Abstract
Purpose
The objective of this work was to explore the involvement of transmembrane domain (TM) 7 of the human apical sodium-dependent bile acid transporter (hASBT) on bile acid (BA) binding/translocation, using two electrophilic BA derivatives as molecular probes.
Methods
Two electrophilic derivatives of chenodeoxycholic acid (CDCA) were designed, synthesized and evaluated for their ability to inactivate hASBT, and the human organic cation/carnitine transporter (hOCTN2) as a control (i.e. a non-BA transporting model). The ability of electrophilic derivatives to interact with hASBT was evaluated by MTSEA-biotin labeling of TM7 cysteine mutants.
Results
Unlike native BA, the electrophilic CDCA derivatives specifically inactivated hASBT, but not hOCTN2, and inhibited hASBT in a time- and concentration-dependent fashion. Preincubation of hASBT Cys-mutants in the exofacial half of TM7 with reactive electrophilic probes blocked transporter biotinylation by MTSEA-biotin, similar to MTSET blocking. This blocking pattern differed from that produced by native BAs, which exposed exofacial TM7 residues, thereby increasing staining.
Conclusion
Kinetic and biochemical data indicate these novel electrophilic BAs are potent and specific irreversible inhibitors of hASBT and offer new evidence about the role of TM7 in binding/translocation of bile acids.
INTRODUCTION
The human apical sodium-dependent bile acid transporter (hASBT; SLC10A2) is a 348 amino acid protein with a molecular weight of 43 kDa in its fully glycosylated form (1, 2). Its physiological function as a solute symporter is characterized by effectively coupling sodium to bile acid translocation with an approximate 2:1 stoichiometry (3). hASBT is a burgeoning pharmaceutical target owing to its central role in cholesterol homeostasis and is primarily expressed in the terminal ileum, kidneys and cholangiocytes (4). Despite the recent crystallization of a prokaryotic ASBT homologue (5), mechanistic understanding at the molecular level of substrate binding and translocation by mammalian ASBT is hindered by the absence of high-resolution structural data. Nonetheless, recent biochemical and biophysical studies by our group on hASBT structure/function support a seven transmembrane domain (TM) topology (2, 6) and reveal a critical role of amino acid residues in TM7 (7) during bile acid binding and translocation events.
Substrate-like probes that interact irreversibly with proteins may provide unique mechanistic insights into substrate-transporter binding and translocation. For example, Kramer and colleagues (8, 9) synthesized photoreactive derivatives of taurocholic acid (TCA) to demonstrate that the bile acid binding site of rabbit ASBT was restricted to the C-terminal portion of the protein. However, this approach relied on 7-azo derivatives which, upon activation with light, generate highly reactive carbene, that can react non-specifically with ASBT residues via nucleophilic, electrophilic, and free radical mechanisms. The present work aimed to apply electrophilic CDCA derivatives, which may interact with ASBT protein through a specific and more controlled reaction, as molecular probes to further understand hASBT function. First, we designed 3β-chloro- and 7α-mesyl derivatives of CDCA to assess their potential as irreversible inhibitors of hASBT. We hypothesized that an electrophilic carbon could be selectively attacked by nucleophilic amino acid residues within the binding site of hASBT, thereby forming covalent bonds that would inactivate the transporter. To the best of our knowledge, such an alkylating approach to elucidate transporter function has not been reported previously. Functional assay data, involving time- and concentration-dependent kinetic studies indicate that electrophilic CDCA derivatives selectively and irreversibly inhibit hASBT.
We next aimed to employ electrophilic bile acid derivates to further examine the reported role of TM7 amino acid residues in bile acid binding and translocation events. We have previously shown that exofacial residues within TM7 (Phe287-Gln297) are most sensitive to modification by methanethiosulfonate (MTS) reagents (7). Since these molecules are also electrophilic in nature, we hypothesized that bile acids bearing electron-withdrawing substituents would display similar reactivity patterns. To test this hypothesis we performed a series of biochemical studies to test whether electrophilic bile acid analogs can bind to ASBT and react with nucleophilic cysteine residues engineered within the binding site. Results from these studies offer novel mechanistic insights regarding the role of TM7 in binding and/or translocation of bile acids via hASBT protein.
MATERIALS AND METHODS
Materials
[3H]-Taurocholic acid (10 μCi/mmol), and [3H]-L-carnitine (66 μCi/mmol) were purchased from American Radiolabeled Chemicals, Inc, (St. Louis, MO). Taurocholic acid (TCA), glyco-chenodeoxycholic acid (GCDCA), and glyco-deoxycholic acid (GDCA) were obtained from Sigma Aldrich (St. Louis, MO). Glyco-ursodeoxycholic acid (GUDCA) was purchased from Calbiochem (San Diego, CA). Chenodeoxycholate (CDCA) was obtained from TCI America (Portland, OR). [2-(trimethylammonium)ethyl]-methanethio-sulfonate (MTSET) and 2-((biotinoyl)amino)-ethyl-methanethiosulfonate (MTSEA-biotin) were acquired from Toronto Research Chemicals, Inc, (North York, ON, Canada). Geneticin®, fetal bovine serum (FBS), trypsin, and DMEM were purchased from Invitrogen (Rockville, MD). All other reagents and chemicals were of the highest purity commercially available.
Synthesis of electrophilic CDCA derivatives
The synthesis of the electrophilic bile acids 3β-chloro-7α-hydroxy-5β-cholan-24-oic acid (3β-Cl-CDCA) and 3α-hydroxy-7α-mesyloxy-5β-cholan-24-oic acid (7α-Ms-CDCA) as described in the Supplementary Material section (Schemes 1 and 2, respectively). Identities of electrophilic derivatives were confirmed 1D 1H NMR and 13C NMR spectra recorded with a Varian Inova 500 MHz (Varian Inc., Palo Alto, CA) (Supplemental Material, Figs. S1 and S2, panels A and B).
