMR spectrum of aminosulfonate (not shown) confirm the presence of an isopropyl group also in the long-chain base moiety. Integration of the signals relative to alkyl protons is in agreement with the proposed structure shown in the inset in Fig. 5.
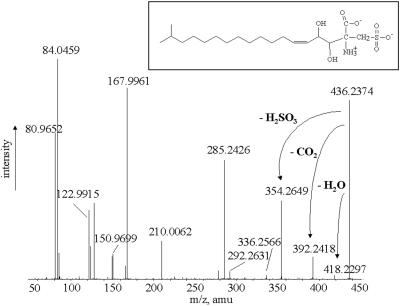
FIG. 5.
High-resolution product ion spectrum of the molecular ion at m/z 436, i.e., aminosulfonate, obtained by strong alkaline hydrolysis. amu, atomic mass unit.
Accurate ESI-MS measurements of aminosulfonate, obtained by alkaline hydrolysis of intact sulfonolipid and by calibrating the averaged spectrum with two ions of known molecular structure present in same spectrum, revealed the [M-H]− to have an m/z of 436.2369, corresponding to the C20H38NO7S molecular formula (calculated m/z, 436.2374; error, 1.3 ppm).
The high-resolution ESI-MS-MS spectrum (Fig. 5) of the [M-H]− ion of m/z 436.2369 displayed fragments at m/z 418.2297 (loss of H2O), 392.2418 (loss of CO2), and 354.2649 (loss of H2SO3). Fragments in the low-mass range (m/z 80.9652, 121.9915, 150.9699, and 167.9961) are identical to those of the ESI-MS-MS spectrum of intact sulfonolipid and of its product ion of m/z 418 (see product ion spectrum of [M-H]− ions of m/z 660 and 418 in Fig. 3). By accurate mass measurements of the fragment ions shown in the spectrum in Fig. 5, we obtained the corresponding molecular formula of all fragment ions originating from the aminosulfonate (Table 2).
The position of the double bond was confirmed by periodate-permanganate oxidation of the long-chain base sulfonic acid obtained by strong alkaline hydrolysis of the intact sulfonolipid. Periodate-permanganate oxidation was performed as previously described (2, 11), employing about 2 mg of aminosulfonate. Fatty acid released by periodate-permanganate oxidation was analyzed by ESI-MS. The spectrum showed a single oxidation product of m/z 227.2017, consistent with the molecular formula C14H27O2 (calculated m/z, 227.2016; error, 0.2 ppm), thus confirming the presence of a double bond between carbons 5 and 6 (counting from the methylene linked with the sulfonate). On the basis of the above data we propose the molecular structure reported in Fig. 6 for the m/z 660 lipid of S. ruber.
The novel sulfonolipid as biomarker for the presence of Salinibacter in hypersaline environments.
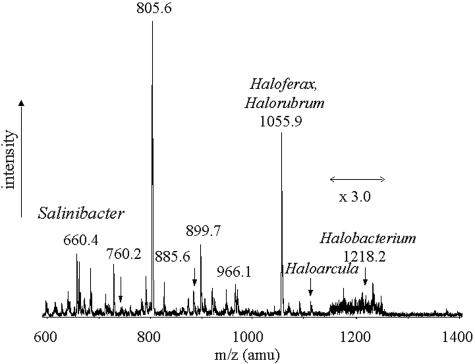
We have recently shown that ESI-MS analysis of the biomass lipid profile is a powerful experimental approach to obtain information on the types of halophilic microorganisms that inhabit saltern brines (17). Figure 7 shows the ESI-MS (−) spectrum of the total lipid extract obtained from the biomass of a crystallizer pond of the Margherita di Savoia (Italy) salterns. The main peaks in the spectrum can be attributed to the molecular ions of different kinds of polar lipids (phospholipids and glycolipids) present in the membranes of various halophilic microorganisms. In particular, certain glycolipid peaks are indicative of the presence of various genera of the Halobacteriaceae family (16, 19, 20). As previously shown (17), the peaks in the spectrum in Fig. 7 are indicative of the presence in the biomass of microorganisms representative of the genera Halobacterium (peak at m/z 1218, S-TGA-1), Haloarcula (peak at m/z 1138, TGA-2), and Haloferax and/or Halorubrum (different sulfated diglycosyl diethers, m/z 1056). A peak at m/z 660 is also evident in the ESI-MS profile of the lipid extract from the biomass of the Margherita di Savoia saltern crystallizer sample. This peak can now be attributed to the novel sulfonolipid here described, indicating the presence of Salinibacter within the halophilic community of the crystallizer ponds of Margherita di Savoia.
