Abstract
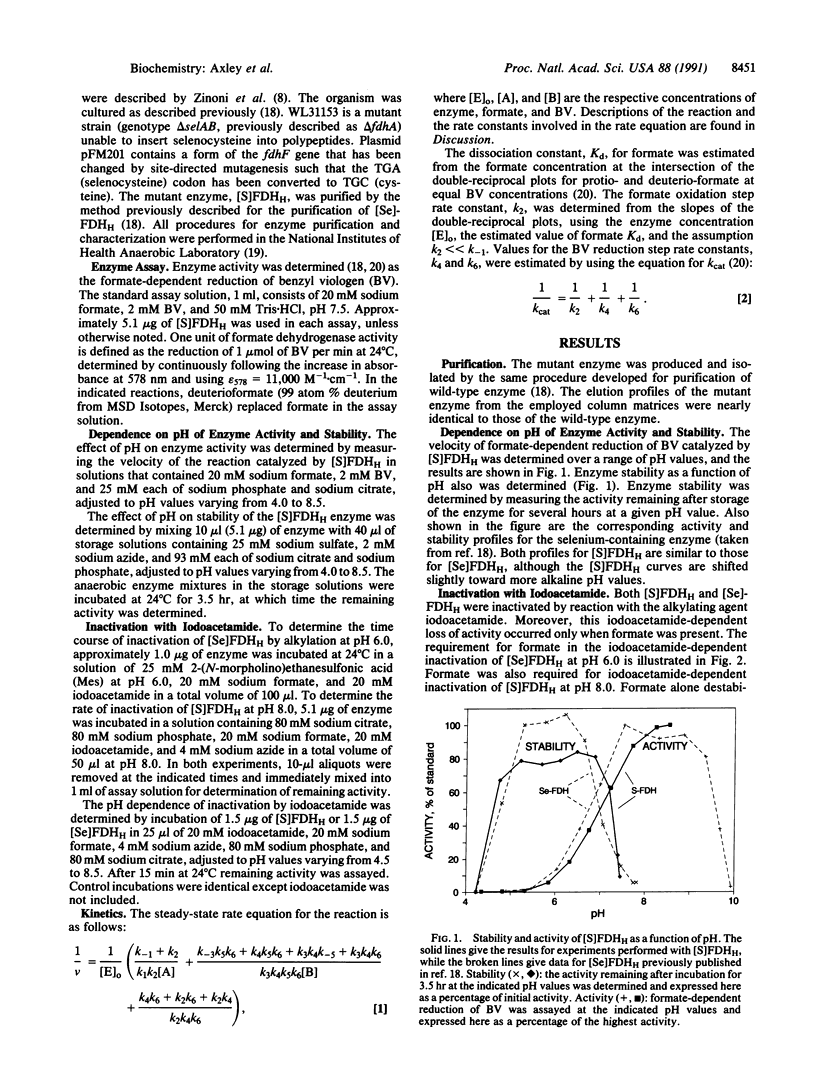
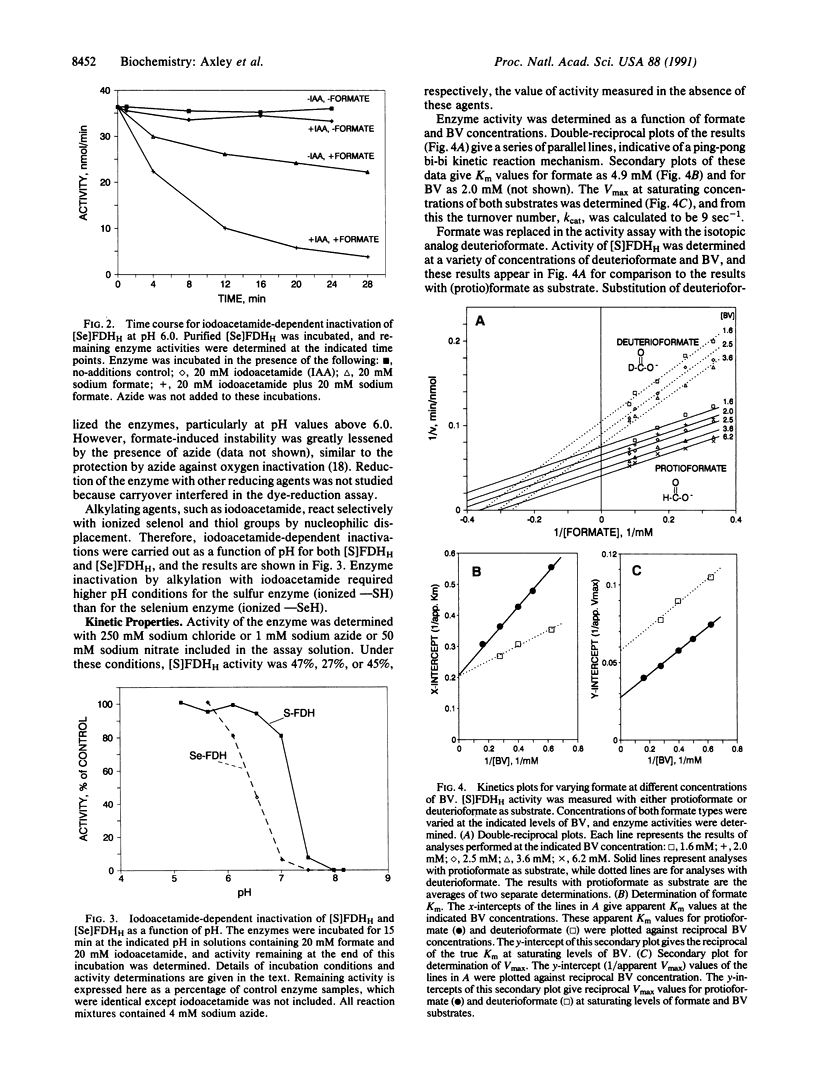
Formate dehydrogenase H of Escherichia coli contains selenocysteine as an integral amino acid. We have purified a mutant form of the enzyme in which cysteine replaces selenocysteine. To elucidate the essential catalytic role of selenocysteine, kinetic and physical properties of the mutant enzyme were compared with those of wild type. The mutant and wild-type enzymes displayed similar pH dependencies with respect to activity and stability, although the mutant enzyme profiles were slightly shifted to more alkaline pH. Both enzymes were inactivated by reaction with iodoacetamide; however, addition of the substrate, formate, was necessary to render the enzymes susceptible to alkylation. Alkylation-induced inactivation was highly dependent on pH, with each enzyme displaying an alkylation vs. pH profile suggestive of an essential selenol or thiol. Both forms of the enzyme use a ping-pong bi-bi kinetic mechanism. The mutant enzyme binds formate with greater affinity than does the wild-type enzyme, as shown by reduced values of Km and Kd. However, the mutant enzyme has a turnover number which is more than two orders of magnitude lower than that of the native selenium-containing enzyme. The lower turnover number results from a diminished reaction rate for the initial step of the overall reaction, as found in kinetic analyses that employed the alternative substrate deuterioformate. These results indicate that the selenium of formate dehydrogenase H is directly involved in formate oxidation. The observed differences in kinetic properties may help explain the evolutionary conservation of selenocysteine at the enzyme's active site.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axley M. J., Grahame D. A. Kinetics for formate dehydrogenase of Escherichia coli formate-hydrogenlyase. J Biol Chem. 1991 Jul 25;266(21):13731–13736. [PubMed] [Google Scholar]
- Axley M. J., Grahame D. A., Stadtman T. C. Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase component. J Biol Chem. 1990 Oct 25;265(30):18213–18218. [PubMed] [Google Scholar]
- Axley M. J., Stadtman T. C. Selenium metabolism and selenium-dependent enzymes in microorganisms. Annu Rev Nutr. 1989;9:127–137. doi: 10.1146/annurev.nu.09.070189.001015. [DOI] [PubMed] [Google Scholar]
- Berry M. J., Banu L., Larsen P. R. Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature. 1991 Jan 31;349(6308):438–440. doi: 10.1038/349438a0. [DOI] [PubMed] [Google Scholar]
- Böck A., Stadtman T. C. Selenocysteine, a highly specific component of certain enzymes, is incorporated by a UGA-directed co-translational mechanism. Biofactors. 1988 Oct;1(3):245–250. [PubMed] [Google Scholar]