Cell culture and transient transfection
Stably transfected hASBT-MDCK and hOCTN2-MDCK cells were cultured as previously described (10). Briefly, cells were grown at 37 °C, 90% relative humidity, 5% CO2 atmosphere and media replenished every other day. Culture media consisted of Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, 50 units/mL penicillin, and 50 μg/mL streptomycin. Geneticin® was added at 1 mg/mL to maintain selection pressure. Cells were passaged after reaching 90% confluency. COS-1 cells (ATCC CRL-1650) were maintained in DMEM containing 10% fetal calf serum, 4.5g/L glucose, 100 units/ml penicillin and 100 μg/ml streptomycin (Life Technologies, Inc, Rockville, MD) at 37 °C in a humidified atmosphere with 5% CO2. Transient transfections were performed as previously described (2, 7).
Co-incubation studies
To determine whether 3β-Cl-CDCA or 7α-Ms-CDCA inhibit the uptake of endogenous hASBT substrates, cis-inhibition studies that assess the uptake of radiolabeled TCA in the presence of varying inhibitor concentrations were conducted. Stably transfected hASBT-MDCK cells were grown on 12-well plates (3.8 cm2, Corning, Corning, NY) as described above. Briefly, cells were seeded at a density of 1.5×106 cells/well and induced with 10 mM sodium butyrate 12–15 h at 37 °C prior to study on day 4. Cells were washed thrice with Hank’s balanced salt solution (HBSS) prior to co-incubation with inhibitors. Cells were exposed to donor solutions containing 2.5 μM TCA with trace [3H]-TCA (0.5 μCi/ml), along with inhibitor (1–200 μM) at 37 °C and 50 rpm. After 10 min, donor solution was removed. Cells were washed three times with chilled sodium-free buffer, lysed, and counted for internalized radiolabeled TCA.
Mechanistic inhibition studies
To determine the mechanism and specificity by which electrophilic bile acids inhibit hASBT, studies were conducted to assess time-dependent transport inhibition in both hASBT and hOCTN2-transfected cell lines. hOCTN2 was selected as a representative sodium-dependent transporter protein in the SoLute Carrier (SLC) superfamily that is predominantly expressed in the small intestine (11). hOCTN2 was further selected since it was not expected to translocate TCA. Briefly, stably transfected hASBT-MDCK or hOCTN2-MDCK cells were grown, seeded, and induced as described above. Cells were washed thrice with HBSS prior to pretreatment (0–20 min) with inhibitor (i.e. either 3β-Cl-CDCA, 7α-Ms-CDCA or GCDCA) in HBSS at concentrations ranging from 0–50 μM at 37 °C, 50 rpm stirring rate (n=3). Pre-incubation was terminated by washing the cell monolayer thrice with HBSS at 37 °C. Subsequently, cells were assayed for residual hASBT or hOCTN2 activity by measuring their ability to transport TCA or L-carnitine, respectively. Uptake buffer consisted of HBSS, which contains 137 mM NaCl (pH 6.8). Remaining hASBT activity was assessed by measuring TCA uptake at 200 μM (spiked with 0.5 μCi/mL of [3H]-TCA) for 10 min, whereas remaining hOCTN2 activity was determined by monitoring the uptake of L-carnitine (200 μM spiked with 0.5 μCi/mL of [3H]-L-carnitine). Since both hASBT and hOCTN2 are sodium dependent, studies using sodium-free buffer allowed for the measurement of passive permeability (10, 11). Pre-incubation studies in the absence of sodium showed identical results to sodium-present studies (data not shown). At the end of the assay, active uptake was stopped by washing the cells thrice with chilled sodium-free buffer. Cells were then lysed with 0.25 mL of 1 N NaOH overnight, allowing for complete evaporation, and reconstituted with 0.50 mL of 0.5 N HCl. Cell lysate was counted for associated radioactivity using an LS6500 liquid scintillation counter (Beckmann Instruments, Inc., Fullerton, CA). hASBT (or hOCTN2) transport activity after pre-incubation was analyzed using a variety of kinetic models, including one for irreversible inhibition that was adapted from the classical Kitz-Wilson model (12) (derived in Appendix 1, Supplementary Material).
Inhibition of C270A hASBT mutant
Site-directed mutations were incorporated into hASBT cDNA as described previously (7). Cysteine reactivity studies assessed the specificity of 3β-Cl-CDCA and 7α-Ms-CDCA for Cys-270 in hASBT. The thiol modifying-agent MTSET was included as positive control. COS-1 cells transiently transfected with either native hASBT protein or hASBT C270A mutant were pre-incubated with each CDCA derivative (50 μM) for 10 min at RT with shaking (50 rpm), followed by three washes with modified HBSS (MHBSS; Ca2+, Mg2+, and phenol red free). Next, cells were incubated in MHBSS, pH 6.8 uptake buffer containing 200 μM (spiked with 5.0 μM [3H]-TCA) at 37 °C for 12 min (n=3). This uptake period ensures linear steady-state kinetics in conjunction with an optimal signal-to-noise ratio for subsequent analysis via liquid scintillation counting (13). Uptake was halted by a series of washes with ice-cold Dulbecco’s phosphate buffered saline (DPBS), pH 7.4 containing 0.2% fatty acid free bovine serum albumin (BSA) and 0.5 mM TCA. Cells were lysed in 350μl of 1N NaOH and subjected to liquid scintillation counting using an LS6500 liquid scintillation counter (Beckmann Coulter, Inc., Fullerton, CA) and total protein quantification using the Bradford protein assay (Bio-Rad, Hercules, CA).
