Abstract
Analysis of short sequence repeats of Mycobacterium avium subsp. paratuberculosis isolated from Crohn's disease patients identified two alleles, both of which clustered with strains derived from animals with Johne's disease. Identification of a limited number of genotypes among human strains implies the existence of human disease-associated genotypes and strain sharing with animals.
Crohn's disease, a chronic inflammation of the intestine, results in substantial morbidity and medical costs in the United States (3). The etiology of Crohn's disease is complex and appears to be associated with multiple factors for its pathological presentation (9; http://www.foodstandards.gov.uk/multimedia/pdfs/mapcrohnreport.pdf; http://gpvec.unl.edu/avc/IssuePapers/out38_en.pdf). The most prevalent theory of a microbial etiology of Crohn's disease is infection with Mycobacterium avium subsp. paratuberculosis, the cause of Johne's disease in animals (8, 18; http://books.nap.edu/openbook/0309086116/html/index.html). Although several studies have associated M. avium subsp. paratuberculosis with a proportion of Crohn's disease cases (5, 19), the evidence for a causal link remains controversial.
The primary objective of this report was to study the clonal distribution and degree of diversity of M. avium subsp. paratuberculosis isolated from humans with Crohn's disease. Short sequence repeats (SSRs) have been successfully used as markers to understand the clonality and distribution of subtypes in several bacterial species (1, 7, 11). Preliminary SSR analysis of M. avium subsp. paratuberculosis isolates derived from a variety of hosts, disease types, and geographic localities suggests an association between allele types and host species (2, 14). Knowledge of strain specificity for the human host and genotypic proximity to isolates responsible for animal disease will be a critical first step in establishing a causal role for M. avium subsp. paratuberculosis in Crohn's disease.
The human M. avium subsp. paratuberculosis isolates analyzed herein included those cultured from breast milk (16) and intestinal tissues (S. A. Naser et al., unpublished data; ATCC 43015, ATCC 43544, ATCC 43545, and ATCC 49164) of patients diagnosed with Crohn's disease. The animal M. avium subsp. paratuberculosis isolates used included ATCC 700535, ATCC 19851, M. avium subsp. paratuberculosis K10, and those obtained from several animal host species with Johne's disease (Table 1) from different geographic localities (Fig. 1) and represented the extent of genotypic diversity determined by multiplex PCR of IS900 loci (15). M. avium subsp. paratuberculosis isolates from animals and birds in close proximity to cattle herds with multiple confirmed cases of Johne's disease were also analyzed (Table 1). All isolates were characterized by using well-defined molecular markers (14, 15) to confirm identity.
TABLE 1.
SSR analysis results by host species and targets
| Host | No. of M. avium subsp. paratuberculosis isolates | G repeat allele(s) | GGT repeat allele(s) |
|---|---|---|---|
| Cowa | 28b | 7, 8, 9, 10, 12, 14, 15 | 4, 5 |
| Sheepc | 17 | 7, p7,d 10, 14, 15 | 3, p3,e 4, 5 |
| Goatf | 20 | 7, 10, 11, 12, 15 | 4, 5 |
| Deer | 2 | 7 | 4 |
| Humang | 11h | 7 | 4, 5 |
| Mouse | 1 | >15 | 5 |
| Raccoon | 5 | 7, 14 | 5 |
| Cat | 1 | 14 | 5 |
| Starlingi | 7 | 7, 12, 13, 14, 15 | 5, 6 |
| Shrew | 1 | 15 | 5 |
| Armadillo | 1 | 7 | 4 |
GenBank accession no. AY587703, AY587706, AY587707, AY587712, AY587714, AY587715, AY587717, AY587724, and AY587728.
Includes 25 clinical strains, 2 animal ATCC strains, and K10.
GenBank accession no. AY587699, AY587704, AY587708, AY587716, AY587718, AY587719, AY587720, AY587721, AY587725, and AY587729.
Polymorphic 7-G repeats.
Polymorphic 3-GGT repeats.
Includes seven clinical strains and four human ATCC strains.
GenBank accession no. AY587731.
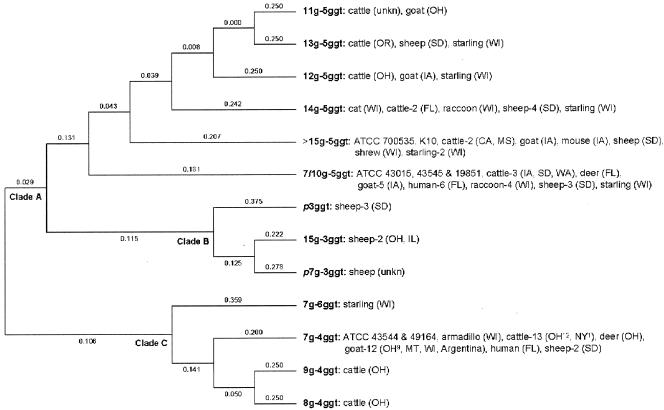
FIG. 1.