FIG. 7.
ESI-MS (negative ion) profile of the lipid extract obtained from the biomass of crystallizer brines of Margherita di Savoia (Italy). amu, atomic mass unit.
DISCUSSION
Salinibacter is one of the most halophilic organisms known within the domain Bacteria. Phylogenetically, Salinibacter belongs to the order Cytophagales (3), but when its physiology is examined, with particular reference to the mode of haloadaptation, there is a surprising similarity between this extremely halophilic bacterium and the Archaea of the family Halobacteriaceae (23).
In this paper we report the first detailed analysis of the membrane lipids of S. ruber. Apart from the phospholipids typical for the bacterial domain (PC, PG, BPG, etc.), membranes of S. ruber also contain significant amounts of a novel sulfonolipid. Studies of its chemical structure indicate that the new sulfonolipid is 2-carboxy-2-amino-3-O-(13′-methyltetradecanoyl)-4-hydroxy-18-methylnonadec-5-ene-1-sulfonic acid. This lipid represents about 10% of the total cellular lipids. The molecule carries a methylmyristic acid moiety. Methylmyristic acid is one of the dominant fatty acids in the lipids of S. ruber (unpublished results), and it is also a component of salinixanthin, the novel carotenoid acyl glycoside that colors the organism red (18).
The novel lipid appears to be an interesting structural variant of the sulfonolipids found as main components of the cell envelope of bacteria of the genus Cytophaga and closely related genera (11) and of diatoms (2). The first evidence for the presence of these unusual sulfonolipids, collectively called capnoids, in prokaryotes was reported by Godchaux and Leadbetter (9). The most common capnoid is the N-acylated version (N-acylcapnine). The parent compound, to which the trivial name “capnine” was assigned, was identified to be 2-amino-3-hydroxy-15-methylhexadecane-1-sulfonic acid. Capnine is structurally related to sphingosine. Its N-acyl derivatives are formed through amide linkage of a fatty acyl group to the free amino group of capnine, and they thus resemble ceramides. It is worth noting that, unlike sphingolipids produced by eukaryotic organisms, capnines and other bacterial sphingolipids contain mainly saturated long-chain bases which not infrequently have methyl branches. At variance from the previously characterized structures, the Salinibacter sulfonolipid presents a novel feature: an extra acidic residue, i.e., the carboxylate group at carbon 2 and an O-acyl group at carbon 3.
The capnoids were found in large quantities in gliding bacteria of the genera Cytophaga, Capnocytophaga, Sporocytophaga, and Flexibacter (10), while they have been found to be absent in numerous related nongliding bacteria. Furthermore N-acylcapnines have recently been isolated from two other members of the Cytophaga-Flavobacterium-Bacteroides phylum: the gram-negative bacteria Cyclobacterium marinus (5) and Flectobacillus major (6). Two other species in which capnoids have been detected, Empedobacter brevis and Myroides odoratus (12), share other Cytophaga-like characteristics. It would be very interesting to know if the Salinibacter sulfonolipid is synthesized from cysteate, as are the cytophagal sulfonolipids (26).
To our knowledge, the only occurrence of a sulfonolipid similar to capnine in the eukaryotic domain is in the diatom Nitzschia alba (2). This lipid is an N-acylated 2-amino-3-hydroxy-4-trans-octadecene-1-sulfonic-acid; i.e., the aminosulfonate moiety has the structure of sphingosine typical of eukaryotes, except for the substitution of the sulfonate group for the 1-hydroxyl group of that compound.
Interest in the novel Salinibacter sulfonolipid stems not only from the fact that it appears to be a very unusual biological compound but also from the properties it may confer to the cells. As suggested for the capnoids (9), such a sulfonolipid may confer some unusual properties upon the cell surface. For example, a high content of O-acylaminosulfonate might result in a membrane with a high intrinsic surface negative charge. As reported, the maintenance of a highly negative charge surface density by a high concentration of acidic lipids in the membrane is a common strategy adopted by the archaeal extreme halophiles for survival in media of high salt concentration (14). Such a highly negative surface charge density would be shielded by the high Na+ ion concentration of the external medium, thus preventing the disruption of the lipid bilayer due to charge-charge repulsion (7, 24). The O-acylaminosulfonate might also contribute to membrane fluidity, via the isostructures of both fatty acid and long-chain base and via mutual repulsion of the sulfonate groups.