MTSEA-biotin labeling studies of cysteine mutants in TM7
In order to determine the potential involvement of TM7 in the recognition of electrophilic CDCA derivatives by hASBT, residues in the extracellular domain of TM7 were mutated to cysteines using hASBT C270A as scaffold. COS-1 cells transiently expressing each mutant were washed twice with MHBSS pH 7.4, and incubated with 0.25 mL MTSEA-biotin (500 μM) prepared in MHBSS, pH 7.4 for 30 min at RT with stirring. For studies evaluating the effect of CDCA-derived irreversible inhibitors on biotin labeling, cells were pre-incubated with each compound (50 μM) in the presence of sodium for 10 min at RT with shaking, washed twice in MHBSS, pH 7.4 buffer followed by MTSEA-biotin labeling as described above. A similar protocol was used for the positive control MTSET, wherein cells were first pre-incubated with MTSET (500 μM) prepared in MHBSS, pH 7.4 for 10 min at RT, then washed twice with MHBSS, pH 7.4 buffer followed by MTSEA-biotin labeling. For studies evaluating the effects of native bile acids GDCA and GUDCA on labeling, MTSEA-biotin (500 μM) was co-incubated in the presence of each bile acid (250 μM) for 30 min at RT with stirring. GDCA, and GUDCA were selected as representative substrates since they: 1) are high affinity hASBT inhibitors (Ki range from 1–3.5 μM); 2) represent different hydroxylation patterns; 3) display low passive permeability (14); and 4) do not exhibit time-dependent inhibition of hASBT. For each experiment, a 200 mM stock solution of MTSEA-biotin was prepared in DMSO and kept cold and dark until appropriately diluted with MHBSS buffer just before use. Following labeling, cells were washed three times with PBSCM containing 100 mM glycine and lysed, after which biotinylated proteins were recovered overnight at 4 °C using 50 μl of streptavidin agarose beads. Biotinylated proteins were resuspended in 30 μL SDS-PAGE buffer and boiled for 5 min, followed by separation on a 12.5% SDS-polyacrylamide gel and transferred onto an Immuno-Blot PVDF membrane (Bio-Rad, Hercules, CA). Blots were probed with rabbit anti-ASBT primary antibody (1:1000) and visualized using goat anti-rabbit IgG/HRP conjugated secondary antibody (1:15,000) with chemiluminescent detection (ECL Plus Western Blot kit, Amersham Biosciences, Piscataway, NJ).
Data analysis
Remaining hASBT activity data were analyzed by fitting TCA uptake into hASBT-MDCK monolayer after pre-incubation using
| (Eq. 1) |
where J is remaining TCA flux after pre-incubation (i.e. remaining hASBT activity), J0 is TCA flux without pre-incubation, k3 is inactivation rate, Ki is binding affinity, I is the irreversible inhibitor concentration, and t is pre-incubation time (i.e. pre-incubation duration). Appendix 1 (Supplemental Material) shows the derivation of eqn 1, which is a new form of the Kitz and Wilson model for irreversible inhibition (12). The active component of TCA flux versus time each concentration of inhibitor were fitted to eqn 1 in WinNonlin 5.2 (Pharsight, Mountain View, CA) using nonlinear regression to obtain global parameters k3 (inactivation rate, min−1) and Ki (binding affinity, μM).
Additionally, the traditional data linearization and analysis procedure of the model was preformed, consisting on the double reciprocal plot 1/Kapp versus [I] −1 (Eqn 2). In this analysis, an irreversible inhibitor will show an intercept different from zero, provided specific recognition of the inhibitor by the protein (i.e. hASBT transporter) prior to inactivation.
| (Eq. 2) |
Statistical analysis
Data analysis was performed with GraphPad Prism 4.0 (GraphPad Software, San Diego, CA) using Student’s t-test. Data were considered statistically significant at p ≤ 0.05.
RESULTS
Co-incubation studies
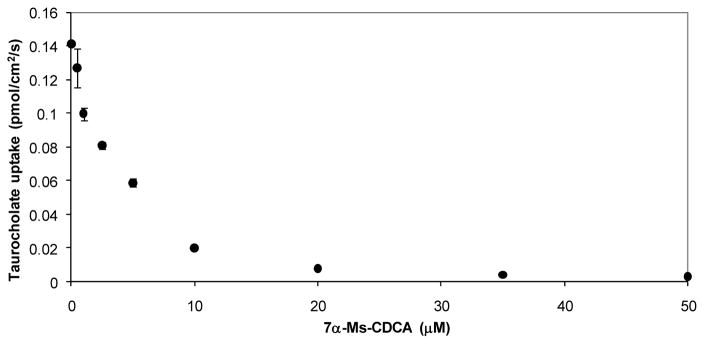
3β-Cl-CDCA and 7α-Ms-CDCA were found to be potent inhibitors of hASBT as judged by the strong inhibition of TCA uptake. Results from these preliminary experiments showed that nearly complete inhibition of hASBT-mediated [3H]-TCA uptake was obtained by 20 μM of 7α-Ms-CDCA (Fig. 1) and 50 μM of 3β-Cl-CDCA (Fig. S3, Supplementary Material).
Figure 1.
7α-Ms-CDCA inhibition of TCA uptake into hASBT-MDCK monolayers. Inhibition profile was obtained by co-incubation TCA (2.5 μM) with 7α-Ms-CDCA for 10 min (n=3). Similar plot was obtained for 3β-Cl-CDCA (Fig. S3, Supplementary Material).
Mechanistic inhibition studies
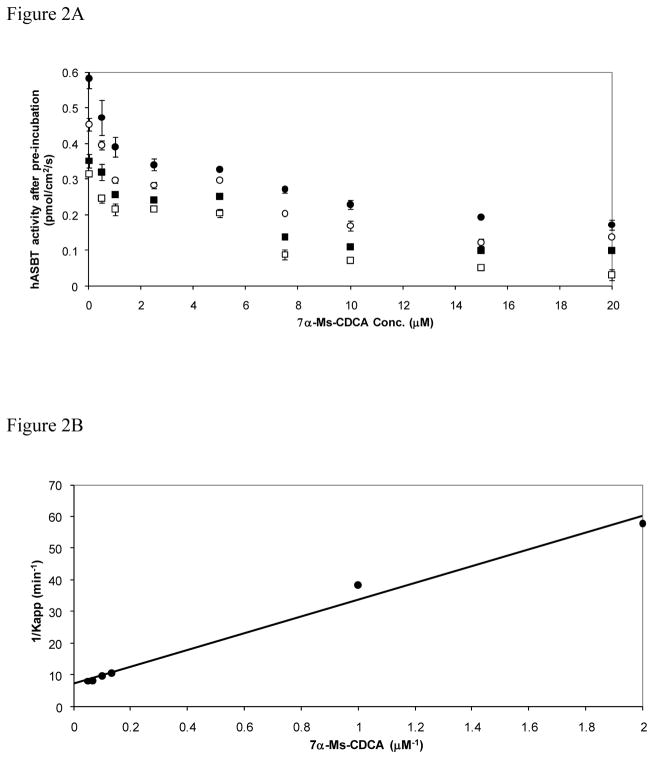
In order to evaluate electrophilic CDCA derivatives as potential irreversible inhibitors of hASBT, pre-incubation experiments were performed to assess for time-dependent inhibition, which is a hallmark feature of irreversible inhibitors (15). hASBT-MDCK monolayers were pre-incubated with increasing concentrations of 3β-Cl-CDCA or 7α-Ms-CDCA for various durations. Figure 2A depicts the remaining hASBT activity after pre-incubation with 7α-Ms-CDCA. A similar plot for 3β-Cl-CDCA pre-incubation can be found in the Supplementary Material section (Fig. S4A). 7α-Ms-CDCA inhibition of hASBT increased for longer pre-incubation times (Fig. 2A), consistent with irreversible inactivation of the transporter as the inhibitory mechanism. Moreover, hASBT activity did not return to basal levels after several washes, supporting the hypothesis that electrophilic CDCA derivatives form a stable adduct with the transporter, either via a covalent or non-covalent association between these bile acid derivatives and hASBT.