Dendrogram showing the distribution of strains by the number of G and GGT repeats. Shown are the number of G repeats followed by the number of GGT repeats and the host species. Host species not followed by a number indicate one isolate. The geographic locations (U.S. postal abbreviations) are in parentheses. A superscript following a geographic location indicates the number of isolates from that location. Also shown at the clade origins are the pairwise distances generated by the neighbor-joining model. unkn, unknown.
Results of a recently described multilocus SSR analysis (2) for M. avium subsp. paratuberculosis strain differentiation indicated that mononucleotide G repeat (GenBank accession no. AAK46234) and trinucleotide GGT repeat loci (GenBank accession no. CAB06859) were most discriminatory of the 11 SSRs analyzed. Hence, these loci were selected for fingerprinting strains in this study as previously described (2, 14). All chromatograms were manually edited and aligned with EditSeq and MegAlign, respectively (DNASTAR, Madison, Wis.). Alleles were identified by the number of G and GGT residues (Table 1). An allele containing a polymorphism within the repeat region was indicated by the letter p. Cluster analysis was performed with the molecular evolutionary genetic analysis program (version 2.1; www.megasoftware.net) by the neighbor-joining method (12). The distance matrix for input into the molecular evolutionary genetic analysis program was created from the allele data with ETDIV and ETMEGA (http://foodsafe.msu.edu/whittam/programs/). Simpson's diversity index was calculated with the equation 1 − ∑(allele frequency)2 (10).
Of the 94 M. avium subsp. paratuberculosis isolates analyzed, 92 had a detectable G repeat amplicon (approximately 400 bp) while all 94 had a detectable GGT repeat product (approximately 425 bp). Cluster analysis divided the isolates into three distinct clades and a total of 13 distinct alleles. Cattle (n = 28, including ATCC strains) and goat (n = 20) isolates were classified into nine and five alleles, respectively (Fig. 1). The sheep isolates (n = 17) were classified into eight alleles, three of which formed a distinct clade. Two sheep strain-specific nucleotide polymorphisms (GGGGGGG → GGCGGGG and GGTGGTGGT → AGTGGTGGT [the nucleotides in question are in italics]) were identified within the repeat regions. Isolates with either of these polymorphic alleles had several additional substitutions throughout the sequenced region (GenBank accession numbers are in the footnotes to Table 1). Two distinct alleles were identified among the 11 human isolates (including ATCC strains), and each clustered with cattle, sheep, and goat isolates. Isolates derived from free-ranging or feral nonruminant host species (n = 18) were dispersed evenly in clades A and C (Fig. 1). Simpson's diversity index for the analysis was 0.78 (D = 0.217), indicative of a strain discrimination capability much higher than that of other markers or methods reported to date (4, 15, 17).
While the number of alleles identified for cattle and goat isolates was greater than those in sheep isolates, 52% of the cattle and goat strains carried a single SSR genotype (7g-4ggt). This suggests a more recent association of M. avium subsp. paratuberculosis with cattle and goats compared to sheep. The presence of nucleotide polymorphisms, exclusive to sheep isolates, further supports the contention that sheep strains may be ancestral. This speculation is consistent with the data from a recently published study that indicated that the sheep strains of M. avium subsp. paratuberculosis were an evolutionary intermediate between the cattle strain of M. avium subsp. paratuberculosis and M. avium (6).
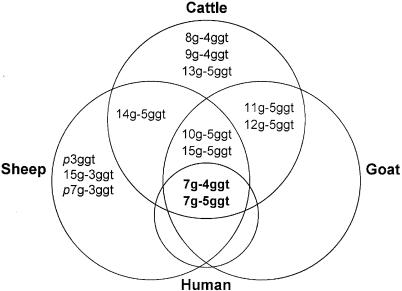
The restricted allelic variation identified within the human M. avium subsp. paratuberculosis strains analyzed may be indicative of the ability of a few animal genotypes to be associated with the pathobiology of Crohn's disease (Fig. 2). The precedent for this line of thought lies in the acknowledgment that Johne's disease in sheep is mostly caused by a distinct group of M. avium subsp. paratuberculosis strains. However, the presence of the same two alleles in 52% of the animal isolates is suggestive of strain sharing and interspecies transmission. Since the human M. avium subsp. paratuberculosis strains analyzed in this study (other than ATCC strains) were isolated from patients residing in Florida, the probability that they were exposed to a genetically and geographically restricted set of strains certainly exists. However, identification of the same alleles in ATCC strains obtained from diverse localities and the presence of disparate alleles in animal isolates (n = 4) from Florida strengthen the possibility that a limited set of strains are associated with Crohn's disease. This inference is also reflected in the presence of several nonsynonymous single-nucleotide mutations unique to human M. avium subsp. paratuberculosis strains (X. Zhu and S. Sreevatsan, unpublished data). On the other hand, it is possible that the human M. avium subsp. paratuberculosis isolates from Florida represent a much broader geographic area than just the peninsula as many people retire to Florida from other parts of the country.
FIG. 2.
Allele distribution across various host species strongly suggests strain sharing and interspecies transmission among M. avium subsp. paratuberculosis strains.