It has been suggested that the capnoids may be involved in some aspect of the gliding motility of Capnocytophaga spp. and other gliding bacteria (10). Interest in this potential role for the sulfonolipids is stimulated by the fact that diatoms, the only other group of organisms of which at least one member contains a sulfonic analog of ceramides, exhibit a form of motility which resembles, at least superficially, that of the gliding bacteria (13). Motility in Salinibacter has not been reported, but absence of gliding motility has never been rigorously ascertained.
Results from this study suggest that the novel sulfonolipid may be used as a chemotaxonomic marker for the detection of Salinibacter within the halophilic microbial community in saltern crystallizer ponds and other hypersaline environments. We have recently shown that ESI-MS analysis of the biomass lipid profile is a powerful experimental approach to obtain information on the types of halophilic microorganisms that inhabit saltern brines (17). As glycolipids of extremely halophilic Archaea might serve as chemotaxonomic markers for classification of these organisms, the presence of certain glycolipid peaks in the mass spectrum of the total lipid extract of the biomass can be considered indicative of the presence of various genera of the Halobacteriaceae family (16, 19, 20). As shown in Fig. 1A, the novel sulfonolipid gives rise to a characteristic peak at m/z 660 in the ESI-MS (−) spectrum of a total lipid extract. Since this sulfonolipid has never been observed in any other known extreme halophilic microorganism, we can consider the peak at m/z 660 the lipid signature of Salinibacter. Thus, by analyzing the lipid profile of the biomass by ESI-MS in negative-ion mode, it is possible to detect the presence of microorganisms of the genus Salinibacter. Moreover, as ESI-MS can also give reliable quantitative results, we conclude that the novel sulfonolipid may represent a useful biomarker to obtain both qualitative and quantitative information about the distribution of this genus in hypersaline environments.
Acknowledgments
This work was supported by PRIN 2003-2004 of Ministero Italiano dell'Università e della Ricerca Scientifica (MIUR), IPCF-CNR (sezione di Bari) Italy, and the Israel Science Foundation (grant 504/03 to A.O.).
REFERENCES
- 1.Alfredsson, G. A., J. K. Kristjansson, S. Hjorleifsdottir, and K. O. Stetter. 1988. Rhodothermus marinus gen. nov., a thermophilic bacterium from submarine hot springs in Iceland. J. Gen. Microbiol. 134:299-306. [Google Scholar]
- 2.Anderson, R., M. Kates, and B. E. Volcani. 1978. Identification of the sulfonolipids in the non-photosynthetic diatom Nitzschia alba. Biochim. Biophys. Acta 528:89-106. [DOI] [PubMed] [Google Scholar]
- 3.Antón, J., R. Rosselló-Mora, F. Rodríguez-Valera, and R. Amann. 2000. Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl. Environ. Microbiol. 66:3052-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antón, J., A. Oren, S. Benlloch, F. Rodríguez-Valera, R. Amann, and R. Rosselló-Mora. 2002. Salinibacter ruber gen. nov., sp. nov., a new species of extremely halophilic Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 52:485-491. [DOI] [PubMed] [Google Scholar]
- 5.Batrakov, S. G., A. E. Mosezhnyi, A. O. Ruzhitsky, V. I. Sheichenko, and D. I. Nikitin. 2000. The polar-lipid composition of the sphingolipid-producing bacterium Flectobacillus major. Biochim. Biophys. Acta 1484:225-240. [DOI] [PubMed] [Google Scholar]
- 6.Batrakov, S. G., D. I. Nikitin, V.I. Sheichenko, and A. O. Ruzhitsky. 1998. A novel sulfonic-acid analogue of ceramides is the major extractable lipid of the gram-negative marine bacterium Cyclobacterium marinus WH. Biochim. Biophys. Acta 1391:79-91. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. S., P. G. Barton, D. Brown, and M. Kates. 1974. Osmometric and microscopic studies on bilayers of polar lipids from the extreme halophile, Halobacterium cutirubrum. Biochim. Biophys. Acta 352:202-217. [DOI] [PubMed] [Google Scholar]
- 8.Drijber, R. A., and W. B. McGill. 1997. Cythophaga hutchinsonii ATCC 33406 contains a structural variant of the sulfonolipid N-acylcapnine. Can. J. Microbiol. 43:689-693. [Google Scholar]
- 9.Godchaux, W., and E. R. Leadbetter. 1980. Capnocytophaga spp. contain sulfonolipids that are novel in procaryotes. J. Bacteriol. 144:592-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godchaux, W., and E. R. Leadbetter. 1983. Unusual sulfonolipids are characteristic of the Cytophaga-Flexibacter group. J. Bacteriol. 153:1238-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godchaux, W., and E. R. Leadbetter. 1984. Sulfonolipids of gliding bacteria. Structure of N-acylaminosulfonates. J. Biol. Chem. 259:2982-2990. [PubMed] [Google Scholar]
- 12.Holmes, B., and R. J. Owen. 1980. Emendation of the genus Flavobacterium and the status of the genus, p. 18-26. In H. Reichenbach and O.B. Weeks (ed.), The Flavobacterium-Cytophaga group. Developments after the eighth edition of Bergey's manual. Verlag Chemie, Weinheim, Germany.