Figure 2.
Remaining hASBT activity after pre-incubation with 7α–Ms-CDCA. hASBT-MDCK monolayers were pre-incubated with 7α-Ms-CDCA (0–20 μM) for various durations. Pre-incubation durations were 3 min (closed circles), 6 min (open circles), 12 min (closed squares), and 20 min (open squares). Remaining hASBT activity was measured by TCA uptake (200 μM). Pre-incubation with 7α-Ms-CDCA inhibited hASBT in a concentration- and time-dependent fashion (Panel A). Similar plot was constructed for 3β-Cl-CDCA (Fig. S4.A, Supplementary Material). Panel B: Kitz and Wilson plot of 7α-Ms-CDCA inactivation of hASBT. Remaining hASBT activity after pre-incubation with 7α-Ms-CDCA was subjected to classical Kitz and Wilson model analysis. Reaction rate k3 was 0.136 min−1 and inhibition constant Ki was 3.59 μM (r2 = 0.99). k3 and Ki for 3β-Cl-CDCA were 0.400 min−1 and 2.17 μM, respectively (r2 = 0.99; Fig. S4.B, Supplementary Material).
Data analysis of remaining hASBT activity after pre-incubation with CDCA-derived electrophilic bile acids was first performed using the classical procedure described by Kitz and Wilson (12). This model describes the irreversible inactivation of a transporter by a reactive inhibitor as a two-step process where 1) the inactivator binds reversibly to the active site of the transporter with affinity Ki and; 2) the inactivator “reacts” with residues in the binding site leading to formation of a bound inhibitor-transporter complex that is inactive. The inactivation rate is characterized by k3 (Eqn 2). Figure 2B shows the double-reciprocal plot for 7α-Ms-CDCA (similar plots for 3β-Cl-CDCA in Fig. S4 panels A and B, Supplementary Material). An intercept different from zero suggests that inactivation of hASBT by electrophilic CDCA derivatives occurs only after specific recognition by the transporter. Classical data analysis (double-reciprocal plot) yielded k3 values of 0.399 and 0.136 min−1 for 3β-Cl-CDCA and 7α-Ms-CDCA, respectively; estimated Ki values were 2.17 μM and 3.59 μM for 3β-Cl-CDCA and 7α-Ms-CDCA, respectively. Of note, this irreversible inhibition model is not different from the Michaelis-Menten transport model for a substrate. The difference lies in that the translocation step (k2) in the Michaelis-Menten model is replaced by the inactivation rate k3 in the irreversible inhibition model.
Motivated by the apparent applicability of the Kitz and Wilson model, we revisited the derivation of this model due to the originally made assumptions. Appendix 1 (Supplemental Material) shows the derivation of the irreversible inhibition model, including a form amenable to nonlinear regression. Unlike the original Kitz and Wilson derivation, the present derivation makes no assumptions, other than those already inherent in the Michaelis-Menten model. Additionally, the use of nonlinear form of the irreversible inhibition model allows for all data points to be used in model analysis, and does not require linearization of data. Remaining hASBT activity data after pre-incubation was fitted to Eqn 1 by using nonlinear regression. Parameters Ki and k3 for 3β-Cl-CDCA and 7α-Ms-CDCA are shown in Table I. Values in Table I are in good agreement with those obtained by the classical data analysis.
Table I.
hASBT inactivation rate and inhibitory affinity of 3β-Cl-CDCA and 7β-Ms-CDCA obtained from Eqn 1.
| Parameter | 3β-Cl-CDCA | 7α-Ms-CDCA |
|---|---|---|
| k3 (min−1) | 0.39 ± 0.03 | 0.23 ± 0.01 |
| Ki (μM) | 0.81 ± 0.19 | 1.19 ± 0.06 |
hASBT specificity: electrophilic bile acids do not inhibit hOCTN2 transport activity
In order to assess the specificity of CDCA-derived irreversible inhibitors towards hASBT, pre-incubation experiments were performed on MDCK monolayers stably transfected with the organic cation transporter hOCTN2. hOCTN2 was chosen as a model transporter to address specificity of electrophilic CDCA derivatives since: 1) hOCTN2 is, like hASBT, a sodium-dependent transporter; 2) L-carnitine displays high affinity for hOCTN2 (Kt ≈ 5 μM) (16), almost identical to TCA affinity for hASBT (5.03 μM) (14); 3) hOCTN2 presents nucleophilic amino acid residues in transmembrane regions and extracellular loops that could serve as reactive sites (29 serines, 7 cysteines, 3 lysines, and 20 threonines) (11, 16); 4) both transporters are stably transfected in the same cell line, facilitating interpretation of the data; 5) both systems showed saturation of active transport at 200 μM.; and 6) TCA does not inhibit L-carnitine transport by hOCTN2 (data not shown).
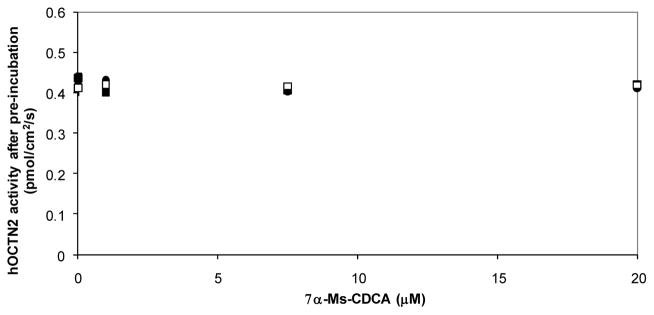
Remaining hOCTN2 activity after pre-incubation with 7α-Ms-CDCA suggests that hOCTN2 was not inhibited by electrophilic CDCA derivatives (Fig. 3), regardless of concentration and pre-incubation time (analogous plot for 3β-Cl-CDCA in Fig. S5, Supplementary Material). This result further suggests that hASBT inactivation by CDCA-derived irreversible inhibitors is the result of a highly specific interaction between these compounds and hASBT.