Isolation of M. avium subsp. paratuberculosis from tissues of other nonruminant animal species (cat, raccoon, shrew, armadillo, and rat) and birds found in the vicinity of infected farm animals suggests that M. avium subsp. paratuberculosis is able to infect and colonize nonruminants. Identification of common strain types between these animals and farm ruminants indicates strain sharing. Although we identified a greater representation of the 7g-5ggt allele in the raccoon isolates, there was no evidence for an association between other nonruminant host species and a particular M. avium subsp. paratuberculosis allele such as that observed for human isolates.
It has been suggested that both genetic susceptibility (e.g., NOD2/CARD15 polymorphisms) (13) and exposure to M. avium subsp. paratuberculosis are necessary for manifestation of Crohn's disease. The existence of possible modes of exposure and/or transmission has been indicated by documentation of the presence of M. avium subsp. paratuberculosis in retail pasteurized milk and its long-term survival in soil and water (http://gpvec.unl.edu/avc/IssuePapers/out38_en.pdf). However, the prevalence of Crohn's disease is low despite the suggested widespread exposure of people to M. avium subsp. paratuberculosis and isolates from humans do not reflect the strain diversity seen in animal M. avium subsp. paratuberculosis isolates. This further supports the idea that a few distinct M. avium subsp. paratuberculosis genotypes are associated with the pathobiology of Crohn's disease. The present study presents evidence for the existence of both human disease-associated genotypes and strain sharing with animals. We were unable to establish if the patients carried the NOD2/CARD15 mutation. Larger studies on M. avium subsp. paratuberculosis strains isolated from well-defined Crohn's disease patient and healthy human populations from diverse geographic locations and time points are warranted to fully elucidate the association of specific genotypes of M. avium subsp. paratuberculosis with Crohn's disease.
Acknowledgments
This study was supported by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, including an Ohio Agricultural Research and Development Center Competitive Research Enhancement seed grant awarded to S. Sreevatsan.
REFERENCES
- 1.Adair, D. M., P. L. Worsham, K. K. Hill, A. M. Klevytska, P. J. Jackson, A. M. Friedlander, and P. Keim. 2000. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 38:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amonsin, A., L. L. Li, Q. Zhang, J. P. Bannantine, A. S. Motiwala, S. Sreevatsan, and V. Kapur. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42:1694-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashford, D., B. Imhoff, F. Angulo, and The FoodNet Working Group. 2001. Over a half-million persons may have Crohn's disease in the United States. Infectious Diseases Society of America, San Francisco, Calif.
- 4.Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146(Pt. 9):2185-2197. [DOI] [PubMed] [Google Scholar]
- 5.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dohmann, K., B. Strommenger, K. Stevenson, L. de Juan, J. Stratmann, V. Kapur, T. J. Bull, and G. F. Gerlach. 2003. Characterization of genetic differences between Mycobacterium avium subsp. paratuberculosis type I and type II isolates. J. Clin. Microbiol. 41:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gascoyne-Binzi, D. M., R. E. Barlow, R. Frothingham, G. Robinson, T. A. Collyns, R. Gelletlie, and P. M. Hawkey. 2001. Rapid identification of laboratory contamination with Mycobacterium tuberculosis using variable number tandem repeat analysis. J. Clin. Microbiol. 39:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermon-Taylor, J. 2001. Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:755-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugot, J. P., M. Chamaillard, H. Zouali, S. Lesage, J. P. Cezard, J. Belaiche, S. Almer, C. Tysk, C. A. O'Morain, M. Gassull, V. Binder, Y. Finkel, A. Cortot, R. Modigliani, P. Laurent-Puig, C. Gower-Rousseau, J. Macry, J. F. Colombel, M. Sahbatou, and G. Thomas. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411:599-603. [DOI] [PubMed] [Google Scholar]
- 10.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 13.McGovern, D. P., D. A. Van Heel, T. Ahmad, and D. P. Jewell. 2001. NOD2 (CARD15), the first susceptibility gene for Crohn's disease. Gut 49:752-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motiwala, A. S., A. Amonsin, M. Strother, E. J. Manning, V. Kapur, and S. Sreevatsan. 2004. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis isolates recovered from wild animal species. J. Clin. Microbiol. 42:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motiwala, A. S., M. Strother, A. Amonsin, B. Byrum, S. A. Naser, J. R. Stabel, W. P. Shulaw, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 41:2015-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naser, S. A., D. Schwartz, and I. Shafran. 2000. Isolation of Mycobacterium avium subsp. paratuberculosis from breast milk of Crohn's disease patients. Am. J. Gastroenterol. 95:1094-1095. [DOI] [PubMed] [Google Scholar]
- 17.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, M. R. du, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38:155-167. [DOI] [PubMed] [Google Scholar]
- 18.Quirke, P. 2001. Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz, D., I. Shafran, C. Romero, C. Piromalli, J. Biggerstaff, N. Naser, W. Chamberlin, and S. A. Naser. 2000. Use of short-term culture for identification of Mycobacterium avium subsp. paratuberculosis in tissue from Crohn's disease patients. Clin. Microbiol. Infect. 6:303-307. [DOI] [PubMed] [Google Scholar]