- Chambers I., Frampton J., Goldfarb P., Affara N., McBain W., Harrison P. R. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the 'termination' codon, TGA. EMBO J. 1986 Jun;5(6):1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone J. E., Del Río R. M., Davis J. N., Stadtman T. C. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2659–2663. doi: 10.1073/pnas.73.8.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstrom J. W., Zakowski J. J., Tappel A. L. Identification of the catalytic site of rat liver glutathione peroxidase as selenocysteine. Biochemistry. 1978 Jun 27;17(13):2639–2644. doi: 10.1021/bi00606a028. [DOI] [PubMed] [Google Scholar]
- Huber R. E., Criddle R. S. Comparison of the chemical properties of selenocysteine and selenocystine with their sulfur analogs. Arch Biochem Biophys. 1967 Oct;122(1):164–173. doi: 10.1016/0003-9861(67)90136-1. [DOI] [PubMed] [Google Scholar]
- Leinfelder W., Forchhammer K., Zinoni F., Sawers G., Mandrand-Berthelot M. A., Böck A. Escherichia coli genes whose products are involved in selenium metabolism. J Bacteriol. 1988 Feb;170(2):540–546. doi: 10.1128/jb.170.2.540-546.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinfelder W., Zehelein E., Mandrand-Berthelot M. A., Böck A. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988 Feb 25;331(6158):723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- Oikawa T., Esaki N., Tanaka H., Soda K. Metalloselenonein, the selenium analogue of metallothionein: synthesis and characterization of its complex with copper ions. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3057–3059. doi: 10.1073/pnas.88.8.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PECK H. D., Jr, GEST H. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J Bacteriol. 1957 Jun;73(6):706–721. doi: 10.1128/jb.73.6.706-721.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman T. C. Selenium biochemistry. Annu Rev Biochem. 1990;59:111–127. doi: 10.1146/annurev.bi.59.070190.000551. [DOI] [PubMed] [Google Scholar]
- Stephenson M., Stickland L. H. Hydrogenlyases: Bacterial enzymes liberating molecular hydrogen. Biochem J. 1932;26(3):712–724. doi: 10.1042/bj0260712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söll D. Genetic code: enter a new amino acid. Nature. 1988 Feb 25;331(6158):662–663. doi: 10.1038/331662a0. [DOI] [PubMed] [Google Scholar]
- Young P. A., Kaiser I. I. Aminoacylation of Escherichia coli cysteine tRNA by selenocysteine. Arch Biochem Biophys. 1975 Dec;171(2):483–489. doi: 10.1016/0003-9861(75)90057-0. [DOI] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Leinfelder W., Böck A. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc Natl Acad Sci U S A. 1987 May;84(10):3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Stadtman T. C., Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Heider J., Böck A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]