Figure 3.
Remaining hOCTN2 activity after pre-incubation with 7α-Ms-CDCA. Stably-transfected hOCTN2-MDCK monolayers were pre-incubated with 7α-Ms-CDCA (0–20 μM). Pre-incubation durations were 3 min (closed circles), 6 min (closed squares), and 12 min (open squares). Remaining hASBT activity was measured by L-carnitine uptake (200 μM). 7α-Ms-CDCA did not inhibit hOCTN2 (p > 0.20), regardless of pre-incubation concentration and duration. Essentially the same plot was obtained for 3β-Cl-CDCA (Fig. S5, Supplementary Material).
Inhibition mechanism of electrophilic bile acids compared to native bile acid
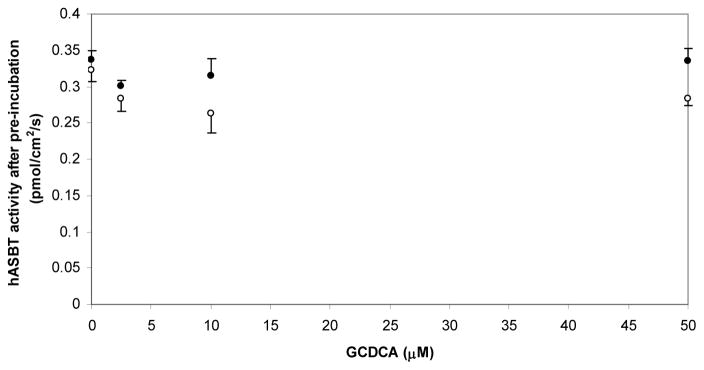
To further explore the mechanistic role of the electrophilic substituents on CDCA derivatives, identical pre-incubation and assessment of activity was performed using glyco-chenodeoxycholate (GCDCA) as inhibitor. GCDCA was chosen as a model reversible inhibitor since it is actively transported by hASBT with high affinity (Kt < 1 μM), and also serves as a potent inhibitor (Ki < 1 μM) (14). GCDCA pre-incubation in the presence of electrophilic bile acid inhibitors did not decrease hASBT activity compared to buffer pre-incubation, regardless of duration of pre-incubation and concentration (p > 0.05, Fig. 4), suggesting that the mechanism of inhibition of electrophilic CDCA derivatives is different from GCDCA, a known potent inhibitor and substrate.
Figure 4.
Remaining hASBT activity after pre-incubation with the native bile acid GCDCA (0–50 μM). Pre-incubation durations were 3 min (closed circles) and 6 min (open circles). GCDCA did not inactivate/inhibit hASBT after pre-incubation regardless of pre-incubation duration and concentration (p > 0.05).
MTSEA-biotin labeling studies of cysteine mutants in TM7
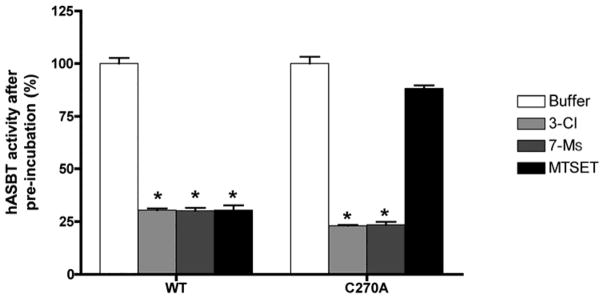
To determine whether electrophilic bile acids specifically recognize hABST amino acids previously shown to interact with native bile acids, we first studied the potential involvement of Cys270 in reaction with CDCA-derived irreversible inhibitors. Our previous studies have shown that Cys270, located in the extracellular loop 3 of hASBT, is the most solvent-accessible and therefore, most reactive cysteine in hASBT (13). Wild-type and C270A hASBT were transiently transfected in COS-1 cells and pre-incubated with buffer, 3β-Cl-CDCA or 7α-Ms-CDCA and the membrane-impermeable thiol-modifying agent MTSET (positive control). Results (Fig. 5) demonstrate that electrophilic compounds and MTSET both inactivate wt-hASBT. Interestingly, the C270A-hASBT construct, here and previously (13) shown to be insensitive to MTS modification, is significantly inhibited by electrophilic CDCA derivatives, suggesting that these analogues do not specifically target native Cys residues.
Figure 5.
COS-1 were transfected with 200 ng/ml DNA encoding wt or C270A hASBT and pre-incubated for 10 min with buffer, 3β-Cl-CDCA (50 μM), 7α-Ms-CDCA (50 μM), or the thiol modifying-agent MTSET (1mM). Remaining hASBT activity was measure by TCA uptake. Unlike MTSET, electrophilic CDCA derivatives were able to inactivate both wt and C270A hASBT (n=3, p < 0.01).