- 13.Jarosch, R. 1962. Gliding, p. 573-581. In R.A. Lewin (ed.), Physiology and biochemistry of algae. Academic Press, New York, N.Y.
- 14.Kamekura, M., and M. Kates. 1999. Structural diversity of membrane lipids in members of Halobacteriaceae. Biosci. Biotechnol. Biochem. 63:969-972. [DOI] [PubMed] [Google Scholar]
- 15.Kates, M. 1986. Techniques of lipidology, p. 100-110, 163-164, 251-253. In R. H. Burdon and P. H. van Knippenberg (ed.), Laboratory techniques in biochemistry and molecular biology, vol. 3, part 2. Elsevier, Amsterdam, The Netherlands.
- 16.Kates, M. 1993. Membrane lipids of Archaea, p. 261-295. In M. Kates, D. J. Kushner, and A. T. Matheson (ed.), The biochemistry of Archaea (Archaebacteria). Elsevier, Amsterdam, The Netherlands.
- 17.Lattanzio, V. M. T., A. Corcelli, G. Mascolo, and A. Oren. 2002. Presence of two novel cardiolipins in the halophilic archaeal community in the crystallizer brines from the salterns of Margherita di Savoia (Italy) and Eilat (Israel). Extremophiles 6:437-444. [DOI] [PubMed] [Google Scholar]
- 18.Lutnæs, B. F., A. Oren, and S. Liaaen-Jensen. 2002. New C(40)-carotenoid acyl glycoside as principal carotenoid in Salinibacter ruber, an extremely halophilic eubacterium. J. Nat. Prod. 65:1340-1343. [DOI] [PubMed] [Google Scholar]
- 19.Oren, A. 25. July 2001, revision date. The order Halobacteriales. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.2. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 20.Oren, A., and P. Gurevich. 1993. Characterization of the dominant halophilic archaea in a bacterial bloom in the Dead Sea. FEMS Microbiol. Ecol. 12:249-256. [Google Scholar]
- 21.Oren, A., and L. Mana. 2002. Amino acid composition of bulk protein and salt relationships of selected enzymes of Salinibacter ruber, an extremely halophilic bacterium. Extremophiles 6:217-223. [DOI] [PubMed] [Google Scholar]
- 22.Oren, A., M. Heldal, S. Norland, and E. A. Galinski. 2002. Intracellular ion and organic solute concentrations of the extremely halophilic bacterium Salinibacter ruber. Extremophiles 6:491-498. [DOI] [PubMed] [Google Scholar]
- 23.Oren, A., F. Rodríguez-Valera, J. Antón, S. Benlloch, R. Rosselló-Mora, R. Amann, J. Coleman, and N. J. Russell. 2004. Red, extremely halophilic, but not archaeal: the physiology and ecology of Salinibacter ruber, a bacterium isolated from saltern crystallizer ponds, p 63-76. In A. Ventosa (ed.), Halophilic microorganisms. Springer-Verlag, Berlin, Germany.
- 24.Quinn, P. J., A. P. R. Brain, L. C. Stewart, and M. Kates. 1986. The structure of membrane lipids of the extreme halophile, Halobacterium cutirubrum, in aqueous systems studied by freeze-fracture. Biochim. Biophys. Acta 863:213-223. [Google Scholar]
- 25.Russell, N. J. 1993. Lipids of halophilic and halotolerant microorganisms, p 163-210. In R. H. Vreeland and L. I. Hochstein (ed.), The biology of halophilic bacteria. CRC Press, Boca Raton, Fla.
- 26.White, R. H. 1984. Biosynthesis of the sulfonolipid 2-amino-3-hydroxy-15-methylexadecane-1-sulfonic acid in the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 159:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]