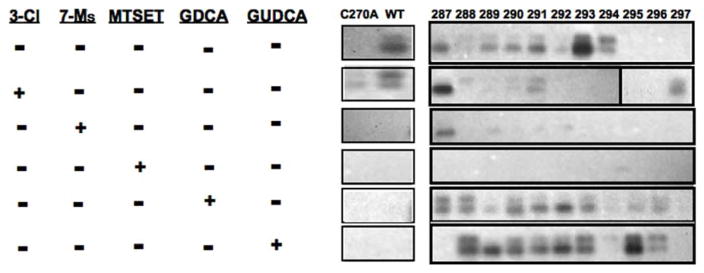
A putative hydrophilic cleft lining the exofacial half of TM7 has been identified for ASBT protein (7), extending from the origin of the TM (Phe287) to approximately the midpoint of the helix (Gln297). Based on this it was inferred that bile acids likely interact or bind along this TM7 region (5). Since 3β-Cl-CDCA and 7α-Ms-CDCA represent a minor chemical modification from the original substrate, we hypothesized they interact with ASBT protein in a similar manner, i.e. interact with this hydrophilic cleft region along the exofacial half of TM7. To evaluate this possibility, we used an MTSEA-biotin derivative, which is electroneutral and enables visualization of thiol modification events using streptavidin capture of the biotin-protein complex for subsequent immunoblotting. hASBT incubation with MTSEA-biotin (500 μM) alone produces visually evident labeling of cysteine residues along the exofacial half of the TM7 helix (Fig. 6, lane 1), indicative of a hydrophilic solvent accessible region. Pre-incubation with the positively charged and membrane-impermeable MTSET (500 μM, control) followed by MTSEA-biotin incubation blocks biotinylation (Fig. 6, lane 4), suggesting that these physicochemically different thiol modifiers can similarly access this hydrophilic cleft. Pre-incubation with the electrophilic CDCA derivates (50 μM) followed by biotin labeling produces essentially similar protection as compared to MTSET pre-incubation, suggesting that these CDCA-derived irreversible inhibitors also access the TM7 cleft region. However, co-incubation of MTSEA-biotin with native bile acids GDCA and GUDCA (250 μM) appears to increase biotin labeling of TM7 regions.
Figure 6. Western blot from MTSEA-biotin labeling studies of cysteine mutants in TM7.
MTSEA-biotin was reactive towards cysteines, particularly hASBT’s Cys-270, which is solvent-accessible. hASBT or mutants were labeled with MTSEA-biotin, before (line 1) and after pre-incubation with 3β-Cl-CDCA (line 2), 7α-Ms-CDCA (line 3), MTSET (line 4), or native bile acids GDCA (line 5) or GUDCA (line 6). Results from 3β-Cl-CDCA or 7α-Ms-CDCA pre-incubations were similar to MTSET pre-incubation, reducing biotinylation, suggesting that these chlorinated bile acids react with cysteines and limit any subsequent MTSEA-biotin labeling. In particular, the chlorinated bile acids reduced biotinylation of C270A mutants with cysteines introduced in TM7. Meanwhile, the native bile acids GDCA and GUDCA generally increased biotinylation of TM7 via alternating access, rather than inhibiting biotinylation.
DISCUSSION
Substrate-like probes that act as irreversible inhibitors can provide important new mechanistic insights into substrate-protein interactions. Whereas, the application of substrate analogues as irreversible inhibitors has been applied previously in the enzyme research field, this approach has found limited application as a tool in transporter research. For example, Wang and colleagues (17) synthesized a series of fluorinated GABA analogs which functioned as irreversible inhibitors of GABA aminotransferase. In the present study, we selected for chloro- and mesyl- groups since they are better leaving groups compared to fluoride. As the objective of our study was to generate bile acids with enhanced residence time within the binding domain(s), we selected against fluorines since they are isosteres of hydroxyl groups, thereby increasing the chance that these compound would remain hASBT substrates.
We designed and synthesized two electrophilic bile acid derivatives where hydroxyl groups on either the 3- or 7- positions of the cholane skeleton have been replaced with electron-withdrawing groups (i.e. 3β-Cl-CDCA and 7α-Ms-CDCA). Both compounds strongly inhibited uptake of the probe substrate [3H]-TCA (Fig. 1), indicating their high affinity for the transporter. To determine inhibition mechanism, we pre-incubated hASBT-expressing cells with the natural substrate GCDCA as well as the electrophilic CDCA analogs. We observed a time- and concentration-dependent hASBT inhibition by electrophilic CDCA analogs (Fig. 2), but not GCDCA (Fig. 4). To confirm the specificity of binding to hASBT and exclude the possibility of nonspecific binding to proteins that may affect ASBT function, we performed identical studies with another sodium-dependent solute carrier, hOCTN2, which does not recognize bile acids as substrates. hOCTN2-mediated L-carnitine uptake was not affected by pre-incubation with electrophilic CDCA-derivatives (Fig. 3), thus excluding non-specific binding events from impacting hASBT function. Combined, these data strongly suggest that the electrophilic bile acid analogs synthesized in this study are irreversible inhibitors of hASBT protein.
Kramer and colleagues (8, 9) identified a potential ligand-binding site in rabbit ASBT by using photoreactive derivatives of TCA acid. Incubation of ileal brush-border vesicles with radiolabeled 7-azo-TCA (activated by UV-light), followed by enzyme digestion and blotting, revealed labeling of the last 67–56 residues in the C-terminus region. Based on a previously validated 7 transmembrane domain topology (2, 6), these residues are located in TM7 and the cytosolic C-terminus. Unlike the two electrophilic CDCA derivatives described here, 7-azo-TCA is a carbene-generating compound that can react as an electrophile, nucleophile, or radical, depending on its singlet or triplet electronic state (18). hASBT inactivation by electrophilic CDCA derivatives may involve reactions with nucleophilic residues in the ligand binding/translocation domain(s) such as cysteines, serines, threonines, and lysines. Site-directed alkylation of consecutively introduced cysteine residues can provide a powerful tool for determining the solvent-accessibility profile of specific protein regions. Since cysteines contain a thiol group, chemical modification can be performed using methanethiosulfonate (MTS) reagents. Moreover, MTS reagents react 109 times more readily with the ionized thiolate species (i.e. thiolate anion) and since the dielectric constant of aqueous, but not lipid or internal protein environment promotes formation of the thiolate anion, any subsequent modification of introduced cysteines translates into an indirect but effective means of determining the solvent-accessibility of a given protein region (19).
hASBT contains 13 Cys residues of which Cys270 located on extracellular loop 3 (EL3) is most solvent-accessible and readily alkylated by MTS reagents. In order to elucidate whether electrophilic CDCA-derivatives react with Cys270, we tested their ability to inactivate the C270A mutant and the wild-type transporter. Unlike MTS reagents the CDCA-analogs effectively reduced TCA uptake by both native and mutant transporter (Fig. 5), suggesting that the inactivation of hASBT by these bile acid analogs is not mediated by direct alkylation of native Cys270. Next, we determined whether electrophilic bile acids could react with Cys-residues engineered within a key hASBT binding region. Using a panel of experiments involving 11 exofacial hASBT-Cys mutants we determined whether MTSEA-biotin labeling could be prevented by pre-incubation with electrophilic bile acids as well as the native substrates GDCA and GUDCA. Pre-incubation with the positive control MTSET completely abolished MTSEA-biotin labeling, indicating the specificity of the Cys-mediated alkylation reaction. Compared to negative control (buffer alone), electrophilic CDCA derivatives revealed a unique labeling pattern similar to that of positive control MTSET than that of native substrates GDCA and GUDCA. Close examination of Fig. 6, reveals various distinct features. First, residue Phe287 located at the extracellular limit of TM7 was accessible to MTSEA-biotin after incubating with either buffer, the substrate GDCA, or electrophilic-CDCA derivatives, but “protected” from labeling by incubation with GUDCA. Second, the TM7-section spanning from Phe289-Ser294 was freely biotinylated after incubation with buffer or substrates but “shielded” from chemical cross-linking by both irreversible inhibitors, with the exception of Leu291 which was faintly labeled after pre-incubation with 3β-Cl-CDCA. Third, co-incubation with the 7β-hydroxylated substrate GUDCA yields a very strong labeling of Ile295 while the center of the TM (i.e. Glu297) was only labeled after pre-incubation with 3-Cl-CDCA. These results suggest that electrophilic bile salts can access and modify the same residues within hASBT mutants compared to MTSET. In contrast, native bile acids were largely unable to afford protection against MTSEA-biotin labeling when co-incubated.
Mechanism of inhibition
Increasing MTSEA reactivity in the putative cleft region by native substrates would be predicted by an alternating access mode of transport, as described by Kaback and colleagues for the Lac permease (20). But why do electrophilic bile acid derivatives behave differently? We speculate that CDCA-derived irreversible inhibitors follow a similar recognition pathway along the TM7 cleft, as compared to native bile acids. However, once situated within the cleft region, these electrophilic probes are either tightly bound to hASBT, aided by their enhanced lipophilicity, or attacked by nucleophilic residues. As an example of non-covalent binding, buprenorphine is an opioid whose greater binding affinity and longer duration of action are attributed to its greater lipophilicity, compared to morphine (21, 22). Buprenorphine is a pseudo-irreversible inhibitor of μ-opioid receptors, but not chemically amenable to forming a covalent bond with the receptor (23). Its t-butyl and the cyclopropylmethyl groups impart lipophilicity to cause pseudo-irreversibility with the receptor. Similarly, the 14-amino-dehydromorphinone derivatives, such as methoclocinnamox and clocinnamox, showed irreversible binding to μ-opioid receptors due to their greater lipophilicity (24). In particular, 4′-chloro and 4′-methyl substitution enhanced lipophilicity and irreversible binding. Alternatively, electrophilic CDCA derivatives may react chemically with hASBT protein. Intimate contact between the inactivator and hASBT could position the electrophilic carbons C-3 or C-7 within appropriate distance from nucleophilic residues in the binding site (i.e. 15 Å, approximately the distance between C-24 and C-3 or C-7 in bile acid molecule). Alkylated hASBT would be “trapped” in an inactive alkylated form, presumably with irreversible inhibitor halfway within the hydrophilic cleft defined by TM7, TM1 and TM3. It is also possible that covalent bonding between 3β-Cl-CDCA or 7α-Ms-CDCA and candidate residues occurs at alternate protein sites exposed during translocation steps downstream of initial interaction with the hydrophilic TM7 cleft. According to alternating access, the outward-facing conformation (i.e. the TM7 cleft region exposed) is followed by an intermediate conformer, wherein substrate is enclosed within the protein core with access to either side of the membrane blocked. Subsequently, the inward-facing conformation is assumed and substrate follows an exit route, presumably lining a cytosolic hydrophilic cleft region. Thus, covalent interactions between the electron-withdrawing groups on CDCA derivatives and candidate nucleophilic residues may occur along these last two steps. Even though both binding scenarios are plausible, additional experiments would be required to determine the definitive chemical nature of the interaction of ASBT with electrophilic probes.
CONCLUSION
A single-point chemical modification in the native bile acid CDCA, where the hydroxyl groups in C-3 or C-7 were substituted by a chloro or a mesyl group, transformed an otherwise high-affinity substrate into a highly potent and specific hASBT irreversible inhibitor. Based on functional and biochemical evidence, we propose electrophilic CDCA derivatives bound to TM7 with high affinity. Binding was effectively irreversible, due to either tight binding aided by derivative lipophilicity or to alkylation of hASBT as a result of electrophilic character. 3β-Cl-CDCA and 7α-Ms-CDCA are excellent biochemical tools to probe the involvement of specific amino acid residues in bile acid binding/translocation by hASBT.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from Fondo Nacional de Desarrollo Científico Tecnológico de Chile (Fondecyt No 11090199 to PMG) and by the National Institutes of Health of USA (grants DK67530 to JEP, and DK61425 to PWS).
ABBREVIATIONS
- 3β-Cl-CDCA
3β-chloro-7α-hydroxy-5β-cholan-24-oic acid
- 7α-Ms-CDCA
3α-hydroxy-7α-mesyl-5β-cholan-24-oic acid
- TM
transmembrane domain
- hASBT
human apical sodium-dependent bile acid transporter
- hOCTN2
human novel organic cation/carnitine transporter 2 (SLC22A5)
- TCA
taurocholic acid
- CDCA
chenodeoxycholic acid
- GCDCA
glyco-chenodeoxycholic acid
- GDCA
glyco-deoxycholic acid
- GUDCA
glyco-ursodeoxycholic acid
- MTSET
[2-(trimethylammonium)ethyl]-methanethio-sulfonate
- MTSEA
2-(aminoethyl)-methanethiosulfonate
- Kt
Michaelis-Menten-type constant
- Ki
inhibition constant
- k3
inactivation rate
- J
flux
References
- 1.Wong MH, Oelkers P, Craddock AL, Dawson PA. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J Biol Chem. 1994 Jan 14;269(2):1340–7. [PubMed] [Google Scholar]
- 2.Zhang EY, Phelps MA, Banerjee A, Khantwal CM, Chang C, Helsper F, et al. Topology scanning and putative three-dimensional structure of the extracellular binding domains of the apical sodium-dependent bile acid transporter (SLC10A2) Biochemistry. 2004 Sep 14;43(36):11380–92. doi: 10.1021/bi049270a. [DOI] [PubMed] [Google Scholar]
- 3.Weinman SA, Carruth MW, Dawson PA. Bile acid uptake via the human apical sodium-bile acid cotransporter is electrogenic. J Biol Chem. 1998 Dec 25;273(52):34691–5. doi: 10.1074/jbc.273.52.34691. [DOI] [PubMed] [Google Scholar]
- 4.Balakrishnan A, Polli JE. Apical sodium dependent bile acid transporter (ASBT, SLC10A2): a potential prodrug target. Mol Pharm. 2006 May-Jun;3(3):223–30. doi: 10.1021/mp060022d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu NJ, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011 Oct 20;478(7369):408–11. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee A, Swaan PW. Membrane topology of human ASBT (SLC10A2) determined by dual label epitope insertion scanning mutagenesis. New evidence for seven transmembrane domains. Biochemistry. 2006 Jan 24;45(3):943–53. doi: 10.1021/bi052202j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussainzada N, Banerjee A, Swaan PW. Transmembrane domain VII of the human apical sodium-dependent bile acid transporter ASBT (SLC10A2) lines the substrate translocation pathway. Mol Pharmacol. 2006 Nov;70(5):1565–74. doi: 10.1124/mol.106.028647. [DOI] [PubMed] [Google Scholar]
- 8.Kramer W, Nicol SB, Girbig F, Gutjahr U, Kowalewski S, Fasold H. Characterization and chemical modification of the Na(+)-dependent bile-acid transport system in brush-border membrane vesicles from rabbit ileum. Biochim Biophys Acta. 1992 Oct 19;1111(1):93–102. doi: 10.1016/0005-2736(92)90278-t. [DOI] [PubMed] [Google Scholar]
- 9.Kramer W, Girbig F, Glombik H, Corsiero D, Stengelin S, Weyland C. Identification of a ligand-binding site in the Na+/bile acid cotransporting protein from rabbit ileum. J Biol Chem. 2001 Sep 21;276(38):36020–7. doi: 10.1074/jbc.M104665200. [DOI] [PubMed] [Google Scholar]
- 10.Balakrishnan A, Sussman DJ, Polli JE. Development of stably transfected monolayer overexpressing the human apical sodium-dependent bile acid transporter (hASBT) Pharm Res. 2005 Aug;22(8):1269–80. doi: 10.1007/s11095-005-5274-8. [DOI] [PubMed] [Google Scholar]
- 11.Ramsay RR, Gandour RD, van der Leij FR. Molecular enzymology of carnitine transfer and transport. Biochim Biophys Acta. 2001 Mar 9;1546(1):21–43. doi: 10.1016/s0167-4838(01)00147-9. [DOI] [PubMed] [Google Scholar]
- 12.Kitz R, Wilson IB. Esters of methanesulfonic acid as irreversible inhibitors of acetylcholinesterase. J Biol Chem. 1962 Oct;237:3245–9. [PubMed] [Google Scholar]
- 13.Banerjee A, Ray A, Chang C, Swaan PW. Site-directed mutagenesis and use of bile acid-MTS conjugates to probe the role of cysteines in the human apical sodium-dependent bile acid transporter (SLC10A2) Biochemistry. 2005 Jun 21;44(24):8908–17. doi: 10.1021/bi050553s. [DOI] [PubMed] [Google Scholar]
- 14.Balakrishnan A, Wring SA, Polli JE. Interaction of native bile acids with human apical sodium-dependent bile acid transporter (hASBT): influence of steroidal hydroxylation pattern and C-24 conjugation. Pharm Res. 2006 Jul;23(7):1451–9. doi: 10.1007/s11095-006-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coppeland RA. Enzymes : a practical introduction to structure, mechanism, and data analysis. new York: J. Wiley; 2000. [Google Scholar]
- 16.Koepsell H, Endou H. The SLC22 drug transporter family. Pflugers Arch. 2004 Feb;447(5):666–76. doi: 10.1007/s00424-003-1089-9. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Yuan H, Nikolic D, Van Breemen RB, Silverman RB. (+/−)-(1S,2R,5S)-5-Amino-2-fluorocyclohex-3-enecarboxylic acid. A potent GABA aminotransferase inactivator that irreversibly inhibits via an elimination-aromatization pathway. Biochemistry. 2006 Dec 5;45(48):14513–22. doi: 10.1021/bi061592m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mieusset J-L, Brinker U. The carbene reactivity surface: A classification. Journal of Organic Chemistry. 2008;73(4):1153–558. doi: 10.1021/jo7026118. [DOI] [PubMed] [Google Scholar]
- 19.Karlin A, Akabas MH. Substituted-cysteine accessibility method. Methods Enzymol. 1998;293:123–45. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- 20.Kaback HR, Dunten R, Frillingos S, Venkatesan P, Kwaw I, Zhang W, et al. Site-directed alkylation and the alternating access model for LacY. Proc Natl Acad Sci U S A. 2007 Jan 9;104(2):491–4. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casy AF, Parfitt RT. Opioid analgesics: Chemistry and receptors. 1. New York: Plenum Press; 1986. [Google Scholar]
- 22.Kajiwara M, Aoki K, Ishii K, Numata H, Matsumiya T, Oka T. Agonist and antagonist actions of buprenorphine on three types of opioid receptor in isolated preparations. Japanese Journal of Pharmacology. 1986 Jan;40(1):95–101. doi: 10.1254/jjp.40.95. [DOI] [PubMed] [Google Scholar]
- 23.Husbands SM, Lewis JW. Opioid ligands having delayed long-term antagonist activity: Potential pharmacotherapies for opioid abuse. Mini Reviews in Medicinal Chemistry. 2003;3:137–44. doi: 10.2174/1389557033405395. [DOI] [PubMed] [Google Scholar]
- 24.Nieland NP, Moynihan HA, Carrington S, Broadbear J, Woods JH, Traynor JR, et al. Structural determinants of opioid activity in derivatives of 14-aminomorphinones: effect of substitution in the aromatic ring of cinnamoylaminomorphinones and codeinones. J Med Chem. 2006 Aug 24;49(17):5333–8. doi: 10.1021/jm0604777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.